Abstract
Stress has complex effects on hippocampal structure and function, which consequently affects learning and memory. These effects are mediated in part by circulating glucocorticoids (GC) acting via the intracellular GC receptor (GR) and mineralocorticoid receptor (MR). Here, we investigated GC regulation of Krüppel-like factor 9 (KLF9), a transcription factor implicated in neuronal development and plasticity. Injection of corticosterone (CORT) in postnatal d 6 and 30 mice increased Klf9 mRNA and heteronuclear RNA by 1 h in the hippocampal region. Treatment of the mouse hippocampal cell line HT-22 with CORT caused a time- and dose-dependent increase in Klf9 mRNA. The CORT induction of Klf9 was resistant to protein synthesis inhibition, suggesting that Klf9 is a direct CORT-response gene. In support of this hypothesis, we identified two GR/MR response elements (GRE/MRE) located −6.1 and −5.3 kb relative to the transcription start site, and we verified their functionality by enhancer-reporter, gel shift, and chromatin immunoprecipitation assays. The −5.3-kb GRE/MRE is largely conserved across tetrapods, but conserved orthologs of the −6.1-kb GRE/MRE were only detected in therian mammals. GC treatment caused recruitment of the GR, histone hyperacetylation, and nucleosome removal at Klf9 upstream regions. Our findings support a predominant role for GR, with a minor contribution of MR, in the direct regulation of Klf9 acting via two GRE/MRE located in the 5′-flanking region of the gene. KLF9 may play a key role in GC actions on hippocampal development and plasticity.
The hippocampus is an important target for the actions of circulating glucocorticoids (GC). GC actions on the hippocampus are complex: they may facilitate or impair hippocampal function by influencing cell survival, dendritic remodeling, and synaptic transmission (1, 2). The actions of GC are mediated in large part by two nuclear receptors that function as ligand-activated transcription factors, the mineralocorticoid receptor (MR) and the GC receptor (GR) (3); both are highly expressed in the mammalian hippocampus (4, 5). The MR and GR bind to DNA at hormone-response elements comprised of two hexanucleotide half-sites and they recruit coactivator or corepressor complexes that contain histone modifying enzymes (6, 7). The histone modifications promote an open or closed chromatin environment, thereby influencing the rate of transcription (8).
Stress hormones cause biochemical, physiological, and structural changes in neurons (9). Several direct transcriptional targets of the GR have been identified in hippocampal neurons (10–14), but the mechanisms by which GC influence neuronal structure and function are poorly understood. Earlier we found that the transcription factor Krüppel-like factor 9 (KLF9) is a direct thyroid hormone (TH) target gene that is strongly up-regulated during postnatal brain development (15, 16). The Klf9 gene is expressed in several rodent tissues, but highest expression is seen in the hippocampus and cerebellum of the developing brain (17–19). Klf9 expression was detected in the hippocampus at postnatal day (PND) 1, which increased through PND 5, at which time there was distinct expression in the pyramidal cell layer; strong expression in the cortical layer of the cerebellum was detected at PND 7 (19). Expression in the hippocampus peaked at PND 15, remained elevated through PND 30, but then declined in the adult to levels seen at PND5 (16, 19, 20). The developmental increase in Klf9 expression parallels the postnatal rise in plasma TH seen in rodents (15, 16, 19–21), which supports findings that KLF9 functions as a key intermediary in TH actions on neuronal structure and function (15, 22).
KLF9 plays a role in neuronal maturation and morphology, and hippocampal and cerebellar function. KLF9-deficient mice exhibited reduced dendritic arborization of cerebellar Purkinje cells (19), delayed neuronal maturation and reduced neurogenesis-dependent long-term potentiation in the dentate gyrus (20), and behavioral deficits consistent with abnormal functions of the amygdala, hippocampus, and cerebellum (19).
We recently reported that Klf9 mRNA and protein were strongly induced in Xenopus brain by exposure to confinement/shaking stressor, which was blocked by treatment with mifepristone (RU486), or mimicked by injection of corticosterone (CORT) (23). Datson et al. (11) recently found Klf9 to be induced by CORT in primary rat hippocampal neurons, and they provided evidence for two putative GR/MR binding sites in the Klf9 5′-flanking region. In the current study, we investigated the molecular basis for CORT regulation of Klf9 in mouse hippocampal neurons. We looked at whether CORT could induce Klf9 expression in mouse hippocampus in vivo, and in mouse hippocampal tissue culture cells, and we identified two evolutionarily conserved hormone-response elements in the Klf9 5′-flanking region to which the GR/MR bind to regulate Klf9 expression.
Materials and Methods
Animal care
We purchased adult wild-type C57/BL6J mice from The Jackson Laboratory (Bar Harbor, ME), and Dr. Fujii-Kuriyama kindly provided Klf9 mutant mice (Klf9 locus disrupted by insertion of LacZ) (19). Mice were maintained on a 12-h light, 12-h dark photoperiod, given food and water ad libitum, and bred in the laboratory. We conducted histochemical staining for β-galactosidase using brains from 7-wk-old heterozygous Klf9 mutant mice. Wild-type mice of mixed sex at PND 6 or 30 were given ip injections of CORT (Sigma Chemical Co., St. Louis, MO) dissolved in corn oil vehicle at 14 mg/kg body weight; this dose is in the midrange of CORT doses reported in the literature to increase plasma CORT (24). Control animals received vehicle or were unhandled. One hour after injection, animals were killed, and blood plasma and brains collected for analysis. We analyzed plasma [CORT] by radioimmunoassay as described (25, 26). We microdissected the hippocampal region for Klf9 mRNA and heteronuclear RNA (hnRNA) analysis by RTqPCR (reverse transcription quantitative PCR). All experiments were conducted in accordance with the guidelines of the University Committee on the Use and Care of Animals at the University of Michigan.
RNA extraction, RT, and quantitative PCR
We extracted total RNA from HT-22 cells or the hippocampal region of PND 6 and 30 mice using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) following the manufacturer's protocol. We treated 1 μg of total RNA with 20 U of ribonuclease-free deoxyribonuclease (DNAse) I (Roche, Indianapolis, IN) (27) before cDNA synthesis with the High Capacity Reverse Transcription kit (Invitrogen) with or without the addition of RT. For quantitative PCR, we designed TaqMan assays and conducted real-time PCR using ABsolute QPCR Low ROX Mix (ABgene Thermo Scientific, Surrey, UK). The Klf9 mRNA TaqMan assay spanned an exon/intron boundary, whereas the Klf9 hnRNA assay targeted the single intron (Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org). Treatment of RNA with DNAse I removed genomic DNA contamination, which was confirmed by the inclusion of −RT controls in the hnRNA analysis. Standard curves were constructed with a cDNA pool from HT-22 cell RNA to compare relative expression levels. Klf9 mRNA was normalized to GAPDH (TaqMan Assay no. 4352662; Applied Biosystems, Carlsbad, CA) mRNA, the expression of which did not change with hormone treatment.
Plasmid constructs
A chromatin immunoprecipitation (ChIP)-promoter DNA chip (ChIP-chip) assay for MR on chromatin from HEK293 cells that express myc-tagged human MR (HEK293-hMR+; for methods see Ref. 28) identified two regions of the upstream region of human Klf9 (Ziera, T., and S. Borden, unpublished data). Bioinformatic analysis revealed that both loci contain putative GR/MR response elements (GRE/MRE). These regions were isolated from human genomic DNA by PCR (Supplemental Table 2); one was 632 bp from −5771 to −5139 bp (fragment 1, centered at ∼5.5kb), a second was 336 bp from −4211 to −3875 bp (fragment 2, centered at ∼4 kb) relative to the transcription start site (TSS). DNA fragments were subcloned into the pGL4.23 vector (Promega, Madison, WI) at the XhoI and HindIII sites for testing in HEK293-hMR+ cells (Supplemental Materials and Methods). The two orthologous regions of the mouse Klf9 gene, a 634-bp fragment (fragment 1) located −5776 to −6410 bp relative to the TSS and a 336-bp fragment (fragment 2) located −4566 to −4892 bp relative to the TSS, were isolated from mouse genomic DNA by PCR and subcloned into pGL4.23. An evolutionarily conserved 179-bp mouse genomic region (−5333 to −5154 bp relative to the TSS) that contained a GRE/MRE identified by DNAse I footprinting (described below) was also isolated by PCR and subcloned into pGL4.23. We also isolated and subcloned into pGL4.23 a 1269-bp fragment of the mouse Klf9 gene (−6409 to −5140 relative to the TSS) spanning fragment 1 and the 179-bp evolutionarily conserved region. This construct included the two GRE/MRE at −6.1 and −5.3 kb described below. Plasmid constructs containing genomic DNA fragments with mutated GRE/MRE half-sites were generated using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA) following the manufacturer's protocol (Supplemental Table 3).
Cell culture and transient transfection assays
HT-22 is a cell line derived from mouse hippocampus immortalized with the simian virus 40 T antigen (29, 30) with properties of differentiated neurons (29–32). HT-22 expresses functional GR but has no detectable MR, because [3H]-CORT binding in HT-22 lysates is completely displaced by RU486 (Rozeboom, A., and A. Seasholtz, unpublished data). HT-22/MR16 cells were engineered to express mouse MR by stably transfecting HT-22 cells with cytomegalovirus (CMV)-MR and pPGKpuro plasmids, and selection in 2.5 μg/ml puromycin. The CMV-MR construct contains the full-length mouse MR cDNA [clone pMR-2A (33) in pCMVneo]. The HT-22/MR16 cells exhibit MR binding and transcriptional activation of GRE-luciferase constructs with 1–10 nm aldosterone (ALDO) (Rozeboom, A., and A. Seasholtz, unpublished data).
We cultured HT-22 and HT-22/MR16 cells in DMEM (Invitrogen) supplemented with sodium bicarbonate (2.2 g/liter), penicillin G (100 U/ml), streptomycin sulfate (100 μg/ml), and 10% fetal bovine serum (FBS) stripped of thyroid (34) and steroid hormones (35). Cells were cultured under a humidified atmosphere of 5% CO2 at 37 C. For gene expression analysis, we seeded cells at 2.5 × 106 per well in six-well plates; for transient transfection assays, we seeded cells at 6.5 × 104 per well in 24-well plates. At 80% confluency, and immediately before hormone treatments, we replaced the growth medium with serum-free DMEM. CORT (Sigma C2505), dexamethasone (DEX) (Sigma D1756), or ALDO (Sigma A9477) was dissolved in 100% ethanol and added to media to the final concentrations indicated below. RU486 (a GR antagonist; Sigma M8046) was dissolved in ethanol and added to wells to a final concentration of 0.5 or 1 μm 1 h before hormone addition. Spironolactone (SPR) (a MR antagonist; Sigma S3378) was dissolved in ethanol and added to wells to a final concentration of 0.5 μm 1 h before hormone addition. T3 (Sigma T2752) was dissolved in dimethylsulfoxide. Control treatments received an equivalent concentration of vehicle (0.001% ethanol and 0.03% dimethylsulfoxide). All hormone treatments were continued for 4 h before cell harvest. Hormone treatment experiments were conducted at least two times with comparable results.
To determine whether hormone-dependent induction of Klf9 mRNA requires ongoing protein synthesis, we treated cells with 100 μg/ml of cycloheximide (CHX) (Sigma Chemical Co.). This dose of CHX reduced protein synthesis by 98% in HT-22 cells (Bagamasbad, P., and R. Denver, unpublished data). We treated cells with CHX for 1 h before and during hormone treatment.
For luciferase reporter assays, we plated HT-22 cells in DMEM with hormone-stripped FBS and transfected at 50–60% confluency with 300 ng of the pGL4.23 reporter constructs plus 20 ng of pRenilla plasmid using the FuGENE 6 transfection reagent (Roche). Immediately before transfection, the growth medium was replaced with medium containing hormone-stripped FBS without penicillin/streptomycin, and cells were incubated with the transfection mixture overnight. Twenty-four hours after transfection, cells were changed to serum-free medium containing 100 nm CORT or vehicle for 4 h before harvest for luciferase activity using the dual luciferase assay kit (Promega). Each transfection experiment was conducted at least three times with four to five wells per treatment.
Electrophoretic mobility shift assay
We conducted EMSA as described (36) with recombinant human GR (hGR) generated using the TnT SP6 Quick Coupled Translation System (Promega). We programmed the reaction with 1 μg of CMV-hGR expression plasmid (OriGene, Rockville, MD) and confirmed protein expression (36). One microgram of duplex oligonucleotides (corresponding to the predicted GRE/MRE at −6.1 and −5.3 kb upstream of the TSS) (Supplemental Table 2) was labeled with 32P-dCTP by Klenow fill-in for use as probes (50 K cpm/reaction) in EMSA. We conducted competitive EMSA with 1.5 μm radioinert (∼1000-fold excess) wild-type or mutant GRE/MRE oligonucleotides (Supplemental Table 2). A 32P-labeled mouse mammary tumor virus (MMTV) probe containing a well-characterized GRE was used as positive control for GR binding (Supplemental Table 3).
Fluorescent DNAse I footprinting
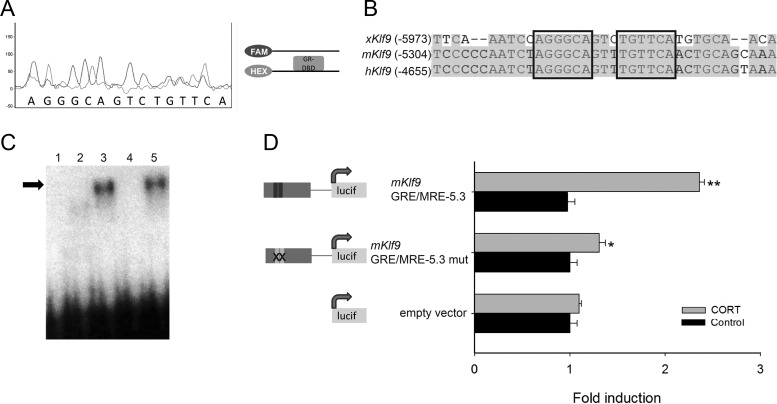
We conducted DNAse I footprinting as described (37) using the Promega Core Footprinting System protocol. We generated FAM or HEX fluorescent-labeled double-stranded DNA probes corresponding to the mouse Klf9 5′-flanking region (−5333 to −5154 bp; 179 bp). The region corresponding to the DNA binding domain (DBD) of the hGR was PCR amplified and subcloned into TOPO/pET151/D (Invitrogen) to make TOPO/pET151/d-hGR-DBD. Bacterially expressed GR-DBD was purified using ProBond (Invitrogen). We combined 1 μg of GR-DBD with 150 ng of HEX (green)-labeled probe in a 50-μl digestion reaction. The FAM (blue)-labeled probe (150 ng) was digested in a 50 μl of reaction without added GR-DBD. The two probe digestion reactions were then combined, extracted with phenol/chloroform, concentrated by ethanol precipitation, and analyzed by Big Dye Terminator Cycle sequencing (Applied Biosystems). Peaks in the chromatograms were assigned to specific nucleotides using FAM-labeled ddATP, ddCTP, ddGTP, or ddTTP in a manual sequencing reaction.
ChIP assay
We plated HT-22 cells in 100-mm plates in DMEM with hormone-stripped FBS. Cells at 90% confluency were treated with 100 nm CORT or ethanol vehicle in serum-free medium for 4 h before harvest for chromatin extraction. We also isolated chromatin from the hippocampal region of PND 6 mice 1 h after injection of CORT (14 mg/kg); control animals were left unhandled. Five micrograms of sheared chromatin were used for each reaction, and ChIP assay was conducted as described (16). Polyclonal rabbit antiserum to mouse GR (5 μg, M-20X; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), histone 3 (H3) (07-690, 5 μl; Millipore, Billerica, MA), or antiacetylated H3 (AcH3) (06-599, 5 μl; Millipore) was used for ChIP assay. ChIP samples were analyzed by quantitative real-time PCR using TaqMan primer/probe sets (Supplemental Table 1) targeted to different regions of the mouse Klf9 gene.
Data analysis and statistics
We analyzed data by one-way ANOVA followed by Tukey's pairwise comparisons test, or by Student's t test using the SYSTAT computer program (version 10; SPSS, Inc., Chicago, IL). Data from dual luciferase assays (firefly luciferase counts divided by Renilla luciferase counts) and ChIP assays (the ratio of ChIP signal to input) were log10 transformed before statistical analysis. P < 0.05 was accepted as statistically significant. Gene expression data are reported as the mean ± sem. To predict putative GRE/MRE sites, we used the online programs Transcription Element Search System (TESS) (www.cbil.upenn.edu) and Match (www.gene-regulation.com/pub/programs.html#match) using a library of matrices from TRANSFAC 6.0 (www.gene-regulation.com) with core and matrix similarity of transcription factor binding sites set higher than 0.85. The conservation of the identified GRE/MRE and flanking sequences was assessed using BLAT (http://genome.ucsc.edu/). Extracted sequences were aligned using ClustalW. The percent similarity to mouse was calculated for the GRE/MRE half-sites, and nucleotide frequency diagrams were produced with Weblogo (http://weblogo.berkeley.edu/logo.cgi).
Results
Exogenous CORT increased Klf9 mRNA and hnRNA in mouse hippocampus in vivo
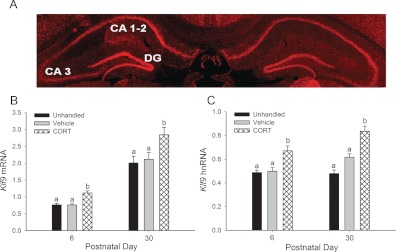
The Klf9 gene is expressed in all regions of the mouse hippocampus (Fig. 1A) (19). Intraperitoneal injection of CORT in PND 6 or 30 mice increased Klf9 mRNA [PND 6: F(2,26) = 17.084, P < 0.0001; PND 30: F(2, 14) = 6.937, P < 0.01; ANOVA] (Fig. 1B) and hnRNA [PND 6: F(2,26) = 10.71, P < 0.001; PND 30: F(2,14) = 27.427, P < 0.01] (Fig. 1C) in the region of the hippocampus 1 h after injection compared with unhandled or vehicle-injected controls. CORT injection elevated plasma [CORT] above controls [F(2,13) = 7.353, P < 0.013]; the vehicle-injected and unhandled groups were not significantly different from each other (unhandled, 48.9 ± 10.2 ng/ml; vehicle, 33.5 ± 9.9 ng/ml; and CORT, 1996 ± 830 ng/ml).
Fig. 1.
Injection of CORT increased Klf9 mRNA and hnRNA levels in the hippocampal region of the postnatal mouse brain. A, A representative coronal section of the hippocampal region of a LacZ-Klf9 heterozygous mouse generated by insertion of LacZ within the coding region of the Klf9 gene (19). Histochemistry was conducted for β-galactosidase with immunofluorescent detection using Cy3. B, PND 6 and 30 wild-type C57/BL6J mice were injected with vehicle (oil), CORT at a dose of 14 mg/kg bodyweight, or left unhandled. One hour after injection, animals were killed, the hippocampal region was dissected, and RNA extracted for measurement of Klf9 mRNA by RTqPCR (n = 8 at PND 6 and n = 5 at PND 30). C, PND 6 mice were injected with CORT as described above, and Klf9 hnRNA was measured by RTqPCR (n = 8). Klf9 mRNA and hnRNA were normalized to the mRNA level of the housekeeping gene GAPDH. Bars represent the mean ± sem, and letters above the means indicate significant differences among treatments (means with the same letter are not significantly different; Tukey's pairwise comparisons test; P < 0.05). DG, Dentate gyrus.
CORT caused a rapid and dose-dependent increase in Klf9 mRNA in HT-22 cells
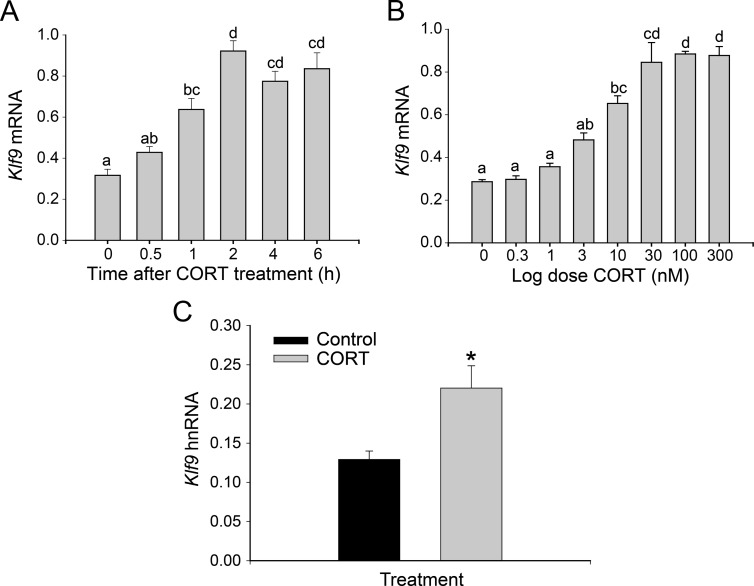
CORT increased Klf9 mRNA in HT-22 cells in a time- and dose-dependent manner [time: F(5,23) = 22.06, P < 0.0001; dose: F(7,30) = 37.69, P < 0.0001; ANOVA] (Fig. 2, A and B); Klf9 hnRNA was also increased by CORT (P < 0.05; Student's t test) (Fig 2C). The mean Klf9 mRNA level was higher at 30 min after CORT exposure, was statistically significant at 1 h and was maximal by 2 h. The EC50 (measured at 4 h) was 5.7 nm, and maximal response was at 30 nm. Klf9 is a direct thyroid hormone receptor target gene in rodent and frog (15, 16, 36, 38). For comparison with the CORT response, we show that Klf9 is regulated by T3 in a time- and dose-dependent manner in HT-22 cells (Supplemental Fig. 1).
Fig. 2.
Treatment with CORT caused dose- and time-dependent increases in Klf9 mRNA in the mouse hippocampal-derived cell line HT-22. HT-22 cells were treated with 100 nm CORT for different times (A) or with increasing doses of CORT for 4 h (B) before harvest and analysis of Klf9 mRNA. C, HT-22 cells were treated with 100 nm CORT for 4 h before harvest and analysis of Klf9 hnRNA. In each experiment, HT-22 cells were treated with the hormone doses for the times indicated before cell harvest, RNA extraction, and analysis by RTqPCR. Klf9 mRNA and hnRNA were normalized to the mRNA level of the housekeeping gene GAPDH. Bars represent the mean ± sem (n = 4), and letters above the means indicate significant differences among hormone concentrations or time point (means with the same letter are not significantly different; Tukey's pairwise comparison test; P < 0.05). *, P < 0.05 (Student's t test).
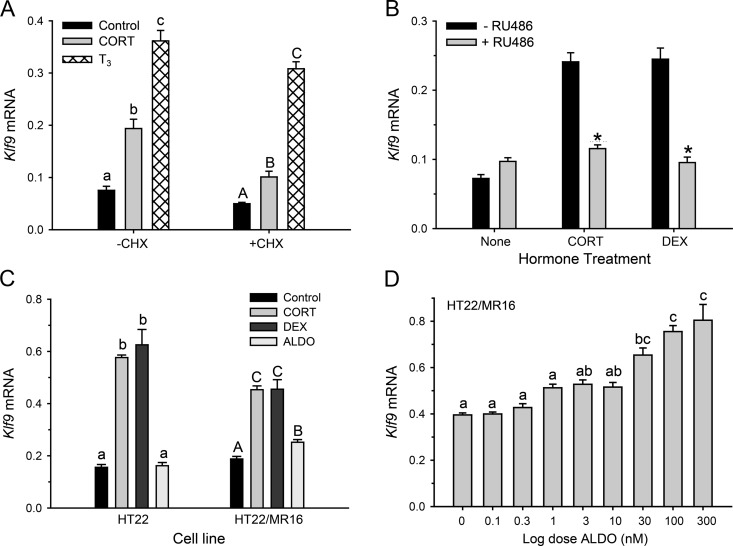
Hormone induction of Klf9 mRNA in HT-22 cells was resistant to protein synthesis inhibition, supporting that Klf9 is a direct hormone-response gene [control (−CHX): F(2,11) = 81.63, P < 0.0001; CHX: F(2,11) = 151.8; P < 0.001; ANOVA] (Fig. 3A).
Fig. 3.
Induction of Klf9 mRNA by CORT in HT-22 cells is resistant to protein synthesis inhibition. Klf9 can be regulated by the GR or by the MR. A, HT-22 cells were treated with or without CHX (100 μg/ml) for 30 min before addition of CORT (100 nm) or T3 (30 nm). Treatment with CHX plus hormones was continued for 4 h before cell harvest for analysis of Klf9 mRNA. We used T3 as a positive control, because earlier we showed that Klf9 is a direct TR target gene (16). B, HT-22 cells were cultured in the presence or absence of the GR-selective antagonist RU486 (1 μm) for 1 h before the addition of vehicle (100% EtOH), 100 nm CORT, or 10 nm of the GR-selective agonist DEX. Hormone treatment was continued for 4 h before cell harvest and analysis of Klf9 mRNA. C, HT-22 and HT-22/MR16 cells were treated with vehicle (100% EtOH), 100 nm CORT, 10 nm DEX, or 10 nm ALDO for 4 h before cell harvest for analysis of Klf9 mRNA. D, HT-22/MR16 were treated with increasing concentrations of the MR-selective agonist ALDO for 4 h before cell harvest for analysis of Klf9 mRNA. In each experiment, cells were treated with the hormone doses for the times indicated before cell harvest, RNA extraction, and gene expression analysis by RTqPCR. Klf9 mRNA was normalized to the mRNA level of the housekeeping gene GAPDH. Bars represent the mean ± sem (n = 4), and letters above the means indicate significant differences among hormone concentrations or time point (means with the same letter are not significantly different; Tukey's pairwise comparison test; P < 0.05). *, P < 0.05 (Student's t test).
Both GR and MR can regulate the Klf9 gene, but the GR may predominate
CORT (100 nm) or the GR-selective agonist DEX (10 nm) caused similar increases in Klf9 mRNA (∼3-fold) measured at 4 h [−RU486: F(2,11) = 42.40, P < 0.001; ANOVA] (Fig. 3B) that could be blocked by cotreatment with the GR receptor antagonist RU486 [+RU486, F(2,11) = 3.128] (Fig. 3B). These data support that Klf9 can be regulated by the GR in HT-22 cells, but they do not exclude a role for MR in Klf9 regulation in other cell types, because HT-22 cells do not express functional MR (Rozeboom, A. M., and A. F. Seasholtz, unpublished data). Consistent with a lack of functional MR, HT-22 cells did not respond to ALDO (10 nm) (Fig. 3C).
To determine whether the MR can mediate corticosteroid actions on Klf9 gene expression, we analyzed hormone responses in HT-22/MR16 cells. A dose of 1 nm ALDO activated a GRE/MRE-containing promoter-reporter construct in these cells [Rozeboom, A., and A. F. Seasholtz, unpublished data; the Kd for ALDO binding to MR is 0.5–2.6 nm (3, 39, 40)]. ALDO (10 nm) caused a small, but statistically significant, increase in Klf9 mRNA [F(8, 27) = 26.45, P < 0.001; ANOVA] (Fig. 3C). The response to ALDO was dose dependent with an EC50 of 25 nm [F(8,35) = 26.45, P < 0.001] (Fig. 3D). This EC50 is approximately five times greater than the EC50 for CORT in HT-22 cells; ALDO concentrations more than 30 nm can activate the GR (41).
To further investigate the contributions of GR vs. MR in the regulation of Klf9, we treated HT-22/MR16 cells with 25 nm CORT or ALDO with or without the GR-specific antagonist RU486 (500 nm), the MR-specific antagonist SPR (500 nm), or RU486 plus SPR (Supplemental Fig. 2). We chose a dose of 25 nm for both hormones, because this dose is the EC50 for ALDO action on Klf9 mRNA in HT-22/MR16 cells. Treatment with RU496 plus SPR in the absence of hormone caused a small, but statistically significant, elevation in Klf9 mRNA [F(3,11) = 8.784, P < 0.01]. Both CORT and ALDO increased Klf9 mRNA [F(2,11) = 27.891, P < 0.0001; ANOVA], although the response to ALDO was significantly lower than for CORT (P < 0.001; Tukey's). RU486 reduced the response to CORT (P < 0.01; Tukey's) but did not affect the ALDO response, supporting that the action of ALDO is via the MR. The mean CORT and ALDO responses were lower in the presence of SPR but were not statistically significant. RU486 plus SPR reduced the response to CORT [F(3,15) = 8.073, P < 0.01], which was significantly lower than SPR alone but was not significantly different from RU486 alone, supporting that the major action of CORT on Klf9 is via the GR. The effect of cotreatment with RU486 plus SPR on the ALDO response was not different from SPR alone.
Identification of functional GRE/MRE in the human and mouse Klf9 genes
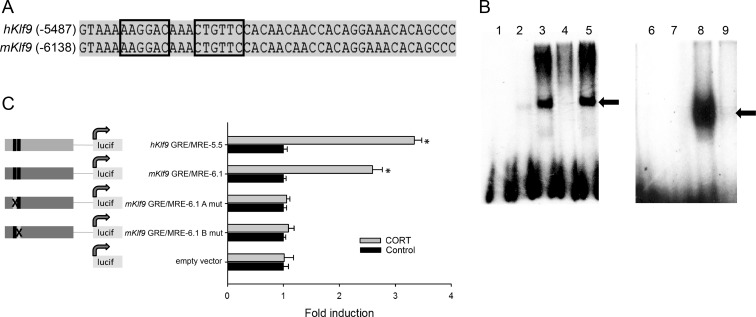
A ChIP-chip assay for MR on chromatin from HEK293-hMR+ cells identified two MR binding sites in the 5′-flanking region of the human Klf9 gene (Ziera, T., and S. Borden, unpublished data). One region (fragment 1) was centered at approximately −5.5 kb and a second (fragment 2) at approximately −4 kb relative to the TSS. Both regions were cloned into pGL4.23 to determine whether these regions support corticosteroid-dependent transcriptional activation. Enhancer-reporter assays in HEK293-hMR+ cells showed that only fragment 1 was activated by ALDO, and this response was blocked by the MR-specific antagonist RU26752 (Supplemental Fig. 3). This genomic region is highly conserved between human and mouse. In silico analysis of the human Klf9 fragment 1 using TESS and Match identified a near perfect palindromic GRE/MRE full site that is completely conserved between mouse (mKlf9 GRE/MRE 6.1) and human (−5474 bp in human, hKlf9 GRE/MRE 5.5; Fig. 4A, partial sequence of the 600-bp fragment shown). The GR bound the mouse Klf9 GRE/MRE-6.1 probe in EMSA, and this binding was competed by radioinert wild-type, but not mutant, GRE/MRE-6.1 oligonucleotide, thus confirming the specificity of binding (Fig. 4B).
Fig. 4.
Identification and functional analysis of a GRE/MRE located at −5.5 kb in the human and −6.1 kb in the mouse Klf9 genes. A, A ChIP-chip promoter assay conducted on chromatin isolated from HEK293-hMR+ cells identified a fragment of approximately 600 bp in the human Klf9 5′-flanking region that contains a functional GRE/MRE (Ziera, T., and S. Borden, unpublished data). The sequence alignment is of a portion of this genomic region of human and mouse Klf9 showing the presence of putative GRE/MRE sites in the two genes (half-sites are boxed) predicted by the TESS and Match programs. The numbers to the left of the alignment indicate the positions upstream of the TSS. The GRE/MRE in the human gene is centered at approximately −5.5 kb, and the GRE/MRE in the mouse gene is centered at approximately −6.1 kb. B, The ability of the GR to bind to the mouse Klf9 GRE/MRE-6.1 in vitro was tested by EMSA. Each reaction contained [32P]-labeled GRE/MRE-6.1 oligonucleotide as probe (lanes 1–5) and 2 μl of in vitro-synthesized luciferase protein (control, lane 2) or hGR protein (lanes 3–5). The GR binding to the probe was competed with 1.5 μm radioinert GRE/MRE-6.1 (lane 4) or mutant GRE/MRE-6.1 oligonucleotides (lane 5). We conducted EMSA with a MMTV GRE probe as a positive control for GR binding (right panel). Each reaction contained [32P]-labeled MMTV oligonucleotide as probe (lanes 6–9) and 2 μl of in vitro-synthesized luciferase protein (negative control, lane 7) or hGR protein (lanes 8 and 9). The GR binding to the MMTV probe was competed with 1.5 μm radioinert MMTV oligonucleotide (lane 9). The supershifted bands indicated by the arrows are the GR-bound probe. C, pGL4.23 reporter constructs containing the human GRE/MRE-5.5, mouse GRE/MRE-6.1, and corresponding half-site mutants (X; A, 5′half-site and B, 3′half-site) of the mouse GRE/MRE-6.1 were transfected into HT-22 cells. Cells were treated with CORT for 4 h before harvest and analysis by dual luciferase assay. Bars represent the mean ± sem (n = 5). Asterisks indicate statistically significant differences between vehicle (control) and CORT-treated cells for each enhancer-reporter construct (P < 0.0001; Student's t test).
We cloned the region of the mouse Klf9 gene (mKlf9 634-bp fragment, −6410 to −5776 bp) orthologous to human fragment 1 and compared the ability of the mouse and human sequences to support CORT-dependent transactivation in transient transfection assays in HT-22 cells. Both the human and mouse Klf9 gene fragments supported CORT-dependent transactivation (hKlf9 GRE/MRE-5.5, 3.3-fold activation, P < 0.0001; mKlf9 GRE/MRE-6.1, 2.6-fold activation, P < 0.0001; Student's t test) (Fig. 4C). Mutation of each half-site of the mouse Klf9 GRE/MRE-6.1 abolished CORT responsiveness (Fig. 4C).
Earlier we found stressor and CORT-dependent regulation of the Klf9 gene in Xenopus (23), but we were unable to identify a GRE/MRE in a similar region of the frog gene (Xenopus tropicalis genome version 4.1). In a separate study looking for CORT-responsive regions of the frog Klf9 gene, we discovered that a region −6 to −5 kb relative to the TSS was CORT responsive (Bagamasbad, P., and R. J. Denver, unpublished data). Sequence comparison revealed an approximately 180-bp sequence within this region that was highly conserved between frog and mammalian Klf9 genes. This region of the frog Klf9 gene was previously reported to possess a T3-response element (T3RE) (42). Through DNAse I footprinting with GR-DBD of the orthologous mouse genomic region (179-bp fragment), we identified a GRE/MRE full site that is highly conserved across tetrapods (centered at −5285 bp in mouse, −4634 bp in human, and −5957 bp in frog Klf9 genes) (Fig. 5, A and B). We designated this mouse GRE/MRE-5.3 and confirmed that GR bound this sequence in EMSA (Fig. 5C). Binding of GR to the GRE/MRE-5.3 probe was competed by radioinert wild-type, but not by mutant, GRE/MRE-5.3 oligonucleotide, thus confirming the specificity of binding (Fig. 5C). The 179-bp fragment of the mouse Klf9 gene supported CORT-dependent transactivation in enhancer-reporter assays in HT-22 cells (Fig. 5D). Mutation of both half-sites of the GRE/MRE-5.3 reduced, but did not eliminate, CORT-dependent transactivation (P < 0.05, Student's t test) (Fig. 5D). The CORT-dependent activity of an enhancer-reporter plasmid containing a 1269-bp Klf9 DNA fragment spanning the two identified GRE/MRE sites was not different from plasmids containing the individual response elements (Supplemental Fig. 4).
Fig. 5.
Identification and functional analysis of an evolutionarily conserved GRE/MRE located at −5.3 kb in the mouse (−4.6 kb in the human) Klf9 gene. A, DNAse I protection assay with the hGR-DBD of the evolutionary conserved 179-bp fragment of the mouse Klf9 gene identified a GRE/MRE located approximately −5.3 kb relative to the TSS (see Materials and Methods). The light gray traces are from the probe incubated with the GR-DBD, whereas the dark gray traces are from the probe incubated without the GR-DBD. Areas where there is a dark gray peak but no light gray peak indicate nucleotides protected from DNAse I digestion by the GR-DBD. The nucleotide sequence is shown beneath the traces. B, Sequence alignment showing conservation of the GRE/MRE (mouse GRE/MRE-5.3) in human, mouse, and frog Klf9 genes (GRE/MRE half-sites are boxed). The numbers to the left of the alignment indicate the positions upstream of the TSS. The GRE/MRE in the human gene is centered at approximately −4.6 kb and in the frog gene at approximately −5.9 kb. C, The ability of the GR to bind to the mouse Klf9 GRE/MRE-5.3 was tested in vitro by EMSA. Each reaction contained [32P]-labeled GRE/MRE-5.3 oligo as probe (lanes 1–5) and 2 μl of in vitro-synthesized luciferase protein (negative control, lane 2) or hGR protein (lanes 3–5). The GR binding to the probe was competed with 1.5 μm radioinert GRE/MRE-5.3 (lane 4) or mutant GRE/MRE-5.3 oligonucleotides (lane 5). The supershifted bands indicated by the arrow are the GR-bound probe. D, pGL4.23 reporter constructs containing the mouse GRE/MRE-5.3 and corresponding GRE/MRE-5.3 mutant (X) were transfected into HT-22 cells. Cells were treated with CORT for 4 h before harvest and analysis by dual luciferase assay. Bars represent the mean ± sem (n = 5). Asterisks indicate statistically significant differences between vehicle (control) and CORT-treated cells for each enhancer-reporter construct (*, P < 0.05; **, P < 0.0001; Student's t test).
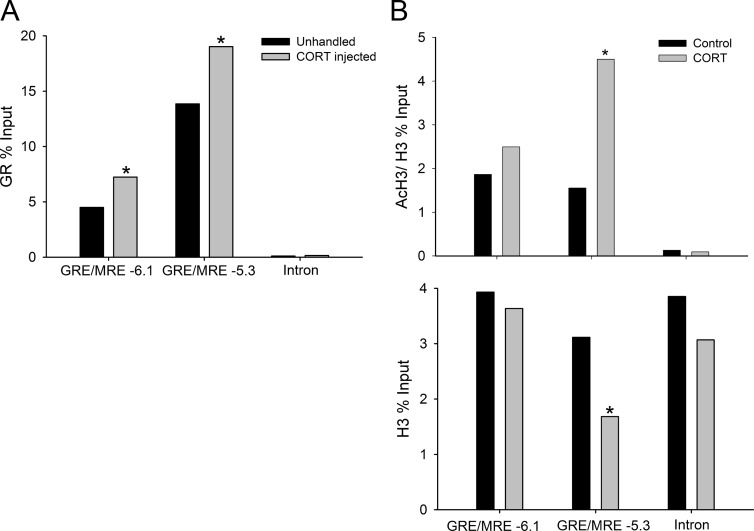
ChIP assay conducted with chromatin extracted from the hippocampal region of PND 6 mice showed GR association at both the GRE/MRE-6.1 and GRE/MRE-5.3 regions, which was increased by CORT injection (P < 0.05, Student's t test) (Fig. 6A). By comparison, there was no significant GR ChIP signal at the Klf9 intronic region (+11 kb from the TSS). In HT-22 cells, AcH3 was substantially greater at the two GRE/MRE regions compared with the intron. Treatment of cells with CORT for 4 h increased H3 acetylation at the GRE/MRE-5.3 (P < 0.01, Student's t test) but not at the GRE/MRE- 6.1 or at the intron (Fig. 6B, top panel). Treatment with CORT decreased H3 association but only at the GRE/MRE-5.3 (P < 0.01, Student's t test) (Fig. 6B, bottom panel).
Fig. 6.
CORT promotes GR association, H3 acetylation, and nucleosome eviction at the 5′-flanking region of the mouse Klf9 gene. A, wild-type PND 6 mice were injected with CORT at a dose of 14 mg/kg bodyweight or left unhandled. One hour after injection, animals were killed, the hippocampal region was dissected, and chromatin extracted for GR ChIP assay. ChIP samples (n = 9) were analyzed by RTqPCR using TaqMan assays that targeted the GRE/MRE-6.1, GRE/MRE-5.3, or a distal intronic region (negative control) of the mouse Klf9 gene. Bars represent the mean ChIP signals expressed as a percentage of input. Statistical analysis was conducted on log10-transformed data, and the asterisks indicate statistically significant differences between unhandled and CORT-injected animals (P < 0.05; Student's t test). B, HT-22 cells were treated with EtOH vehicle or 100 nm CORT for 4 h before harvest for chromatin extraction and ChIP assay for AcH3 (top panel) and H3 (bottom panel). ChIP samples (n = 4) were analyzed by RTqPCR using TaqMan assays that target the GRE/MRE-6.1, GRE/MRE-5.3, or a distal intronic region (negative control) of the mouse Klf9 gene. Bars represent the mean ChIP signals expressed as a percentage of input. Statistical analysis was conducted on log10-transformed data, and the asterisks indicate statistically significant differences between vehicle (control) and CORT-treated cells (P < 0.01; Student's t test).
Evolutionary conservation of the GRE/MRE in vertebrate Klf9 genes
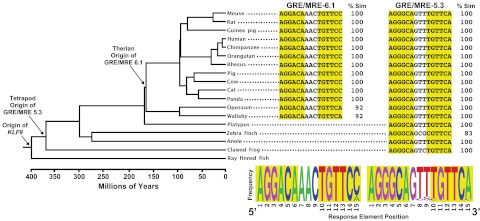
Bioinformatic analysis of the two GRE/MRE identified in this study (−5.3 and −6.1 kb in mouse) showed that these sequences are highly conserved across species. Both half-sites of the mouse GRE/MRE-5.3 are identical across several orders of mammals, a representative lizard, and a frog (Fig. 7). There are two nucleotide substitutions in one of the half-sites in zebra finch. We located an orthologous region in galliform genomes (chicken and turkey), but the putative GRE/MRE sequences were highly divergent and included an insertion mutation in the half-site spacer region. We were unable to identify a region orthologous to GRE/MRE-5.3 in ray-finned fish. The GRE/MRE-6.1 is highly conserved across therian mammals, with only a single base substitution in a half-site between marsupials and eutherians (Fig. 7). This GRE/MRE occurs within an approximately 150-base region that is also mostly conserved across therians. We could not identify an orthologous region, nor a GRE/MRE-6.1 in the genomes of a monotreme (platypus), birds (zebra finch, chicken, and turkey), a lizard (anole), clawed frogs, or fish (puffer fish, zebra fish, or stickle back).
Fig. 7.
Origin and evolution of two GRE/MRE in vertebrate Klf9 genes. Phylogeny of vertebrate relationships with branch lengths scaled to average divergence time estimates (www.timetree.org). Arrows indicate the minimum origin of the Klf9 gene, and Klf9 GRE/MRE-5.3, and 6.1 (numbering based on the distance from the mouse Klf9 TSS). The GRE/MRE sequences for each taxon are shown to the right of the tree (when present). Half-sites are highlighted, and their percent similarities (Sim) to mouse are listed. The frequency of each nucleotide position for the taxa shown is depicted below its respective GRE/MRE alignment.
Discussion
Glucocorticoids exert profound and complex actions on the central nervous system, many of which are mediated by the GR and MR. However, the genes regulated by GR and MR, and roles for the protein products of these genes in central nervous system development and function, are poorly understood. Here, we describe the identification and functional characterization of two GRE/MRE in Klf9, a GC-inducible gene in hippocampal neurons that codes for a transcription factor known to play a role in neuronal structure and function (15, 19, 20, 22, 23, 43). We found that the mouse Klf9 gene is induced by GC in vivo and in the hippocampal-derived cell line HT-22. The GR is recruited to Klf9 upstream regions in a hormone-dependent manner, leading to histone acetylation and nucleosome eviction at the GRE/MRE-5.3, consistent with GC promoting an open chromatin structure at the Klf9 locus. These results agree with our previous findings in Xenopus, where exposure to handling/shaking stressor or injection of CORT caused rapid increases in Klf9 mRNA and protein in frog brain (23).
Increased levels of Klf9 hnRNA in mouse brain in vivo and in HT-22 cells show that CORT increased transcription of the Klf9 gene. Also, the rapid kinetics of CORT-dependent up-regulation of Klf9 mRNA in HT-22 cells, and the resistance to protein synthesis inhibition, support that CORT regulation of Klf9 transcription is direct. Regulation of Klf9 by GR is supported by induction of Klf9 mRNA by the GR-specific agonist DEX and blockade of DEX and CORT responses by the GR-specific antagonist RU486. These results parallel our findings in Xenopus, where stressor-induced increases in Klf9 mRNA in vivo, or CORT-dependent responses in XTC-2 cells, were completely blocked by RU486 (23).
Our findings in mouse and frog support that Klf9 is regulated by the GR. We also provide evidence that mammalian Klf9 genes can be regulated by the MR, although the GR may predominate. ALDO caused a small, but significant, increase in Klf9 mRNA in HT-22/MR16 cells. However, this induction was significantly less than the CORT-mediated increase. In addition, CORT-dependent induction of Klf9 mRNA in HT-22/MR16 cells was blocked by the GR antagonist RU486 but not by the MR antagonist SPR. Taken together, we conclude that although the MR may contribute to the regulation of Klf9, this action is less important than the GR and may be dependent on the sequence of the GRE/MRE, the cell type, and perhaps the MR:GR ratio.
KLF9 is directly regulated by GC in hippocampal neurons via two GRE/MRE located in the 5′-flanking region of the gene. A ChIP-chip assay identified a GRE/MRE located at −5.5 kb in human Klf9, and we confirmed its presence in an orthologous region of the mouse genome (at −6.1 kb) and the functionality of this response element in both species. This sequence, comprising a GRE/MRE of two half-sites with a three base spacer (see Fig. 3B), was earlier found by So et al. (44) to be bound by GR in mouse C3H10T1/2 mesenchymal stem-like cells. Comparative sequence analysis showed that the GRE/MRE-6.1 is conserved among several orders of therian mammals but is not found in monotremes or other vertebrates. Datson et al. (11) recently found that GR binds to this region in rat hippocampal cells. We extend their findings by showing that this sequence supports hormone-dependent transactivation, which is abolished by point mutation of the half-sites; that GR binds to this sequence in vitro; and that GR associates with this genomic region in a hormone-dependent manner in vivo. We also show the therian origin of this response element and conservation over more than 160 million years in mammals (Fig. 7).
We identified a second GRE/MRE located at −5.3 kb in the mouse Klf9 gene (∼800 bp downstream of the GRE/MRE-6.1). This region was not bound by MR in a ChIP-chip assay of chromatin isolated from HEK293-hMR+ cells (Ziera, T., and S. Borden, unpublished data); we identified it in a promoter-reporter scan of the Xenopus Klf9 5′-flanking region. The orthologous region of the mouse gene supported GC-dependent transactivation, so we conducted DNAse I footprinting and subsequently identified a GRE/MRE full site. The GRE/MRE-5.3 half-sites are 100% conserved across several mammalian orders, a lizard, and a frog. Datson et al. (11) recently found evidence for a GRE/MRE in this region of the rat Klf9 gene. We extend their findings by confirming its functionality and high degree of conservation over 370 million years of tetrapod evolution.
The GRE/MRE-5.3 is found within an evolutionarily conserved genomic region of approximately 180 bp located −5 to −6 kb relative to the TSS in vertebrate Klf9 genes. Outside of the coding region and the core promoter, there is no evolutionary conservation between frog and mammal Klf9 genes, suggesting that this approximately 180-bp sequence has been subject to strong stabilizing selection. In Xenopus, this approximately 180-bp region was earlier found to contain a functional T3RE, to which TR binds to induce Klf9 expression during tadpole metamorphosis (42). We confirmed that the mouse has a homologous T3RE [Bagamasbad, P., and R. J. Denver, unpublished data; in addition to a T3RE at −3.8 kb relative to the TSS (16)]. The approximately 180-bp sequence also contains an evolutionarily conserved, predicted GRE/MRE half-site adjacent to the T3RE. However, this region was not protected by GR-DBD in DNAse I footprint assay. It is possible that the GR associates with the region of the T3RE through low-affinity DNA binding, or though protein-protein interactions, which could account for the residual CORT response observed when the GRE/MRE-5.3 was mutated.
ChIP assay showed greater GR association at the mouse GRE/MRE-5.3 compared with the GRE/MRE-6.1. This was accompanied by hormone-induced H3 acetylation at the GRE/MRE-5.3 but not at the GRE/MRE-6.1. H3 acetylation is a marker for transcriptionally active chromatin (8) and is necessary to promote nucleosome eviction and chromatin disassembly (45). Consistent with this, we observed hormone-dependent nucleosome eviction (depletion of H3) at the region of the GRE/MRE-5.3 but not at the GRE/MRE-6.1. Although the GR associates with the GRE/MRE-6.1 in vivo and in vitro, and this sequence can support hormone-dependent transactivation, our findings of GR recruitment and histone modification suggest that the GRE/MRE-5.3 may be a more significant hormone-response element in vivo. This may reflect the location of the GRE/MRE-5.3 within the conserved approximately 180-bp genomic region, where other proteins, such as the liganded TR, bind to cis regulatory elements to promote an open chromatin structure, thereby allowing for greater GR recruitment and action (Bagamasbad, P., and R. J. Denver, unpublished data).
GC actions on the hippocampus require new protein synthesis achieved though transactivation of GC target genes by liganded GR or MR. Recently, some GR/MR genomic targets have been identified in hippocampal neurons (e.g. Ref. 11), but little is known about the function of their gene products in hippocampal cell physiology and structure. Earlier we showed that KLF9 is in the pathway induced by TH that promotes differentiation of neuronal cells (15, 22, 23). Morita et al. (19) found that Klf9 null mice had reduced dendritic arborization of cerebellar Purkinje cells and behavioral abnormalities consistent with defects of the hippocampus, cerebellum, and amygdala. These mice also displayed impaired late-phase adult hippocampal neurogenesis (20), a critical component of hippocampal-dependent learning and memory (46). Given that structural plasticity of the hippocampus is regarded to be the basis for learning and memory (47) and that Klf9 is regulated by corticosteroids and TH, and is highly expressed in the hippocampus, a brain region targeted by corticosteroids and influences neuronal morphology, we hypothesize that KLF9 functions as an important intermediate in the actions of stress and TH on hippocampal plasticity and consequently on learning and memory.
Acknowledgments
We thank Dr. Yoshiaki Fujii-Kuriyama (Japan Science and Technology Agency, Saitama, Japan) for providing the Klf9-LacZ mice and to Dr. David Schubert (Salk Institute, La Jolla, CA) for providing the HT-22 cells.
This work was supported by the National Institute of Neurological Disorders and Stroke, National Institutes of Health Grant 1 R01 NS046690 and the National Science Foundation Grant IOS 0922583 (to R.J.D.). This research used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center funded by the National Institute of Diabetes and Digestive Kidney Diseases Grant NIHP60 DK20572. P.B. was supported by a predoctoral fellowship from the Rackham Graduate School and an Edwin Edwards Scholarship from the Department of Molecular, Cellular, and Developmental Biology, University of Michigan. A.M.R. was supported in part by the National Institutes of Health Cellular and Molecular Biology Training Grant T32-GM007315 at the University of Michigan.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AcH3
- Acetylated H3
- ALDO
- aldosterone
- ChIP
- chromatin immunoprecipitation
- CHX
- cycloheximide
- CMV
- cytomegalovirus
- CORT
- corticosterone
- DBD
- DNA binding domain
- DEX
- dexamethasone
- DNAse
- deoxyribonuclease
- FBS
- fetal bovine serum
- GC
- glucocorticoid
- GR
- GC receptor
- GRE
- GC-response element
- H3
- histone 3
- hGR
- human GR
- hnRNA
- heteronuclear RNA
- KLF9
- Krüppel-like factor 9
- MMTV
- mouse mammary tumor virus
- MR
- mineralocorticoid receptor
- MRE
- mineralocorticoid-response element
- PND
- postnatal day
- RTqPCR
- reverse transcription quantitative PCR
- RU486
- mifepristone
- SPR
- spironolactone
- TESS
- Transcription Element Search System
- TH
- thyroid hormone
- T3RE
- T3-response element
- TSS
- transcription start site.
References
- 1. Joëls M, Pu Z, Wiegert O, Oitzl MS, Krugers HJ. 2006. Learning under stress: how does it work? Trends Cogn Sci 10:152–158 [DOI] [PubMed] [Google Scholar]
- 2. McEwen BS. 2008. Central effects of stress hormones in health and disease: understanding the protective and damaging effects of stress and stress mediators. Eur J Pharmacol 583:174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reul JM, de Kloet ER. 1985. Two receptor systems for corticosterone in rat brain: microdistribution and differential occupation. Endocrinology 117:2505–2511 [DOI] [PubMed] [Google Scholar]
- 4. De Kloet ER, Vreugdenhil E, Oitzl MS, Joëls M. 1998. Brain corticosteroid receptor balance in health and disease. Endocr Rev 19:269–301 [DOI] [PubMed] [Google Scholar]
- 5. Puymirat J. 1992. Thyroid receptors in the rat brain. Prog Neurobiol 39:281–294 [DOI] [PubMed] [Google Scholar]
- 6. Aranda A, Pascual A. 2001. Nuclear hormone receptors and gene expression. Physiol Rev 81:1269–1304 [DOI] [PubMed] [Google Scholar]
- 7. Ribeiro RC, Kushner PJ, Baxter JD. 1995. The nuclear hormone receptor gene superfamily. Annu Rev Med 46:443–453 [DOI] [PubMed] [Google Scholar]
- 8. Aoyagi S, Archer TK. 2008. Dynamics of coactivator recruitment and chromatin modifications during nuclear receptor mediated transcription. Mol Cell Endocrinol 280:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McEwen BS. 2010. Stress, sex, and neural adaptation to a changing environment: mechanisms of neuronal remodeling. Ann NY Acad Sci 1204(Suppl):E38–E59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Datson NA, Morsink MC, Meijer OC, de Kloet ER. 2008. Central corticosteroid actions: search for gene targets. Eur J Pharmacol 583:272–289 [DOI] [PubMed] [Google Scholar]
- 11. Datson NA, Polman JA, de Jonge RT, van Boheemen PT, van Maanen EM, Welten J, McEwen BS, Meiland HC, Meijer OC. 2011. Specific regulatory motifs predict glucocorticoid responsiveness of hippocampal gene expression. Endocrinology 152:3749–3757 [DOI] [PubMed] [Google Scholar]
- 12. Datson NA, Speksnijder N, Mayer JL, Steenbergen PJ, Korobko O, Goeman J, de Kloet ER, Joëls M, Lucassen PJ. 2012. The transcriptional response to chronic stress and glucocorticoid receptor blockade in the hippocampal dentate gyrus. Hippocampus 22:359–371 [DOI] [PubMed] [Google Scholar]
- 13. Datson NA, van der Perk J, de Kloet ER, Vreugdenhil E. 2001. Identification of corticosteroid-responsive genes in rat hippocampus using serial analysis of gene expression. Eur J Neurosci 14:675–689 [DOI] [PubMed] [Google Scholar]
- 14. Morsink MC, Steenbergen PJ, Vos JB, Karst H, Joëls M, De Kloet ER, Datson NA. 2006. Acute activation of hippocampal glucocorticoid receptors results in different waves of gene expression throughout time. J Neuroendocrinol 18:239–252 [DOI] [PubMed] [Google Scholar]
- 15. Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. 1999. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J Biol Chem 274:23128–23134 [DOI] [PubMed] [Google Scholar]
- 16. Denver RJ, Williamson KE. 2009. Identification of a thyroid hormone response element in the mouse Kruppel-like factor 9 gene to explain its postnatal expression in the brain. Endocrinology 150:3935–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Imataka H, Nakayama K, Yasumoto K, Mizuno A, Fujii-Kuriyama Y, Hayami M. 1994. Cell-specific translational control of transcription factor BTEB expression. The role of an upstream AUG in the 5′-untranslated region. J Biol Chem 269:20668–20673 [PubMed] [Google Scholar]
- 18. Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. 1992. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J 11:3663–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morita M, Kobayashi A, Yamashita T, Shimanuki T, Nakajima O, Takahashi S, Ikegami S, Inokuchi K, Yamashita K, Yamamoto M, Fujii-Kuriyama Y. 2003. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol Cell Biol 23:2489–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scobie KN, Hall BJ, Wilke SA, Klemenhagen KC, Fujii-Kuriyama Y, Ghosh A, Hen R, Sahay A. 2009. Kruppel-like factor 9 is necessary for late-phase neuronal maturation in the developing dentate gyrus and during adult hippocampal neurogenesis. J Neurosci 29:9875–9887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dussault JH, Ruel J. 1987. Thyroid hormones and brain development. Annu Rev Physiol 49:321–334 [DOI] [PubMed] [Google Scholar]
- 22. Cayrou C, Denver RJ, Puymirat J. 2002. Suppression of the basic transcription element-binding protein in brain neuronal cultures inhibits thyroid hormone-induced neurite branching. Endocrinology 143:2242–2249 [DOI] [PubMed] [Google Scholar]
- 23. Bonett RM, Hu F, Bagamasbad P, Denver RJ. 2009. Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: implications for neural development and plasticity. Endocrinology 150:1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quadrilatero J, Hoffman-Goetz L. 2005. In vivo corticosterone administration at levels occurring with intense exercise does not induce intestinal lymphocyte apoptosis in mice. J Neuroimmunol 162:137–148 [DOI] [PubMed] [Google Scholar]
- 25. Denver RJ. 1998. Hormonal correlates of environmentally induced metamorphosis in the Western spadefoot toad, Scaphiopus hammondii. Gen Comp Endocrinol 110:326–336 [DOI] [PubMed] [Google Scholar]
- 26. Licht P, McCreery BR, Barnes R, Pang R. 1983. Seasonal and stress related changes in plasma gonadotropins, sex steroids, and corticosterone in the bullfrog, Rana catesbeiana. Gen Comp Endocrinol 50:124–145 [DOI] [PubMed] [Google Scholar]
- 27. Manzon RG, Denver RJ. 2004. Regulation of pituitary thyrotropin gene expression during Xenopus metamorphosis: negative feedback is functional throughout metamorphosis. J Endocrinol 182:273–285 [DOI] [PubMed] [Google Scholar]
- 28. Ziera T, Irlbacher H, Fromm A, Latouche C, Krug SM, Fromm M, Jaisser F, Borden SA. 2009. Cnksr3 is a direct mineralocorticoid receptor target gene and plays a key role in the regulation of the epithelial sodium channel. FASEB J 23:3936–3946 [DOI] [PubMed] [Google Scholar]
- 29. Morimoto BH, Koshland DE., Jr 1990. Induction and expression of long- and short-term neurosecretory potentiation in a neural cell line. Neuron 5:875–880 [DOI] [PubMed] [Google Scholar]
- 30. Morimoto BH, Koshland DE., Jr 1990. Excitatory amino acid uptake and N-methyl-D-aspartate-mediated secretion in a neural cell line. Proc Natl Acad Sci USA 87:3518–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sagara Y, Dargusch R, Chambers D, Davis J, Schubert D, Maher P. 1998. Cellular mechanisms of resistance to chronic oxidative stress. Free Radic Biol Med 24:1375–1389 [DOI] [PubMed] [Google Scholar]
- 32. Maher P, Davis JB. 1996. The role of monoamine metabolism in oxidative glutamate toxicity. J Neurosci 16:6394–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rozeboom AM, Akil H, Seasholtz AF. 2007. Mineralocorticoid receptor overexpression in forebrain decreases anxiety-like behavior and alters the stress response in mice. Proc Natl Acad Sci USA 104:4688–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Samuels HH, Stanley F, Casanova J. 1979. Depletion of L-3,5,3′-triiodothyronine and L-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology 105:80–85 [DOI] [PubMed] [Google Scholar]
- 35. Yao M, Schulkin J, Denver RJ. 2008. Evolutionarily conserved glucocorticoid regulation of corticotropin-releasing factor expression. Endocrinology 149:2352–2360 [DOI] [PubMed] [Google Scholar]
- 36. Hoopfer ED, Huang L, Denver RJ. 2002. Basic transcription element binding protein is a thyroid hormone-regulated transcription factor expressed during metamorphosis in Xenopus laevis. Dev Growth Differ 44:365–381 [DOI] [PubMed] [Google Scholar]
- 37. Blauwkamp TA, Chang MV, Cadigan KM. 2008. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J 27:1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Denver RJ, Pavgi S, Shi YB. 1997. Thyroid hormone-dependent gene expression program for Xenopus neural development. J Biol Chem 272:8179–8188 [DOI] [PubMed] [Google Scholar]
- 39. de Kloet ER, Reul JM, Sutanto W. 1990. Corticosteroids and the brain. J Steroid Biochem Mol Biol 37:387–394 [DOI] [PubMed] [Google Scholar]
- 40. Veldhuis HD, Van Koppen C, Van Ittersum M, De Kloet ER. 1982. Specificity of the adrenal steroid receptor system in rat hippocampus. Endocrinology 110:2044–2051 [DOI] [PubMed] [Google Scholar]
- 41. Arriza JL, Simerly RB, Swanson LW, Evans RM. 1988. The neuronal mineralocorticoid receptor as a mediator of glucocorticoid response. Neuron 1:887–900 [DOI] [PubMed] [Google Scholar]
- 42. Furlow JD, Kanamori A. 2002. The transcription factor basic transcription element-binding protein 1 is a direct thyroid hormone response gene in the frog Xenopus laevis. Endocrinology 143:3295–3305 [DOI] [PubMed] [Google Scholar]
- 43. Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. 2008. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455:1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. 2008. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc Natl Acad Sci USA 105:5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luebben WR, Sharma N, Nyborg JK. 2010. Nucleosome eviction and activated transcription require p300 acetylation of histone H3 lysine 14. Proc Natl Acad Sci USA 107:19254–19259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Toni N, Laplagne DA, Zhao C, Lombardi G, Ribak CE, Gage FH, Schinder AF. 2008. Neurons born in the adult dentate gyrus form functional synapses with target cells. Nat Neurosci 11:901–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leuner B, Gould E. 2010. Structural plasticity and hippocampal function. Annu Rev Psychol 61:111–140:C111–C113 [DOI] [PMC free article] [PubMed] [Google Scholar]