Abstract
TBR1 encodes a transcription factor with critical roles in corticogenesis, including cortical neuron migration and axon pathfinding, establishment of regional and laminar identity of cortical neurons, and control of glutamatergic neuronal cell fate. Based upon TBR1's role in cortical development, we sought to investigate TBR1 hemizygosity in individuals referred for genetic evaluation of intellectual disability and developmental delay. We describe 4 patients with microdeletions identified by molecular cytogenetic techniques, encompassing TBR1 and spanning 2q24.1q31.1, ranging in size from 2.17 to 12.34 Mb. Only the patient with the largest deletion had a possible cortical malformation. Mild ventriculomegaly is the only common brain anomaly, present in all patients; a Chiari I malformation is seen in 2 patients, and mega cisterna magna is seen in a third. Our findings are consistent with Tbr1 mouse models showing that hemizygosity of the gene requires additional genetic factors for the manifestation of severe structural brain malformations. Other syndromic features are present in these patients, including autism spectrum disorders, ocular colobomas, and craniosynostosis, features that are likely affected by the deletion of genes other than TBR1.
Key Words: 2q24, aCGH, Cortical development, Microdeletion, TBR1
TBR1 encodes a T-box family transcription factor expressed in postmitotic projection neurons and functionally significant in embryological corticogenesis [Bulfone et al., 1995; Hevner et al., 2001]. In mice, Tbr1 regulates regional and laminar identity, and the cortex of null mutants displays disturbed laminar organization from defective preplate splitting and Reln downregulation [Hevner et al., 2001; Bedogni et al., 2010a]. Tbr1 also acts as a direct transcriptional repressor to restrict the formation of the corticospinal tract to layer 5 of the cortical plate [Han et al., 2011] while specifying corticothalamic neuron identity in layer 6 [McKenna et al., 2011], and it functions in cortical and thalamic axonal pathfinding [Hevner et al., 2002].
TBR1 is part of the PAX6-TBR2-NEUROD-TBR1 transcription factor cascade that is critical for controlling glutamatergic neuronal cell fate in the cortex, cerebellum, and hippocampus [Englund et al., 2005; Hevner et al., 2006; Mendez-Gomez et al., 2011], and TBR1 also functions in the CASK-TBR1-RELN pathway necessary for proper neuronal migration during corticogenesis [Hevner et al., 2006]. Mutations of these genes in humans are associated with defects in embryological brain development; for example, individuals with heterozygous PAX6 mutations have been reported to have eye malformations and absence or hypoplasia of the anterior commissure, reduced olfaction, absence of the pineal gland, and polymicrogyria [Sisodiya et al., 2001; Mitchell et al., 2003]. Likewise, heterozygous mutation or deletion of CASK results in microcephaly, simplified gyral pattern, thin brainstem with flattening of the pons, and severe cerebellar hypoplasia [Najm et al., 2008]. Homozygous mutation of RELN has been associated with lissencephaly, a neuronal migration disorder [Hong et al., 2000]. Disruption of these pathways in animals demonstrate their crucial function in embryological brain development [Englund et al., 2005; Atasoy et al., 2007; Arnold et al., 2008; Tuoc et al., 2009; Bedogni et al., 2010a], though homozygous Tbr1 deletion is required in mice before brain abnormalities manifest. Heterozygous mice do not display an obvious phenotype [Bulfone et al., 1998]. Given its roles in neurodevelopment, we targeted TBR1 for study to investigate whether hemizygosity would result in cortical malformations and neurological impairment.
Materials and Methods
Patient Ascertainment
Patients 1, 3, and 4 were ascertained by Signature Genomic Laboratories (Spokane, Wash., USA) following referral for clinical microarray-based comparative genomic hybridization (aCGH) testing. Patient 2 was ascertained by Nemours Children's Clinic. Written consent was obtained to publish images using an Institutional Review Board Spokane-approved consent form.
Oligonucleotide-Based aCGH
Oligonucleotide-based aCGH analysis was performed at Signature Genomics on DNA from patients 1, 3, and 4 using a 105K-feature, whole-genome microarray (SignatureChipOS® version 1, custom-designed by Signature Genomics; manufactured by Agilent Technologies, Santa Clara, Calif., USA) as previously described [Ballif et al., 2008]. Results were analyzed and visualized using aCGH analysis and web-based data visualization software (Genoglyphix®; Signature Genomics).
Single Nucleotide Polymorphism Array
A single nucleotide polymorphism (SNP) array (Genome-Wide Human SNP Array 6.0; Affymetrix, Santa Clara, Calif., USA), which consists of 906,600 SNPs and 946,000 probes for analyzing copy number variation, was performed at LabCorp (Burlington, N.C., USA) on patient 2. The mean intermarker distance for the SNP array is 700 bp. In brief, DNA was extracted from whole blood, and 250 ng of patient DNA was digested with restriction enzymes. Next, the fragments were ligated to adaptors that are recognized by a particular primer necessary for polymerase chain reaction amplification within a certain size range. Finally, the amplified DNA was fragmented and labeled prior to array hybridization, and visualization was performed following washing and staining of the array. GeneChip Genotyping Analysis Software (Affymetrix, Santa Clara, Calif., USA) was used to statistically analyze data and obtain breakpoints that were reported via graphic visualization.
FISH
Copy number abnormalities detected by microarray in patients 1–4 were visualized by metaphase FISH using one or more BAC clones located within the abnormal regions as previously described [Traylor et al., 2009]. When available, parental samples were also analyzed using FISH.
Results
Microarray analysis in each of the patients identified a deletion of 2q24, including TBR1 (fig. 1). No additional clinically significant copy number changes were identified in any of the 4 patients. Parental FISH testing for patients 2 and 3 showed the deletions to be apparently de novo in origin. All other parental samples were unavailable for testing (table 1).
Fig. 1.
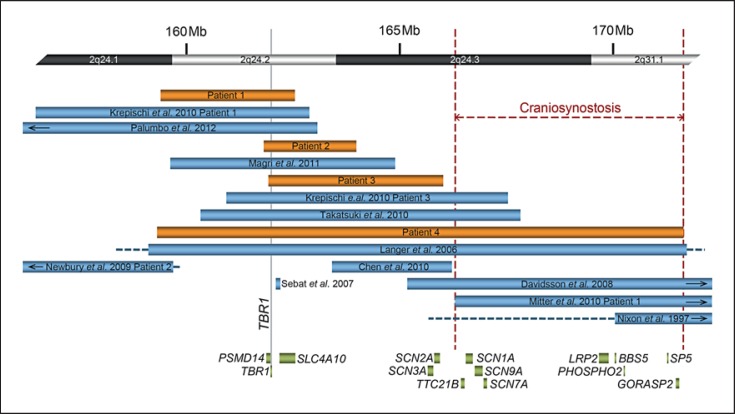
Overview of molecularly defined deletions within 2q24.1q31.1. Schematic of cases in the literature (shown in light blue) with deletions characterized by molecular cytogenetic techniques and those in patients 1–4 (shown in orange). At the top of the figure there is a partial idiogram showing chromosome bands 2q24.1q31.1, with genomic coordinates corresponding to the hg18 build of the human genome. Blue and orange bars represent minimum deletion sizes, and horizontal dashed lines extend to show maximum deletion sizes. Genes of note within the region are represented by green bars. The vertical gray solid line indicates the location of TBR1. The largest possible region of overlap among individuals with craniosynostosis is bordered by red dashed lines.
Table 1.
Features of individuals with deletions including TBR1
| Features | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Langer et al. [2006] | Krepischi et al. [2010] | Krepischi et al. [2010] | Takatsuki et al. [2010] | Magri et al. [2011] | Palumbo et al. [2010] |
|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Patient 3 | |||||||||
| Sex | male | male | male | male | female | female | female | female | male | female |
| Age, years | 11.5 | 8 | 33 months | 16 months | 2 years 8 months | 14 | 5 | 5 months | 3.5 | 3 |
| Minimum deletion coordinates (hg18) | 159,392,858–162,536,161 | 161,813,983–163,980,679 | 161,919,306–166,011,752 | 159,307,955–171,648,560 | 159,108,825–171,723,298 | 156,469,445–162,877,841 | 160,937,947–167,531,521 | 160,332,614–167,824,700 | 159,618,452–164,882,054 | 155,526,470–163,058,894 |
| Deletion size, Mb | 3.14 | 2.17 | 4.09 | 12.34 | 12.61 | 6.41 | 6.59 | 7.49 | 5.26 | 7.53 |
| Number of genes | 15 | 9 | 16; includes | 64; includes | 65; includes | 31 | 27; includes | 33; includes | 22 | 33 |
| SCN2A | SCN1A | SCN1A | SCN1A | SCN1A | ||||||
| Inheritance | unknown | de novo | de novo | unknown | de novo | de novo | de novo | de novo | de novo | de novo |
| Growth features | ||||||||||
| Birth weight | ||||||||||
| percentile | 10th–25th | 25th–50th | 50th–75th | 3rd | 15th | <3rd | 50th | <3rd (−2.4 SD) | 3rd | 50th |
| Weight percentile | <3rd (−4.2 SD) | 95th | <3rd(−2.1 SD) | <3rd (−5 SD) | 80th | 50th–75th | NS | <3rd (−2.3 SD) | NS | 25th |
| Height percentile | 5th–10th | 71st | 3rd–10th | <3rd (−4 SD) | 50th | <3rd (−2.3 SD) | NS | 3rd–10th | 10th | 50th |
| OFC percentile | 2nd | 51st | 10th−25th | <3rd (−7.2 SD) | 3rd | 5th | <3rd (−4.3 SD) | NS | NS | 10th |
| Neurological features | ||||||||||
| DD/ID | moderate | + | + | severe | severe | + | profound | + | severe | + |
| Speech | apraxia; nasal voice | no speech; uses signs | single word | no speech | no speech | no speech | no speech | NA | NS | first words at 3 years |
| Behavior problems | ADHD; some perseverations | PDD; tantrums; hyperactivity | autistic-like | NA | − | autistic | − | NA | − | − |
| Hypotonia | + | + | + | − | + | − | moderate | + | severe | generalized |
| Seizures | staring spells at 5 years | − | − | onset 11 week, infantile spasms and complex partial seizures, intractable | onset 3.5 months, initially 40 daily | onset 10 years, tonic-generalized | onset 2 months, intractable epilepsy, varied seizure types | at 2 months, continuous myoclonic jerks, intractable; at 3 months, tonic-clonic | − | − |
| EEG | normal | abnormal | NA | abnormal | NS | abnormal | abnormal | NS | NA | NA |
| Brain malformations | mega cisterna magna, mild ventriculomegaly | thick CC, mild ventriculomegaly | Chiari I, thick CC, mild ventriculomegaly | Chiari I, possible cortical dysplasia, mild ventriculomegaly | ultrasound: lack of homogeneity around thalamus | normal MRI | normal MRI | NS | normal ultrasound | normal MRI |
| Other | ataxic gait | bruxism | ||||||||
| Ophthalmologic | − | strabismus, | intermittent | bilateral | microphthalmia, | − | bilateral | − | − | − |
| features | surgically | left esotropia | colobomas | coloboma, | coloboma | |||||
| repaired | blepharophimosis | |||||||||
| Dysmorphic features | ||||||||||
| Head | − | facial asymmetry | − | craniosynostosis | bulging metopic suture | − | brachycephaly | − | flat occiput, fine hair, prominent forehead | − |
| Auricular region | − | − | small and low-set ears | low-set and rotated ears | low-set and rotated ears | − | − | low-set ears | − | − |
| Periocular region | − | downslanting PF | epicanthal folds, short PF | small and downslanting PF | short and downslanting PF, hypertelorism | − | − | thick and arched eyebrows, upslanting PF, long eyelashes | sparse eyebrows | − |
| Midface | − | wide alae nasi | − | − | broad nasal root | − | thin nose, depressed and broad nasal bridge, anteverted nares | flat nasal bridge, short nose | − | − |
| Perioral region | − | − | − | retrognathia | − | − | short philtrum | long philtrum, small mouth, micrognathia | large mouth, short philtrum, micrognathia | − |
| Limb features | hyperextensible elbows | turned-in foot | tapered fingers; joint laxity | brachydactyly; STPC | − | small feet and hands, tapered fingers, valgus feet, abduction of upper limbs | tapered fingers | − | camptodactyly of the hallux; joint laxity | − |
| Respiratory features | frequent infections | normal | − | respiratory failure | central apnea | − | recurrent infections due to swallowing difficulties | emphysema caused by viral infection | − | − |
| Gastrointestinal features | dysphagia | GERD | GERD, resolved | G-tube feedings | − | − | − | − | − | − |
| Endocrine features | − | − | − | hypothyroidism | − | − | hypothyroidism | − | − | − |
| Other features | t(7;10)(q22;q26) | wide-spaced nipples, chordee | displaced anus | hypotrophic muscles | hip dislocation |
+ = Feature present; − = feature absent; ADHD = attention deficit hyperactivity disorder; CC = corpus callosum; GERD = gastroesophageal reflux disease; ID = intellectual disability; NA = not applicable; NS = not specified; PDD = pervasive developmental disorder; PF = palpebral fissures; SD = standard deviations; STPC = single transverse palmar crease.
Clinical Summaries
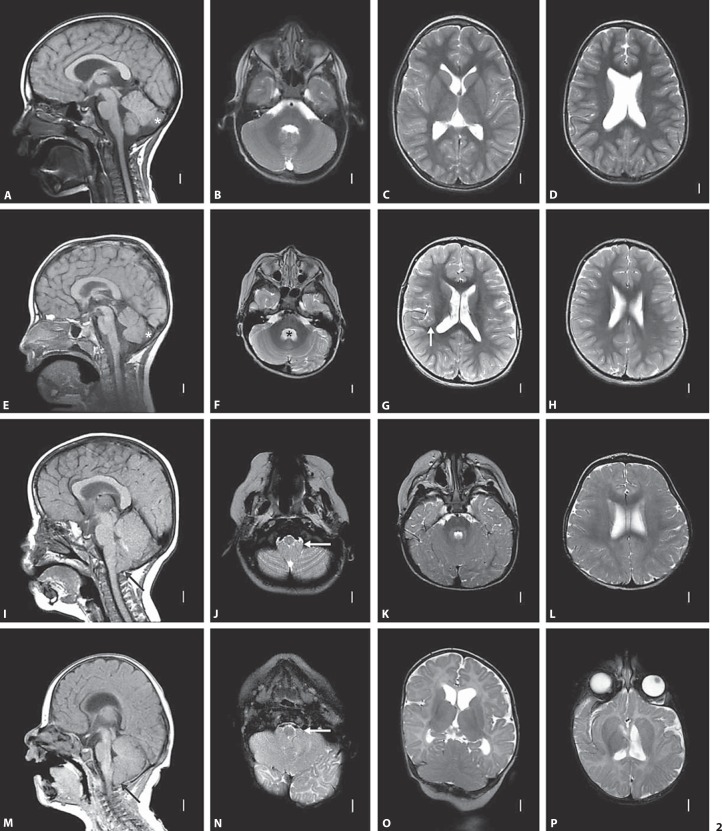
Patient 1 is an 11.5-year-old male with global developmental delay (DD), moderate intellectual disability, attention deficit hyperactivity disorder, speech apraxia, dysphagia, and small size. He was born at 42 weeks gestation to a G2P1 mother following a pregnancy complicated by bleeding at 3 months and a choroid plexus cyst identified by ultrasound. His birth weight was at the 10th–25th percentile, and length was at the 75th–90th percentile. His postnatal course was complicated by a sebaceous cyst that was removed at 6 months of age. DD and constitutional small size were first noted in the early newborn period. Developmentally, he sat at 7 months, stood at 9 months, walked at 1.5 years, and had his first words at 2 years. A formal psychological evaluation performed at 5 years 11 months showed a Pictorial Intelligence Quotient in the range of 52–58 (normal 85–115) and a General Adaptive Composite in the range of 50–63 (normal 85–115). Staring spells prompted an MRI and electroencephalogram (EEG) at 5 years of age, but both were reportedly normal. MRI at 6 years 5 months showed mild ventriculomegaly and mild mega cisterna magna with normal versus borderline small size of the cerebellar vermis (fig. 2). He has a tendency for frequent respiratory infections. He takes methylphenidate daily for attention deficit hyperactivity disorder. At the age of 11.5 years his weight is <3rd percentile, his height is at the 5th–10th percentile, and his occipitofrontal circumference (OFC) is at the 2nd percentile. He has verbal apraxia and hypernasal speech. He speaks in sentences and phrases but is not easily understood. He has both fine motor and gross motor delays. He can write his first name only. He has tight heel cords, decreased muscle tone, foot slap, and does some toe walking. Oligonucleotide aCGH, performed at 11.5 years, revealed a 3.14-Mb, apparently de novo 2q24.1q24.2 deletion (fig. 1).
Fig. 2.
MRI images of patients 1–4. Brain MRI in patients 1 (A–D), 2 (E–H), 3 (I–L) and 4 (M–P) with deletion 2q24 include T1-weighted midline sagittal (left column) images and T2-weighted axial images at multiple levels (all others except 1 coronal image in O). The midline sagittal images in patients 1 (A) and 2 (E) show normal skull shape and mildly prominent extra-axial fluid below and behind the cerebellum, indicating subtle mega cisterna magna (asterisks in A and E overlie the hemispheres, not vermis). The fourth ventricle is mildly enlarged in patient 2 (asterisk in F), and the lateral ventricles are mildly enlarged in both patients (C, D, G, H). Midline sagittal images in patients 3 (I) and 4 (M) show brachycephaly, small posterior fossa with reduced extra-axial spaces surrounding the cerebellum, small ‘pinched’ fourth ventricle, and marked cerebellar tonsillar ectopia consistent with Chiari malformation type 1 (long arrows in I and M). The low lying cerebellar tonsils are also seen on low axial images (arrows in J and N). The coronal image in patient 4 (O) shows mild upward displacement of the anterior (superior) cerebellum, suggesting a small posterior fossa. The lateral ventricles appear normal in patient 3 and mildly enlarged and asymmetric in patient 4. The angle of the axial images in P is nonstandard, lending an unusual appearance to the ventricles.
Patient 2 is an 8-year-old male with significant verbal expressive language delay and possible pervasive developmental disorder. He was born at 39 weeks gestation by Cesarean delivery after his mother developed preeclampsia late in the pregnancy. She suffered from frequent emesis but was otherwise healthy during the pregnancy. Following an abnormal maternal serum screen, fetal karyotyping was performed and showed a normal male result, 46,XY. His birth weight was at the 25th–50th percentile, and his length was at the 50th–75th percentile. At 3 months of age, he was diagnosed with gastroesophageal reflux disease. At 18 months, he was diagnosed with pervasive developmental disorder and placed on a gluten-free, casein-free diet. Examination at 2 years of age indicated DD, hypotonia, and a history of alternating between constipation and diarrhea. While EEG exam was normal, MRI showed a mild dilation of the ventricles and thick corpus callosum (fig. 2). Subsequent EEG revealed abnormal spikes and wave patterns during sleep, but he has never had any overt seizures, and he takes valproic acid, which he tolerates well. He had a tendon transfer due to turning in of his foot and strabismus surgery. He is nonverbal but has about 200 signs and is able to communicate using a tablet computer. He experiences some difficulty interacting with other children, has tantrums, and has some obsessive-compulsive traits. Treatment with guanfacine has helped with focus and decreasing hyperactivity. At 8 years of age, his weight is at the 95th percentile, his height is at the 71st percentile, and his OFC is at the 51st percentile. Minor dysmorphic features include a slight facial asymmetry with the right cheek fuller than the left, mildly downslanting palpebral fissures, and a slight widening of the alae nasi. SNP array analysis revealed a 2.17-Mb, apparently de novo 2q24.2q24.3 deletion (fig. 1).
Patient 3 is a 33-month-old male with DD and hypotonia. He was born at 41 weeks gestation by Cesarean delivery to a 28-year-old G2P1 mother who had a kidney stone during the fourth month of pregnancy. His birth weight was at the 50th–75th percentile, and length was >95th percentile. He was noted to have significant gastroesophageal reflux in the first month of life with recurrent vomiting and poor suck. At 6 months of age, he was noted to have DD as he did not roll over or sit unaided. At 12 months of age, he had tubes placed in his ears for recurrent otitis media. MRI at 12 months showed a moderate to severe Chiari I malformation with cerebellar tonsils herniated ∼8 mm below foramen magnum (fig. 2). Chromosome analysis showed an apparently balanced, de novo translocation, 46,XY,t(7;10)(q22;q26). By 14 months, his reflux had resolved, but he occasionally suffered from constipation. He first walked at 27 months, and at 33 months has a single word (‘hey’). He is also reported to have autistic-like features. At 33 months, his weight is <3rd percentile, his height is at the 3rd–10th percentile, and his OFC is at the 10th–25th percentile. Dysmorphic features include short palpebral fissures, intermittent left esotropia, slight bilateral epicanthal folds, subtle low-set ears, slight taper in the fingers, and overall small size. Oligonucleotide aCGH, performed at 14 months, revealed a 4.09-Mb, apparently de novo 2q24.2q24.3 deletion (fig. 1).
Patient 4 was a male with severe DD, craniosynostosis, epilepsy, colobomas, and growth retardation, who passed away at 16 months. He was born at 39 weeks via induced vaginal delivery to a 30-year-old G2P1 mother following an uncomplicated pregnancy. He was noted to have dysmorphic features and to be small for gestational age. Dysmorphic features included small downslanting palpebral fissures, posteriorly rotated ears, wide-spaced nipples, chordee of penis, brachydactyly, a single transverse palmar crease on the right hand, and a bridged palmar crease on the left hand. At 1 month, ophthalmology exam showed severe uveal coloboma on the left with likely no potential for central visual development and coloboma of the choroid and retina on the right with likely preserved vision. Cytogenetic studies showed 46,XY,inv(9)(p11q13); the inverted 9 is a known population variant. Head CT at 2 months showed craniosynostosis consisting of fusion of the posterior portion of the sagittal suture and superior portion of the left coronal suture, which was repaired at 6 months. At 11 weeks of age, he began having seizures as frequently as 3 times per day that were characterized by staring with a jerking of his hands and feet and nystagmoid movements of the eye preceded by crying. The seizures were temporarily stopped with the use of phenobarbitol but then recurred. He was placed on levetiracetam and oxcarbazepine, and then phenytoin with clonazepam and diazapam daily, but he continued to have 6–20 seizures daily. EEG at 4 months showed mild, diffuse dysfunction in both hemispheres, at 6 months showed frequent left temporal interictal discharges, and at 9 months showed unusual diffusely slow background pattern and multifocal sharp wave discharges that were potentially epileptogenic. MRI at 4 months showed a Chiari I malformation (fig. 2). While his length was following a curve below the third percentile, he was gaining very little weight. He was diagnosed with hypothyroidism and required G-tube feedings. He died at 16 months of age while asleep, most likely due to respiratory failure in relation to a seizure; bronchoscopy at 14 months had shown pharyngeal collapse, glossoptosis, and laryngomalacia. Oligonucleotide aCGH, performed at 1.5 months, revealed a 12.34-Mb 2q24.1q31.1 deletion (fig. 1).
Discussion
Based on its demonstrated role in corticogenesis in mouse models, as well as its involvement in critical developmental pathways, we sought to interrogate TBR1 hemizygosity for association with cortical malformations and neurological impairment. We gathered detailed clinical records and brain images for a cohort of 4 individuals with 2q24.2 deletions encompassing TBR1. Whilst the individuals reported here were referred for genetic evaluation of intellectual disability and DD, other features common to this cohort include hypotonia (3/4), dysphagia (4/4), constitutional small size and/or low weight (3/4), behavior problems (3/3), downslanting palpebral fissures (2/4), and mildly abnormal ears (2/4), all relatively nonspecific features that are common in individuals with chromosomal abnormalities. The absence of a common, distinctive phenotype among the 4 patients leads us to conclude that TBR1 deletions do not result in a recognizable, novel microdeletion syndrome. In addition, syndromic features present in some of these patients are likely attributable to deletion of multiple genes in the region, in combination with other background genetic/environmental factors, such as the de novo translocation in patient 3.
Within this cohort various brain abnormalities were identified by MRI, and the only common finding was mild ventriculomegaly, another relatively nonspecific finding. Only patient 4, with the largest deletion, had a possible malformation in the right perisylvian region of the cortex (fig. 2), which was our hypothesized TBR1-associated malformation, given the role of TBR1 in corticogenesis. Cortical malformations were not seen in the other patients in our study, nor have they been reported in other individuals in the literature with TBR1 deletions. Patient 2, with the smallest deletion, showed only mild changes on MRI, and patient 1 displayed a mega cisterna magna, a feature opposite from the Chiari I malformation observed in patients 3 and 4. Patients 2 and 3 each had a thick corpus callosum, a feature not seen in patient 1, while patient 4 was too young to ascertain this feature. TBR1 hemizygosity may possibly contribute to these structural brain changes, but as seen with human mutations of other genes in the PAX6 transcription factor cascade and the RELN pathway, homozygous loss may be required before major brain malformations manifest [Glaser et al., 1994; Baala et al., 2007; Solomon et al., 2009]. Additionally, the more severe malformations, such as the Chiari I malformations in patients 3 and 4, may be attributed to loss of other gene(s) in their ∼4-Mb shared deletion region. Alternatively, as Chiari I malformations have been observed in association with craniosynostosis, like in patient 4, and are thought to be due to mechanical forces put on the brain [Raybaud and Di Rocco, 2007], the malformations may have different etiologies in the 2 patients.
Deletions of various sizes spanning the 2q21q31 region have been reported in over 100 cases and are associated with a broad spectrum of phenotypic features [Pereira et al., 2004; Langer et al., 2006; Pescucci et al., 2007; Davidsson et al., 2008; Newbury et al., 2009; Chen et al., 2010; Krepischi et al., 2010; Takatsuki et al., 2010; Magri et al., 2011; Palumbo et al., 2012], including seizure disorder [Grosso et al., 2008]. Substantial evidence indicates that the sodium channel (SCN) α subunit genes of the 2q24.3 region, in particular SCN1A, induce the seizure phenotype when mutated or deleted [Davidsson et al., 2008; Escayg and Goldin, 2010], although there is evidence for the contribution of SLC4A10 at 2q24.2 to a milder seizure phenotype when deleted [Krepischi et al., 2010]. In our cohort, patient 4 had deletion of the entire SCN gene cluster and intractable seizures with onset at 11 weeks, while the rest of the patients have deletions that include SLC4A10, but spare SCN1A, and do not have any confirmed seizure activity. Due to deletion of SLC4A10 these patients may still be at risk for later-onset seizures, as previously reported [Gurnett et al., 2008; Krepischi et al., 2010], although the oldest patient remains seizure-free at 11.5 years.
Common features in several of our, and previously reported, patients may aid in other genotype-phenotype correlations for 2q24q31 deletions. For example, an autism spectrum disorder was present in patient 2 and a previously reported patient [Krepischi et al., 2010], and other behavioral problems were seen in patients 1 and 3. Only 3 genes are commonly deleted in these individuals (PSMD14, TBR1, and SLC4A10), which are all involved in proper brain development and function [Hevner et al., 2001; Staropoli and Abeliovich, 2005; Jacobs et al., 2008]. Furthermore, TBR1 interacts with autism susceptibility gene AUTS2 mRNA [Bedogni et al., 2010b], and there has been a report of a female with autism and a de novo deletion of SLC4A10 (fig. 1) [Sebat et al., 2007]. We cannot eliminate the possibility of multiple genes in this region contributing to a behavioral phenotype or a positional effect, as 2 other individuals with autism spectrum disorders have been reported with nonoverlapping deletions, one 940 kb distal to SLC4A10 and the other 2.2 Mb proximal to PSMD14 (fig. 1) [Newbury et al., 2009; Chen et al., 2010]. Colobomas are present in patient 4 and in 4 previously reported individuals with molecularly defined deletions [Nixon et al., 1997; Langer et al., 2006; Krepischi et al., 2010; Mitter et al., 2010], 2 of whom have deletions distal to TBR1 (fig. 1). A possible shared deletion region among these individuals is in the area of the SCN gene cluster, and a candidate gene in this region is TTC21B; in mice, Ttc21b encodes an axonemal protein required for intraflagellar transport that helps to control Shh signaling [Stottmann et al., 2009]. Heterozygous and homozygous mutations in TTC21B are found in some individuals with various ciliopathies, including Joubert syndrome [Davis et al., 2011], of which chorioretinal coloboma can be a feature [Parisi, 2009]. However, as the patient reported by Nixon et al. [1997] did not have any seizures, it is unlikely that the patient's deletion included SCN1A and the more proximally located TTC21B. Instead, deletion of multiple genes in the region may contribute to the formation of colobomas, which is supported by the report of an additional individual with colobomas and a 2q31.1 deletion [Mitter et al., 2010]. Three additional candidate genes for coloboma are present in patient 3's deleted region, including TBR1, which is a candidate due to its role in the PAX6 transcription factor cascade and PAX6's well-established role in ocular development and mutation in coloboma [Azuma et al., 2003; Nallathambi et al., 2006]. Second, BBS5 is also involved in ciliary function and implicated in Bardet-Biedl syndrome, although coloboma is rare in Bardet-Biedl syndrome [Li et al., 2004; Hjortshoj et al., 2008]. Third, LRP2 encodes an endocytic receptor, and autosomal recessive mutations in LRP2 cause Donnai-Barrow or facio-oculo-acoustico-renal syndrome, of which iris colobomas are a feature [Chassaing et al., 2003; Kantarci et al., 2007]. Craniosynostosis is present in patient 4 and has occasionally been reported in individuals with molecularly characterized 2q deletions [Nixon et al., 1997; Davidsson et al., 2008; Mitter et al., 2010], who share a deletion region distal to TBR1 (fig. 1). This region has multiple candidate genes including PHOSPHO2, which may be involved in the generation of phosphate for bone mineralization [Roberts et al., 2005]. SP5 encodes a transcription factor that mediates WNT signaling [Weidinger et al., 2005; Fujimura et al., 2007], and inhibition of WNT signaling can lead to craniosynostosis [Behr et al., 2010]. GORASP2, also in the region, encodes a Golgi-associated protein that is required for activation of MMP14 and MMP16 [Roghi et al., 2010], metalloproteases required for normal bone formation [Holmbeck et al., 2003, 2005; Shi et al., 2008; Zhou et al., 2009], and copy number gain of another metalloprotease gene, MMP23, has been implicated in craniosynostosis [Gajecka et al., 2005].
Our genotype-first approach to establish phenotypic consequences of TBR1 deletion has shown that TBR1 hemizygosity alone does not cause significant brain anomalies. This is consistent with animal models, which are essentially phenotypically normal when missing 1 copy of Tbr1 [Bulfone et al., 1998] but have neocortical and deep cerebellar malformations with homozygous loss [Hevner et al., 2001 2002; Fink et al., 2006]. It is possible that TBR1 hemizygosity is contributing to the neurodevelopmental phenotypes in these patients, given its role in neurodevelopment and high likelihood of being subject to haploinsufficiency [Huang et al., 2010]. However, our patients show additional clinical features that are likely due to deleted genes other than TBR1, and a comparison of our patients to earlier reports has allowed for the identification of some possible critical regions for features including craniosynostosis and colobomas. While this study demonstrates the challenges inherent in the genotype-first approach, when the deletion of the candidate gene may only have mild effects or contribute to a nonspecific phenotype, it also implies potential in mining genomic databases of copy number variations for candidate genes. Through a genotype-first approach, cohorts of patients with loss of developmentally important genes can be evaluated simultaneously to further delineate clinical significance.
Acknowledgements
We thank the patients and their families for their participation in this study. We also thank Beth Torchia (Signature Genomics) for technical assistance and Erin Dodge and A. Michelle Caldwell (Signature Genomics) for editorial assistance.
References
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, et al. Deletion of CASK in mice is lethal and impairs synaptic function. Proc Natl Acad Sci USA. 2007;104:2525–2530. doi: 10.1073/pnas.0611003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma N, Yamaguchi Y, Handa H, Tadokoro K, Asaka A, et al. Mutations of the PAX6 gene detected in patients with a variety of optic-nerve malformations. Am J Hum Genet. 2003;72:1565–1570. doi: 10.1086/375555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baala L, Briault S, Etchevers HC, Laumonnier F, Natiq A, et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet. 2007;39:454–456. doi: 10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, McDonald-McGinn DM, Zackai EH, Hersh JH, et al. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet. 2008;74:469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA. 2010a;107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Nelson BR, Frederick EA, Shiba N, et al. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns. 2010b;10:9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr B, Longaker MT, Quarto N. Differential activation of canonical Wnt signaling determines cranial sutures fate: a novel mechanism for sagittal suture craniosynostosis. Dev Biol. 2010;344:922–940. doi: 10.1016/j.ydbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JL. T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron. 1995;15:63–78. doi: 10.1016/0896-6273(95)90065-9. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Wang F, Hevner R, Anderson S, Cutforth T, et al. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21:1273–1282. doi: 10.1016/s0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- Chassaing N, Lacombe D, Carles D, Calvas P, Saura R, Bieth E. Donnai-Barrow syndrome: four additional patients. Am J Med Genet A. 2003;121A:258–262. doi: 10.1002/ajmg.a.20266. [DOI] [PubMed] [Google Scholar]
- Chen CP, Lin SP, Chern SR, Chen YJ, Tsai FJ, et al. Array-CGH detection of a de novo 2.8 Mb deletion in 2q24.2–>q24.3 in a girl with autistic features and developmental delay. Eur J Med Genet. 2010;53:217–220. doi: 10.1016/j.ejmg.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Davidsson J, Collin A, Olsson ME, Lundgren J, Soller M. Deletion of the SCN gene cluster on 2q24.4 is associated with severe epilepsy: an array-based genotype-phenotype correlation and a comprehensive review of previously published cases. Epilepsy Res. 2008;81:69–79. doi: 10.1016/j.eplepsyres.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escayg A, Goldin AL. Sodium channel SCN1A and epilepsy: mutations and mechanisms. Epilepsia. 2010;51:1650–1658. doi: 10.1111/j.1528-1167.2010.02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, et al. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura N, Vacik T, Machon O, Vlcek C, Scalabrin S, et al. Wnt-mediated down-regulation of Sp1 target genes by a transcriptional repressor Sp5. J Biol Chem. 2007;282:1225–1237. doi: 10.1074/jbc.M605851200. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Yu W, Ballif BC, Glotzbach CD, Bailey KA, et al. Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur J Hum Genet. 2005;13:139–149. doi: 10.1038/sj.ejhg.5201302. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Grosso S, Pucci L, Curatolo P, Coppola G, Bartalini G, et al. Epilepsy and electroencephalographic anomalies in chromosome 2 aberrations. A review. Epilepsy Res. 2008;79:63–70. doi: 10.1016/j.eplepsyres.2007.12.011. [DOI] [PubMed] [Google Scholar]
- Gurnett CA, Veile R, Zempel J, Blackburn L, Lovett M, Bowcock A. Disruption of sodium bicarbonate transporter SLC4A10 in a patient with complex partial epilepsy and mental retardation. Arch Neurol. 2008;65:550–553. doi: 10.1001/archneur.65.4.550. [DOI] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci USA. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Miyashita-Lin E, Rubenstein JL. Cortical and thalamic axon pathfinding defects in Tbr1, Gbx2, and Pax6 mutant mice: evidence that cortical and thalamic axons interact and guide each other. J Comp Neurol. 2002;447:8–17. doi: 10.1002/cne.10219. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hjortshoj TD, Gronskov K, Philp AR, Nishimura DY, Adeyemo A, et al. Novel mutations in BBS5 highlight the importance of this gene in non-Caucasian Bardet-Biedl syndrome patients. Am J Med Genet A. 2008;146A:517–520. doi: 10.1002/ajmg.a.32136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Chrysovergis K, Yamada S, Birkedal-Hansen H. MT1-MMP-dependent, apoptotic remodeling of unmineralized cartilage: a critical process in skeletal growth. J Cell Biol. 2003;163:661–671. doi: 10.1083/jcb.200307061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Pidoux I, Inoue S, Billinghurst RC, et al. The metalloproteinase MT1-MMP is required for normal development and maintenance of osteocyte processes in bone. J Cell Sci. 2005;118:147–156. doi: 10.1242/jcs.01581. [DOI] [PubMed] [Google Scholar]
- Hong SE, Shugart YY, Huang DT, Shahwan SA, Grant PE, et al. Autosomal recessive lissencephaly with cerebellar hypoplasia is associated with human RELN mutations. Nat Genet. 2000;26:93–96. doi: 10.1038/79246. [DOI] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S, Ruusuvuori E, Sipila ST, Haapanen A, Damkier HH, et al. Mice with targeted Slc4a10 gene disruption have small brain ventricles and show reduced neuronal excitability. Proc Natl Acad Sci USA. 2008;105:311–316. doi: 10.1073/pnas.0705487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarci S, Al-Gazali L, Hill RS, Donnai D, Black GC, et al. Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat Genet. 2007;39:957–959. doi: 10.1038/ng2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepischi AC, Knijnenburg J, Bertola DR, Kim CA, Pearson PL, et al. Two distinct regions in 2q24.2–q24.3 associated with idiopathic epilepsy. Epilepsia. 2010;51:2457–2460. doi: 10.1111/j.1528-1167.2010.02742.x. [DOI] [PubMed] [Google Scholar]
- Langer S, Geigl JB, Wagenstaller J, Lederer G, Hempel M, et al. Delineation of a 2q deletion in a girl with dysmorphic features and epilepsy. Am J Med Genet A. 2006;140:764–768. doi: 10.1002/ajmg.a.31141. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Magri C, Piovani G, Pilotta A, Michele T, Buzi F, Barlati S. De novo deletion of chromosome 2q24.2 region in a mentally retarded boy with muscular hypotonia. Eur J Med Genet. 2011;54:361–364. doi: 10.1016/j.ejmg.2010.12.011. [DOI] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Gomez HR, Vergano-Vera E, Abad JL, Bulfone A, Moratalla R, et al. The T-box brain 1 (Tbr1) transcription factor inhibits astrocyte formation in the olfactory bulb and regulates neural stem cell fate. Mol Cell Neurosci. 2011;46:108–121. doi: 10.1016/j.mcn.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Mitchell TN, Free SL, Williamson KA, Stevens JM, Churchill AJ, et al. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann Neurol. 2003;53:658–663. doi: 10.1002/ana.10576. [DOI] [PubMed] [Google Scholar]
- Mitter D, Chiaie BD, Lüdecke HJ, Gillessen-Kaesbach G, Bohring A, et al. Genotype-phenotype correlation in eight new patients with a deletion encompassing 2q31.1. Am J Med Genet A. 2010;152A:1213–1224. doi: 10.1002/ajmg.a.33344. [DOI] [PubMed] [Google Scholar]
- Najm J, Horn D, Wimplinger I, Golden JA, Chizhikov VV, et al. Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat Genet. 2008;40:1065–1067. doi: 10.1038/ng.194. [DOI] [PubMed] [Google Scholar]
- Nallathambi J, Neethirajan G, Shashikant S, Vijayalakshmi P, Sundaresan P. PAX6 missense mutations associated in patients with optic nerve malformation. Mol Vis. 2006;12:236–242. [PubMed] [Google Scholar]
- Newbury DF, Warburton PC, Wilson N, Bacchelli E, Carone S, et al. Mapping of partially overlapping de novo deletions across an autism susceptibility region (AUTS5) in two unrelated individuals affected by developmental delays with communication impairment. Am J Med Genet A. 2009;149A:588–597. doi: 10.1002/ajmg.a.32704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon J, Oldridge M, Wilkie AO, Smith K. Interstitial deletion of 2q associated with craniosynostosis, ocular coloboma, and limb abnormalities: cytogenetic and molecular investigation. Am J Med Genet. 1997;70:324–327. [PubMed] [Google Scholar]
- Palumbo O, Palumbo P, Palladino T, Stallone R, Zelante L, Carella M. A novel deletion in 2q24.1q24.2 in a girl with mental retardation and generalized hypotonia: a case report. Mol Cytogenet. 2012;5:1. doi: 10.1186/1755-8166-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA. Clinical and molecular features of Joubert syndrome and related disorders. Am J Med Genet C Semin Med Genet. 2009;151C:326–340. doi: 10.1002/ajmg.c.30229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, Vieira JP, Barroca F, Roll P, Carvalhas R, et al. Severe epilepsy, retardation, and dysmorphic features with a 2q deletion including SCN1A and SCN2A. Neurology. 2004;63:191–192. doi: 10.1212/01.wnl.0000132844.20654.c1. [DOI] [PubMed] [Google Scholar]
- Pescucci C, Caselli R, Grosso S, Mencarelli MA, Mari F, et al. 2q24–q31 deletion: report of a case and review of the literature. Eur J Med Genet. 2007;50:21–32. doi: 10.1016/j.ejmg.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Raybaud C, Di Rocco C. Brain malformation in syndromic craniosynostoses, a primary disorder of white matter: a review. Childs Nerv Syst. 2007;23:1379–1388. doi: 10.1007/s00381-007-0474-7. [DOI] [PubMed] [Google Scholar]
- Roberts SJ, Stewart AJ, Schmid R, Blindauer CA, Bond SR, et al. Probing the substrate specificities of human PHOSPHO1 and PHOSPHO2. Biochim Biophys Acta. 2005;1752:73–82. doi: 10.1016/j.bbapap.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Roghi C, Jones L, Gratian M, English WR, Murphy G. Golgi reassembly stacking protein 55 interacts with membrane-type (MT) 1-matrix metalloprotease (MMP) and furin and plays a role in the activation of the MT1-MMP zymogen. FEBS J. 2010;277:3158–3175. doi: 10.1111/j.1742-4658.2010.07723.x. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Son MY, Yamada S, Szabova L, Kahan S, et al. Membrane-type MMPs enable extracellular matrix permissiveness and mesenchymal cell proliferation during embryogenesis. Dev Biol. 2008;313:196–209. doi: 10.1016/j.ydbio.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, et al. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet. 2001;28:214–216. doi: 10.1038/90042. [DOI] [PubMed] [Google Scholar]
- Solomon BD, Pineda-Alvarez DE, Balog JZ, Hadley D, Gropman AL, et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A. 2009;149A:2543–2546. doi: 10.1002/ajmg.a.33081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staropoli JF, Abeliovich A. The ubiquitin-proteasome pathway is necessary for maintenance of the postmitotic status of neurons. J Mol Neurosci. 2005;27:175–183. doi: 10.1385/JMN:27:2:175. [DOI] [PubMed] [Google Scholar]
- Stottmann RW, Tran PV, Turbe-Doan A, Beier DR. Ttc21b is required to restrict sonic hedgehog activity in the developing mouse forebrain. Dev Biol. 2009;335:166–178. doi: 10.1016/j.ydbio.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki S, Nakamura R, Haga Y, Mitsui K, Hashimoto T, et al. Severe pulmonary emphysema in a girl with interstitial deletion of 2q24.2q24.3 including ITGB6. Am J Med Genet A. 2010;152A:1020–1025. doi: 10.1002/ajmg.a.33362. [DOI] [PubMed] [Google Scholar]
- Traylor RN, Fan Z, Hudson B, Rosenfeld JA, Shaffer LG, et al. Microdeletion of 6q16.1 encompassing EPHA7 in a child with mild neurological abnormalities and dysmorphic features: case report. Mol Cytogenet. 2009;2:17. doi: 10.1186/1755-8166-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuoc TC, Radyushkin K, Tonchev AB, Pinon MC, Ashery-Padan R, et al. Selective cortical layering abnormalities and behavioral deficits in cortex-specific Pax6 knock-out mice. J Neurosci. 2009;29:8335–8349. doi: 10.1523/JNEUROSCI.5669-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/beta-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. 2005;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Zhou H, Mak W, Kalak R, Street J, Fong-Yee C, et al. Glucocorticoid-dependent Wnt signaling by mature osteoblasts is a key regulator of cranial skeletal development in mice. Development. 2009;136:427–436. doi: 10.1242/dev.027706. [DOI] [PubMed] [Google Scholar]