Abstract
The use of model organisms to provide information on gene function has proved to be a powerful approach to our understanding of both human disease and fundamental mammalian biology. Large-scale community projects using mice, based on forward and reverse genetics, and now the pan-genomic phenotyping efforts of the International Mouse Phenotyping Consortium (IMPC), are generating resources on an unprecedented scale which will be extremely valuable to human genetics and medicine. We discuss the nature and availability of data, mice and ES cells from these large-scale programmes, the use of these resources to help prioritise and validate candidate genes in human genetic association studies, and how they can improve our understanding of the underlying pathobiology of human disease.
Keywords: mouse, genetics, phenotyping, human, ontology, GWAS, CNV, database
Introduction
The use of non-human species to understand normal and patho-biology, and to create models of human diseases tractable to experimental investigation, has been a dominant and successful paradigm in the biomedical sciences for many years [Rosenthal and Brown, 2007]. The combination of this approach with genetics and the ability to manipulate the genome of vertebrates, particularly the mouse, has signaled a shift in that paradigm, making it hugely more powerful and scaleable, and now, in what might be termed the post-genomic phase of biomedical research, we are reaping the fruits of the hypothesis-driven manipulation of genes to provide insights into their function.
Completion of the first draft of the human genome sequence in 2001 [Lander, 2011] was closely followed by that of the C57BL/6J mouse strain in 2002 [Waterston et al., 2002]. In 2011, the sequence of 17 strains of mice using next generation sequencing was reported, capturing the genomic variation from most of the commonly-used strains of laboratory mice. More than 56 M single nucleotide polymorphisms (SNPs) are now catalogued, together with 8.8M unique indels and 0.28M structural variants including 0.07M transposable element insertion sites [Keane et al., 2011]. Provision of common frameworks by the reference sequences of human and mouse genomes coupled with the increasing amounts of information on genetic variation in both species, (for example, provide through the 1000 genomes project (1000_genomes_consortium, 2010]) gives us powerful tools for the discovery of relationships between genes and phenotypes, one of the central goals of the biomedical sciences. The ability to genetically cross-reference across species allows the bridging of the biology of model organisms to that of the human, with the attendant richness of phenotypic information derived from experimental investigation not possible with human subjects, and the potential to test hypotheses by making the targeted mutations in critical regions of the genome.
Vital statistics: mouse genome and phenome
The mouse genome closely mirrors that of the human. It contains 2.6 billion nucleotides assembled in 21 chromosomes (1–19 plus the X and Y sex chromosomes). Analysis of the current reference genome release (Build 37) shows 28,947 genes with nucleotide sequence data and 25,106 genes with protein sequence data. Compared with 30,446 genes in the human. Human and mouse genes contain on average 8.2 and 8.4 exons respectively and there are 17,829 mouse genes with identifiable orthologs in humans (data from Mouse Genome Database (MGD) accessed December 2011 [Blake et al., 2009; Blake et al., 2011].
Of the 28,947 mouse genes, 13,822 have experimentally-based functional annotations, and to date there are 29,318 mutant alleles in 15,331 genes available in mice. The disparity in these latter numbers is due to the availability of multiple alleles for many genes. Extending that to ES cells there are 43,237 targeted alleles in total. There are 3,587 mouse genotypes modeling human diseases as confirmed by direct experimentation, and 1,134 human diseases with one or more mouse models. Each mouse model may contain one or more mutated genes on various background strains, and often a human disease has been modeled several times using different combinations of alleles and backgrounds.
Phenotypic information curated from the literature is available through the MGD for ~8,200 of the 25,000 mouse genes with associated protein sequence data, and is coded using the Mammalian Phenotype ontology (MP). The mouse is only one of several established model organisms and the potential leverage provided by model organism phenotypes, such as those of zebrafish and flies, towards our understanding of human gene function is extraordinary [Bult, 2006]. Currently there are 5,392 model organism genes with phenotype data for which the human ortholog has no phenotype information, and this number will continue to grow. Model organism databases have therefore the potential to serve not just as data repositories, but also as research tools to identify and study human disease.
The corpus of phenotypic data available for the mouse, and that part which is even now known to reflect human phenotype/genotype correlations, together with the complete genome sequences and variation available in laboratory strains, makes the mouse an extremely useful resource for human genetics. High-density polymorphism mapping in humans, together with genome-wide association studies (GWAS) and next generation sequencing (NGS) of normal and diseased populations has greatly accelerated the identification of candidate genes for monogenic and complex diseases. However, despite the accumulation of huge volumes of data on genetic variation and its phenotypic correlates, the normal function of most genes is still unknown, candidate genes for rare diseases often remain unvalidated, and the largest part of the heritability of many complex diseases is yet undefined (see below). The availability of forward and reverse genetic resources for the mouse, together with rapidly increasing volumes of phenotypic data, offer insights into all these problems, and recent developments in large-scale community projects promise to revolutionise our understanding of gene function by taking a pan-genomic and pan-phenomic approach to data gathering and resource generation on an unprecedented scale.
Mouse genetic resources
Reverse genetics – the International Knockout Mouse Consortium
The recent commitment of major funding agencies internationally to the generation of pan-genomic public resources for the mouse has seen a watershed in the utility of the mouse in the biomedical sciences. In 2004, the NIH and European Commission jointly spearheaded an ambitious project, the International Knockout Mouse Consortium (IKMC), to create a knockout strain for every gene in the mouse genome [Auwerx et al., 2004, Austin et al., 2004; Collins et al., 2007]. This was joined by other international funding initiatives from the Wellcome Trust, Genome Canada and the Texas Institute of Genomic Medicine (TIGM). Initially efforts were spread between several strategies for gene knockout – gene targeting and gene traps - which focused down during the early years of the project as it became apparent that the efficiency of gene targeting had become high enough to meet the target number of knockouts in the timeframe of funding. Improvements in automated or semi-automated targeting construct design, recombineering and high efficiencies of targeting and transmission facilitated the production of mutant ES at an unprecedented rate, and the target is now close to being met [Skarnes et al., 2011].
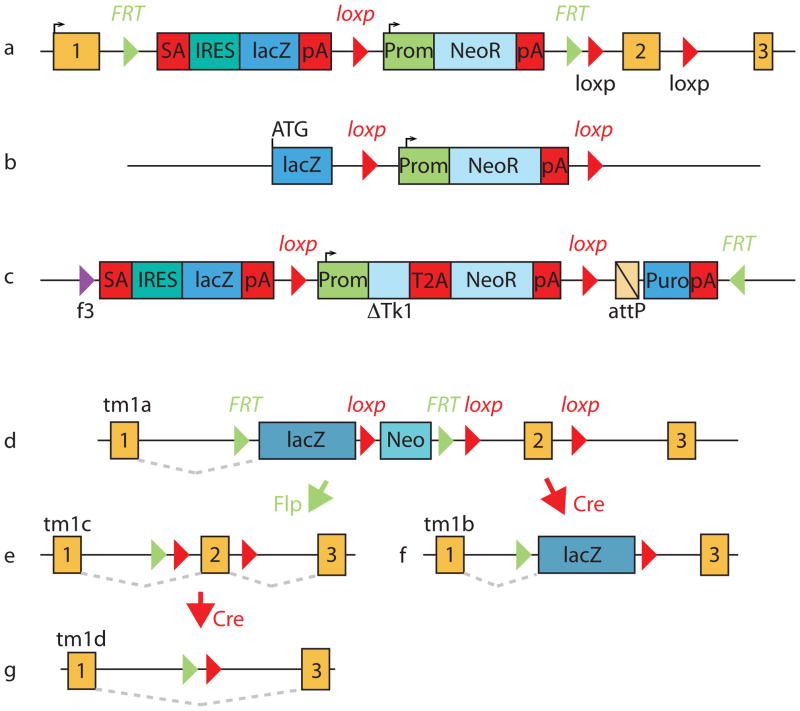
The IKMC alleles are either null/conditional (“knockout first”) or null alleles removing the entire locus (Figure 1). A description of the strategies used by the Consortium can be found on; http://www.knockoutmouse.org/about/targeting-strategies. The “knockout-first” alleles, from which conditional mutations can be obtained following exposure to a site-specific recombinase, are very useful for the study of lethal mutations, where constitutive mutation is incompatible with survival, and for the study of gene function in tissue specific and temporally restricted contexts. The knockout-first/conditional constructs can also act as a landing site for recombinase-mediated cassette exchange, which can be used to insert other coding, sequences into these alleles [Osterwalder et al., 2010; Schebelle et al., 2010; Schnutgen et al., 2011]. In some cases problems with construct design required the different approach offered by Regeneron’s Velocigene (LacZ knock-in) platform. These lacZ-tagged nulls are constructed as large deletions removing all or most of the locus [Valenzuela et al., 2003]; this has been necessary for 3,500 of the 8,500 targets selected for NIH knockout mouse project (KOMP) as part of the IMPC effort. All of the constructs, ES cells and, where made, mice are available at a nominal charge from the consortium, providing a valuable bioresource for the community (see Table 1).
Figure 1.
Knockout strategies used by the IKMC. A. EUCOMM/KOMP-CSD knockout-first allele; B. KOMP-Regeneron null allele; C. NorCOMM promoter-driven targeting vector. Allele tm1a (D) contains an IRES:lacZ cassette and a floxed promoter-driven neo cassette inserted into the intron of the target gene knocking out gene function. Flp converts the tm1a allele to the conditional allele (tm1c), restoring gene function. Cre deletes the promoter-driven selection cassette and floxed exon of the tm1a allele to generate a lacZ-tagged allele (tm1b) or deletes the floxed exon of the tm1c allele to produce a frameshift mutation (tm1d).
Table 1.
Generation of mutant ES cells and mice by each IKMC centre and their repositories
| Centre | Project Goal | ES cells generated | Mutant mice generated | Repository | |
|---|---|---|---|---|---|
| KOMP-CSD | KOMP | 5000 | 5283 | 356 | www.komp.org |
| KOMP-Regeneron | 3500 | 4006 | 303 | www.komp.org | |
|
| |||||
| EUCOMM | 8000 | 6695 | 593 |
www.eummcr.org (ES cells) EMMA (mice) |
|
|
| |||||
| NorCOMM | 500 | 569 | 4 | www.cmmr.ca | |
|
| |||||
| TIGM | - | 9405 | 81 | www.tigm.org | |
|
| |||||
| Total | 17,000 | 25, 958 | 1,337 | ||
IKMC ES cell and mouse resources
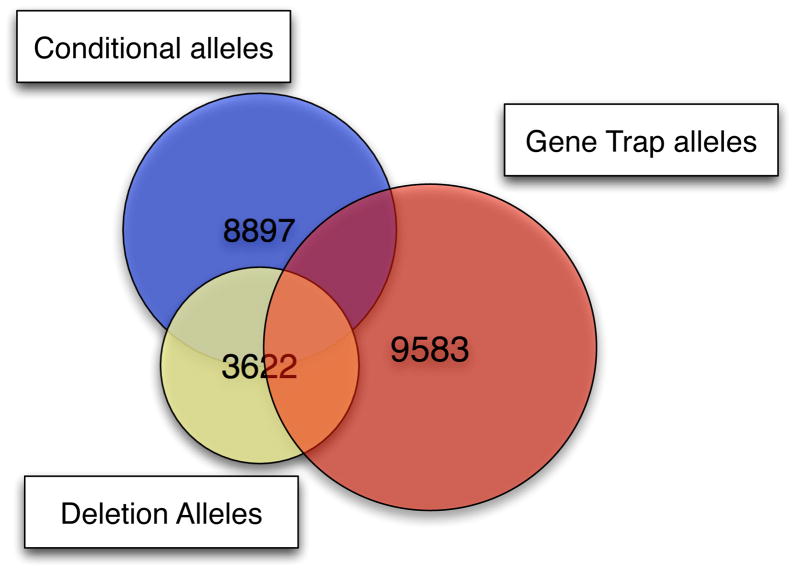
Currently the IKMC ES cell resource contains targeted and trapped alleles for about 17,912 unique genes (Figure 2). The ES cell gene targeted resource contains mutations in approximately 12,189 unique genes of which around 10,500 are conditional. The ES cell gene trap resource contains mutations in about 9,638 unique protein-coding genes of which 4,396 are conditional, Due to the random nature of gene trapping, there is some overlap between the resources and redundancy within each resource with a median of 4 independent mutations per gene which provides useful allelic series and a range of options for further experimentation. To date, more than 1,454 mutated alleles have been established in mice, which are held either live or as germ plasm in the repositories; see Table 1.
Figure 2.
Number of genes for which IKMC ES cells with a given allele type are available. (Redrawn from http://www.knockoutmouse.org/ December 20. 2011)
The generation of mice on this large scale has taken place in centres carrying out systematic phenotyping programmes, including those in EUMODIC (The European Mouse Disease Clinic; http://www.eumodic.org), the Wellcome Trust Sanger Institute’s Mouse Genetics Programme (MGP), and at the University of California at Davis, following funding from NIH to create and characterize 312 unique mutant lines from targeted ES cells developed by the CHORI-Sanger-UC Davis and Regeneron consortium. These coordinated efforts have enabled these genetic resources to be efficiently archived, and the mice are distributed through the European Mutant Mouse Archive (EMMA) and the Mutant Mouse Regional Resource Centers (MMRRC), the European and USA mouse repositories respectively. Large numbers of IKMC strains have already been distributed and, for example, 70% of mice distributed by the European mutant mouse archive [Wilkinson et al., 2011] in 2011 were derived from IKMC resources.
Accessing mice, ES cells and data
Information on the data and resources of the IKMC is available through the official IKMC web portal (http://www.knockoutmouse.org) developed and maintained by the KOMP-DCC (KOMP-Data Coordination Center) and the I-DCC (International-Data Coordination Center) funded by the NIH and the European Commission respectively [Ringwald et al., 2011]. The IKMC web portal and database facilitate nomination of genes by the community for prioritisation and coordination of work and progress tracking of knockout projects within the IKMC. Data is made readily accessible to the research community together with pointers to the repositories which distribute IKMC products. Table 2 provides a list of selected mouse genome and phenotype resources.
Table 2.
Selected Mouse genome and phenome resources
| Resource | URL | Resource content |
|---|---|---|
| Mouse genome informatics (MGI) | http://www.informatics.jax.org/ | This is the international database resource for the laboratory mouse, providing integrated genetic, genomic, and biological data to facilitate the study of human health and disease. Contains information from several projects: Mouse Genome Database (MGD) Project, Gene Expression Database (GXD) Project, Mouse Tumor Biology (MTB) Database Project, Gene Ontology (GO) Project at MGI MouseCyc Project at MGI |
| Wellcome Trust Sanger Institute (WTSI) Mouse Portal | http://www.sanger.ac.uk/mouseportal/ | One-stop access to the different resources available from the WTSI. The resources include: 129S7 and C57BL6/J bacterial artificial chromosomes (BACs), MICER gene targeting vectors, knock-out first conditional- ready gene targeting vectors, embryonic stem (ES) cells with gene targeted mutations or with retroviral gene trap insertions, mutant mouse lines, and phenotypic data generated from the Institute’s primary screen. |
| Europhenome | http://www.europhenome.org/ | Provides access to raw and annotated mouse phenotyping data generated from primary pipelines such as EMPReSSlim and secondary procedures from specialist centres. |
| Mouse Phenome Database (MPD) | http://phenome.jax.org/ | Contains comprehensive phenotypic information on hundreds inbred mouse strains and analytical tools |
| European mouse disease clinic (EUMODIC) | http://www.eumodic.org/ | Project site for EUMODIC which contains projects details and access to information on the EMPReSS slim phenotyping pipeline and Europhenome. |
| CREATE (Coordination of resources for conditional expression of mutated mouse alleles project) | http://www.creline.org | Unified portal for worldwide access to Cre mice with expression patterns and gene details |
| MGI Cre Portal | http://www.creportal.org/ | Collection and annotation of expression and activity data for recombinase-containing transgenes and knock-in alleles by MGI at the Jackson Laboratory. |
| International Knockout Mouse Consortium (IKMC) | http://www.knockoutmouse.org | Contains all the details for the IKMC (Eucomm, Norcomm, KOMP and TIGM) ES cells, knockout alleles, vectors and mice. Pointers to ordering resources. |
| International Knockout Mouse consortium (IMPC) | http://www.knockoutmouse.org | The web site of the IMPC. Information on the project, news and in the future links to the phenotype data as it is acquired. |
| International Mouse Strain Resource (IMSR) | http://www.findmice.org/ | A searchable online database of mouse strains and stocks available worldwide, including inbred, mutant, and genetically engineered mice. |
| NCBI guide to mouse genome resources | http://www.ncbi.nlm.nih.gov/genome/guide/mouse/ |
Further mouse on-line resources can be found in [Peters et al., 2007], [Zouberakis et al., 2010] and on the mouse resource browser, (http://bioit.fleming.gr/mrb/).
Going conditional - developing the Cre zoo
The ability to carry out conditional knockouts from ES cells carrying the tm1a “knockout-first” allele (Figure 3) provides valuable opportunities to analyze knockouts which are constitutively developmental lethal or where tissue or time-specific analysis is required. The scope of such studies depends on the Cre driver strains available for breeding with mice carrying the allele. Major efforts are currently underway to catalog, locate and make available all Cre driver strains currently extant, and to identify and create strains that have desirable patterns of temporal or tissue-specific expression.
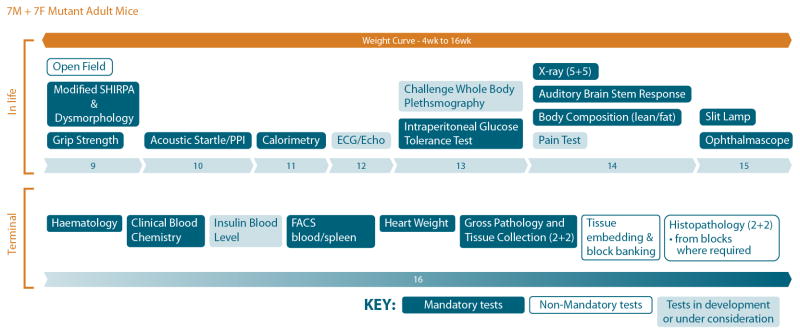
Figure 3.
Draft in vivo and post mortem phenotyping pipelines for the IMPC. Mandatory tests will be carried out by all of the mouse clinics, with some non-mandatory tests such as histopathology being carried out in only a few centres. The details, order and density of assays may change as discussions progress. Details can be found on http://www.mousephenotype.org/workgroups/impc-phenotyping-work-group.
Three Cre databases are currently available and likely to converge on a single or linked resource [Smedley et al., 2011]. The Cre-X-mice database (http://www.mshri.on.ca/nagy/) was originally developed in Andras Nagy’s laboratory and contains information on more than 500 Cre lines. The CREATE portal (www.creline.org) was developed as part of the CREATE project funded by the European Commission, and provides an aggregating data resource through a BioMart, integrating data from several distributed Cre databases including the MGI Cre portal (www.creportal.org), [Blake et al., 2010], which uses the resources of GXD and MGI (GXD; [Finger et al., 2010] to provide a comprehensive catalogue of all Cre alleles currently available, and where possible the precise patterns of reporter gene deletion or Cre expression.
Mice may be obtained from a variety of resources using the FIMRe (Federation of International Mouse REsources) portal (http://www.fimre.org/; [Davisson, 2006] but a specific Cre mouse repository has also been established at the Jackson Laboratory which currently distributes more than 250 Cre strains ( http://cre.jax.org/).
Forward Genetics - Genetic reference populations and the Collaborative Cross
Panels of mouse recombinant inbred lines (RIL) have been used for some time to investigate the genetics of complex phenotypic traits, but their power has been restricted by the lack of variation present in laboratory mice as a consequence of the historical development of the inbred strains. It was proposed in 2002 to address the need for a genetic reference panel with a large genetic diversity and high resolution for the analysis of complex traits, by creating a large multiparental RIL panel using 8 founder strains – five inbred laboratory lines and three wild-derived strains. The number and combination of founders allows the capture of a much greater level of genetic diversity than existing mouse RIL panels, and variation is much more evenly spread across the genome than in previous panels or reference populations. The resulting Collaborative cross (CC) project was set up in 2005 and is now being carried out in the US, Australia and Israel [Threadgill et al., 2011; Threadgill and Churchill, 2012; Collaborative Cross Consortium, 2012]. Diversity is generated and then captured in the CC population by a special funnel breeding program where in the first three generations all eight founder genomes are mixed together then, in a second step, progressive inbreeding by sibling mating is performed to generate hundreds of inbred lines capturing novel allelic combinations. Around 700 inbred lines are planned, and the CC is now at the stage where the first lines are completely inbred and therefore beginning to be of use for genetic mapping studies [Callaway, 2011].
The variation captured in these inbred lines allows the identification of individual genes and variants contributing to complex traits, QTLs and diseases. The genotyping of the individual lines has to be performed only once and then the haplotype structure allows determination of the association between the genotype and the phenotype of interest. The phenotypic traits to be studied can be as diverse as predisposition to cancers, infectious disease [Durrant et al., 2011] physiology [Mathes et al., 2011], behaviour [Philip et al., 2011] or anatomy (e.g. tail length], and reproduction. The phenotype in question can be first measured in a limited number of strains -250 lines permits accurate high resolution mapping using small groups of mice per line - then the mapping resolution can subsequently be increased by selecting lines with recombinations in the critical intervals for additional phenotype analysis.
The CC strains model the complexity of the human population [Threadgill et al 2011], and will provide insights into common human diseases with complex etiologies, especially those originating from interactions between combinations of alleles and the environment. This will impact on approaches to personalised medicine as well as our understanding of complex diseases, such as cancer, diabetes and cardiovascular disease, and will extend our knowledge of both normal physiology and gene function. For example pre-CC lines are already being used to look at gene/environment interactions in asthma and to dissect the complex genetic contributions to human psychiatric disease. The huge advantage of the CC approach is that it provides an unbiased forward genetics approach to the relations between genotype and phenotype, allowing the discovery of the contributions of many genes and alleles to complex phenotypes and underlying molecular networks. This is truly “systems genetics’ providing an integrated view of the genome and phenome, which has enormous potential. For example as the CC strains are characterized, transcriptomics, metabolomics and proteomics profiling data will become available which will build a reference resource linking tissue-specific systems data to genetic variation. Importantly it is a genetically stable resource, allowing replication of experiments on unique combinations of alleles in different laboratories and facilitating the discovery of correlations between phenotypes measured in separate studies.
A recently developed approach which makes use of the CC resource is the use of a “Diversity Outcross” population (DO) for high resolution mapping [Svenson et al. 2012]. The DO population is an eight-way heterogeneous stock population derived from incipient CC lines at early stages of inbreeding. This population is then maintained by a randomized outbreeding strategy, making each DO animal a unique individual carrying one of an enormous number of possible combinations of the segregating alleles derived from the original CC founder strains.
The combination of the IKMC with the IMPC phenotyping efforts (see below) and the CC panel is potentially extremely powerful, allowing fine mapping of phenotypes together with direct validation of candidate genes through the knockout phenotype. The allelic variants in the CC are likely to be much less deleterious than engineered null alleles with the result that more extreme phenotypes, for example those giving rise to embryonic lethality, do not reduce the phenotypic variation available for study. The additional advantages of the mouse as an experimental organism open up the possibilities of examining the impact of environment on multi-gene traits and of experimental challenge to expose underlying phenotypes.
Using mouse genetic and phenotype data for human genetics
Three methods are currently in widespread and increasing use to identify the relationship between genotype and phenotype. Dominant are classical linkage mapping which examines genetically well-characterized populations, such as individuals related through a known pedigree, to identify loci that contain mutations contributing to the phenotype, and genome-wide association studies (GWAS) in which thousands of single-nucleotide polymorphisms (SNPs) are tested for association with a disease, using less well characterized populations, to identify common loci or variants associated with a phenotype. To date many hundreds of GWAS and pedigree studies have been carried out, resulting in the identification of thousands of loci associated with phenotypes and diseases [Feero et al., 2010; Hindorff et al., 2009; Manolio, 2010].
Despite obvious successes, both pedigree and GWAS approaches suffer from intrinsic limitations [Cantor et al., 2010]. Population structure, selection of patient and control groups, and accurate phenotyping are problems for many GWAS, while linkage disequilibrium and limited genetic diversity are challenges for many QTL studies. Many QTL studies lack the statistical power to narrowly define implicated loci, often resulting in poor resolution and association with intervals which contain contain large numbers of candidate genes. Discriminating causative genes within such intervals is often deeply problematical, and the amount of heritable variation explained by these loci tends to be small. Similarly, regions of copy number variation (CNV) might include large numbers of genes within the minimum region of overlap between patients, and it is difficult to attribute changes in dosage of specific genes within the interval to aspects of the overall phenotype. Consequently association studies of individual rare CNVs generally have insufficient power to discriminate benign from pathological variants. High resolution microarrays and sequencing approaches have identified up to 600–900 CNVs in a single individual, apparently unassociated with pathology [Pinto et al., 2007], and the assumption that common and inherited CNVs are benign, and rare and de novo CNVs are causative is no longer sustainable as discriminating criterion for pathogenicity [Hehir-Kwa et al., 2011].
The third method relies on novel “next-generation sequencing” (NGS) technology such as whole-exome sequencing (WES) which is, for the first time, enabling comprehensive screening of genetic variants to identify disease candidates [Robinson et al., 2011]. As with the interpretation of classical GWAS studies, however, a major problem is that all individuals have been found to carry large numbers of missense, nonsense, and insertion/deletion variants, many of which are not known common variants. Initial reports suggesting that every genome carries around 165 homozygous protein-truncating or stop-loss variants in a wide range of genes [Pelak et al., 2010] have been recently revised by MacArthur and co-workers [MacArthur et al 2012]. In this study they systematically investigated the presence of loss of function (LoF) mutations in normal individuals using pilot data from the 1000 genomes project and a single european individual. They concluded that each normal individual probably contains between 103–121 LoF alleles with around 20% being homozygous. Overall they identified around 1000 “natural knockout” alleles in the human genome, many of which are in protein coding genes whose normal function is completely uncharacterized, but whose bearer is apparently disease-free. The insights into gene function provided by the IMPC results will be invaluable in identifying the normal function and the role of such genes in disease, and will assist in the discovery of potential new phenotypes in the human population.
Finally, an additional problem with establishing pathogenicity is the existence of pathogenic synonymous “silent mutations’; those which are seemingly innocuous to gene function but carry serious functional consequences [Katsnelson, 2011]. Evidence of such pathogenic “silent” mutations is now accumulating, and more than 50 disorders including depression, schizophrenia, Crohn’s disease and cystic fibrosis and have now been linked to synonymous mutations.
Mouse genetic data help interpret human GWAS
Mouse QTL studies on well-characterized phenotypes are extremely helpful in prioritization of candidate genes identified in humans. For example the use of nested congenic strains has been exploited to decompose the genetic complexity of orthologous gene-rich regions associated in mouse and human with regulation of bone mineral density [Beamer et al., 2011]. Further examples abound; in a recent elegant study the mouse QTL map has been used to interpret results from a GWA study for genes associated with plasma HDL cholesterol levels [Leduc et al., 2011], and in a cross-species genetic mapping strategy, comparing data from gene mapping in human patients with functional data obtained by QTL mapping in highly diverse mouse genetic reference panels, a single locus associated with the development of the corpus callosum was identified [Poot et al., 2011].
Combination of the mouse QTL map and the IKMC knockout resource has proved to be very powerful in accelerating the discovery of gene/phenotype associations for complex diseases. In atherosclerosis 21 QTLs have been identified in the mouse. Among the 27 human atherosclerosis QTLs reported, 17 (63%) are located in regions homologous (concordant) to mouse QTLs, suggesting that these mouse and human atherosclerosis QTLs share the same underlying genes. A combination of QTL concordance and then phenotypic examination of candidate gene mutants provides a powerful route into causation and the underlying biology [Wang et al., 2005]. The IKMC resource now provides knockout and conditional mutations, which can be used to take such studies rapidly to the identification of causative genes. An excellent summary of the availability of complex trait resources for the mouse can be found in [Peters et al., 2007].
Functional genomic approaches complement those of human genetics
The approaches of functional genomics are now being applied to complement some of the weaknesses in human genetic studies with considerable success. Model organism derived knowledge of the underlying pathobiology of disease is often key information in deciding on the importance of candidate genes for orphan diseases and discriminating between different candidates highlighted by GWAS. There are many examples of this in the literature and it remains one of the most important uses of model organism data in the generation and validation of hypotheses [Cox and Church, 2011]. More sophisticated computation on phenotypic data has been made possible as the model organism, and now human, databases have begun to code their phenotype data using formal ontologies and ontology-based semantics. The adoption of formal phenotype semantics, specifically here focussing on the mouse and human, the mammalian phenotype [MP; Smith and Eppig, 2009] and human phenotype [HPO; Robinson et al., 2008] ontologies, by key databases, MGD [Blake et al., 2011], OMIM [Amberger et al., 2011] and Orphanet [Weinreich et al., 2008] has for the first time allowed a genome-scale phenotypic approach to integrating phenotypic and genotypic data across two species, making use of the rich phenotypic data now available. The problems of coding, integrating and computing on phenotype annotations from multiple species are discussed in [Schofield et al. 2011a].
OMIM and Orphanet are currently the easiest knowledge bases from which to extract high-resolution human genotype/phenotype data. Although there are many other databases and resources with valuable information, this is often not structured or coded in standard ways, and is scattered. The problem of integrating and standardizing data resources in human genetics is currently a topic of lively and important discussion [e.g. Oti et al., 2009]. The data used for the mouse are the gold standard, manually-curated phenotype data set from the mouse genome informatics database (MGD). Currently, for reasons discussed by Kitsios and co-workers [Kitsios et al., 2010], the utility of these data are limited by their derivation from experiments in the literature. Kitsios et al. analyzed human GWAS data from the National Human Genome Research Institute (NHGRI) catalog comparing human against mouse phenotype data and concluded that while there is an excellent concordance between human and mouse phenotypes associated with orthologous loci, a significant number of GWAS associations do not have mouse mutants annotated to equivalent phenotype descriptors. This is almost certainly due to the fact that curated mouse data are derived from reports of hypothesis-driven science, often incomplete, driven by the interests and competence of the investigators, and underlines the need for the systematic, agnostic phenotyping of the mouse genome which will be provided by the IMPC. Despite this, the successes resulting from cross-species phenotype integration and mining are impressive and we will return to this below.
High throughput phenotyping of mutant mice
The development of large-scale phenotyping strategies grew out of the phenotype-driven ENU mutagenesis [Acevedo-Arozena et al., 2008; Munroe, et al., 2000], gene-trapping [Hansen et al., 2008] or insertional mutagenesis [Ivics et al., 2009] functional genomics programmes initiated more than a decade ago. The idea that it would be possible to apply these techniques to a reverse genetics approach had always been regarded as hopelessly ambitious given the scale of resources, the cost and the complexity of the undertaking, but since 2004 the issues of feasibility, reproducibility of distributed phenotyping assays and aggregation and analysis of data have been addressed in a series of international programmes and projects.
The earliest project, EUMORPHIA (European Union Mouse Research for Public Health and Industrial Applications; [Gates et al., 2010]) began the integration of the Mouse Clinics across Europe and established the first phenotyping pipeline (EMPReSS [Brown et al., 2005] involving, amongst others the Mouse Clinics at MRC Mammalian Genetics Unit, UK, GSF - National Research Center for Environment and Health, Germany (now Helmholtz-Zentrum), Institute Clinique de la Souris, France, and the Wellcome Trust Sanger Institute (WTSI), UK. Specialist technologies and contributors were recruited throughput Europe [Brown et al., 2006]. The resulting network formed the basis of the subsequent EUMODIC programme. Integrated with the IKMC, the role of EUMODIC (European Mouse Disease Clinics) was to phenotype 500 mutant lines derived from the IKMC project (EUCOMM) as proof of principle for the full-scale genome-wide phenotyping of all the mutant lines produced. Primary phenotype assessment using EMPReSS-slim (a refined version of the EMPReSS pipeline) was undertaken in the four large-scale phenotyping centres and then made publicly available on the EuroPhenome database; (http://www.europhenome.org/). A distributed network of centres with in-depth expertise in a number of phenotyping domains undertook more complex, secondary phenotyping screens and applied them to a triaged subset of the mice which have shown interesting phenotypes in the primary screen. The EuroPhenome database now contains phenotyping data from 407 strains of mutant mice corresponding to 3, 416 annotations on 25, 500 mice. The database is accessible through a web interface or through the EuroPhenome biomart [Mallon et al., 2008; Morgan et al., 2010] and programmatic access is planned. Complementary phenotype data is available through the database of the WTSI Mouse Genetics Programme (MGP) mouse resource portal (MRP; http://www.sanger.ac.uk/mouseportal/). To date, primary pipeline phenotypes are available for 372 lines and 290 lines have been characterised for their phenotypes under infection challenges. Phenotyping under challenge is an area under active development and one which is likely to be increasingly important in establishing the phenotypic repertoire of mutants [Beckers et al., 2009]. The WTSI MGP phenotyping pipelines contain two challenges, one for infection challenge using Citrobacter and Salmonella, and a second for high fat diet. Data from both MRP and EuroPhenome is being compiled with the knockout gene allele record, and coded to the mammalian phenotype ontology in the MGD database.
Phenotyping pipelines
Phenotyping pipelines comprise a series of assays carried out on the same cohort of mice; these assays are designed to characterise large numbers of animals for a wide range of phenotypic parameters; for example, the German Mouse Clinic measures more than 550 parameters in two cohorts of 10 mice of each sex and genotype [Fuchs et al., 2010; Fuchs et al., 2009; Gailus-Durner et al., 2009; Gailus-Durner et al., 2011]. The breadth of these, predominantly in vivo, assays is key to the pipeline approach, which is designed to detect a wide spectrum of phenotypes ranging, for example, from behaviour to dysmorphology, clinical chemistry and immune responses [Brown et al., 2009; Gates et al., 2010; Justice 2008; Mandillo et al., 2008]. The pipelines currently under discussion for the IMPC are shown in Figure 3 and discussed below. The complexity of phenotypes assayed is directly proportional to the cost and inversely proportional to throughput; as assays become more labour-intensive, the number of mice that can feasibly be examined decreases. This then requires the stratification of assays into primary, secondary and tertiary phenotyping efforts, whereby mice from the primary pipeline are selected for additional in-depth analysis on the basis of phenotypes uncovered in the first battery of assays. In EuroPhenome the current dataset contains significant annotations (phenodeviants) for clinical chemistry (more than 100 strains), haematology (92), body weight (61), behaviour (modified SHIRPA (SmithKline Beecham, Harwell, Imperial College, Royal London Hospital, Phenotype Assessment)) (52), to give some idea of the highest assay hit rates.
The International Mouse Phenotyping Consortium
With the completion of the IKMC project the International Mouse Phenotyping Consortium (IMPC) was launched in September 2012 with the aim of phenotyping 20, 000 mutant lines from the IKMC resource [Abbott, 2010]. The Consortium currently consists of 22 Academic and Governmental Institutes across the world, ranging from North America, through Europe to Australia, China, and Japan. Details of the partners, their funding and distribution can be found on the IMPC website; http://www.mousephenotype.org. The KOMP2 initiative, funded by the United States NIH though the Common Fund (http://commonfund.nih.gov/KOMP2/) includes three aspects of work, mouse production and distribution, phenotyping and informatics. Three consortia have been created, BASH (Baylor College of Medicine, WTSI, Harwell), DTCC (University of California at Davis, Toronto Centre for Phenogenomics, Charles River Inc.), and The Jackson laboratory, with a data coordination centre being provided by MRC Harwell, WTSI and, through additional funding, the Jackson Laboratory based MGI. More than $110M has been provided by NIH with the additional partners and clinics being funded by national and international agencies. In all a growing number of more than 12 clinics will take part, with commitment to funding of additional clinics being made by national agencies in Europe and elsewhere. Efforts will be extensively coordinated with the European Commission funded Infrafrontier ESFRI project for mouse functional genomics infrastructure (http://www.infrafrontier.eu). Infrafrontier is an infrastructure-building project for mouse phenotyping and distribution, which aims at mobilizing resources from national governments and already has commitments of more than 135M from European national governments and more promised. The InfraCOMP project (http://www.infrafrontier.eu/infracomp.php) will provide a coordination framework for the cooperation of Infrafrontier and the IMPC.
The IMPC project will have two phases Phase I (2011–2016): phenotype up to 5,000 lines, and Phase II (2016–2021) to phenotype 15,000 mutants. Primary phenotype data will be made publicly available on the IMPC website as it is created and quality controlled, and will be migrated to MGD to be integrated with all of the other phenotype information already available. Integrated datasets all coded to standard MP semantics will be available from the beginning of the project, and primary data will be available for download.
The primary IMPC phenotyping pipeline is currently under discussion (see Figure 3) and further details may be found on the IMPC website. Derived principally from the EMPReSS pipeline this covers a broad range of phenotypes. Homozygous null mice in cohorts of 7 males and 7 females will be in vivo phenotyped up to 16 weeks of age, followed by terminal phenotyping including, in some centres, detailed necropsy and histopathology. Because primary phenotyping will end at 16 weeks, mainly because of the cost of keeping mice for longer, it will therefore be expected to skew phenotyping results against finding degenerative and neoplastic diseases which are predominantly age-related [Sundberg et al 2011]. Some centres will, however, be undertaking limited aging studies, and it is expected that many of these diseases, important sources of morbidity and mortality in humans, will be detected earlier in life using histopathology [Schofield et al. 2011]. Where there is poor or no viability, heterozygous animals will be phenotyped and subviable lines will be subjected to embryonic phenotyping to determine reasons for embryonic or perinatal lethality. Around 30% of homozygotes are expected to show sub-viability so embryonic phenotyping is going to be an important contributor to our understanding of gene function.
Secondary and tertiary phenotyping are expected to provide detailed descriptions of the phenotypes identified by the primary pipeline. In many cases this will be carried out outside the IMPC partners by groups with special clinical or biological interests. The UK Medical Research Council has created a mouse network (http://www.har.mrc.ac.uk/MRCMouseNetwork/) which will coordinate basic research and training around the IMPC resources in the UK, providing an potentially useful model for the engagement of the whole research community.
Computational use of model organism phenotype/genotype associations
Only about half (3710, corresponding to approximately 2000 genes) of the diseases listed in OMIM have been linked to a causative gene, and most approaches using model organism phenotypes, particularly the mouse, have addressed the problem of identifying causative genes for these rare diseases. This has become a major policy goal of research for the European Commission and the United States NIH, as well as a target for other national and international funding agencies. These aims are represented by the International Rare Disease Research Consortium (IRDiRC; http://ec.europa.eu/research/health/medical-research/rare-diseases/irdirc_en.html ). IRDiRC has two goals by 2020; to produce diagnostic tests for most rare diseases and to develop new therapies for 200 rare diseases. Diseases caused by copy number variation (CNV) are less well documented in OMIM and the more recent resource, Orphanet, and present a subtly different problem in that the region of variation is known but it can contain in some cases many hundreds of genes where one, or a small number of which, might be responsible for the overall phenotype.
Approaches to the identification of candidate genes for human diseases can be broadly separated into two categories; those which are orthology-dependent and those which are purely phenotype based.
Orthology-dependent candidate gene discovery
Inference of gene function and disease implication may be made through orthology, often as part of a wider range of functional and sometimes structural criteria, and often including phenotype information from both species. This includes “guilt-by-association” studies where functions and disease involvement are inferred across functional networks built in both species from a variety of data including, for example, phenotypes, gene expression, or pathway involvement. These approaches are all predicated on the assumption that orthologous genes are involved in orthologous pathways and therefore likely to produce similar phenotypes when mutated; the “phenologs” of McGary et al. [McGary et al., 2010] which represent phenotypes related by the orthology of the associated genes in two organisms. In this model two phenotypes are said to be orthologous if their genetic associations are enriched for the same orthologous genes, even if the phenotypes are superficially dissimilar. Using this method they were able, for example, to predict novel genes involved in angiogenesis from considering yeast, human and mouse phenologs.
Orthology has also been used by Espinosa and Hancock [Espinosa and Hancock, 2011] to map mouse phenotype annotations onto OMIM diseases to create a phenotype-genotype network and is also used for integration of phenotype data in Phenomic DB [Groth et al., 2011] where text-mined phenotype data from the literature is captured into a cross-species database from OMIM and model organism databases. Zhang et al. [Zhang et al., 2010] group human disease and mouse phenotype-associated gene sets into disease and phenotype groups. Using data from the gene association database (GAD) [Becker et al., 2004] and MGD, they generated disease-based genes sets for 1,317 human disease phenotypes as well as 5,142 mouse experimentally determined phenotypes and showed a striking concordance between human disease phenotypes and mouse mutant phenotypes. Orthology-based approaches are also described by Hu et al. [Hu et al., 2010] and an interesting approach by Sardana et al where they establish phenotypic “homology” using UMLS as a bridge between organism phenotype annotations (see below) and compare this with the expected “phenologs” using genetic orthology [Sardana et al., 2010].
Hehir Kwa [Hehir-Kwa et al., 2011] reported a Bayseian tree classification approach, GECCO, using thirteen structural and functional genomic features to establish pathogenicity of human copy number variations. GECCO perfectly classifies CNVs causing known mental retardation-associated syndromes and achieves high accuracy (94%) and negative predictive value (99%) on a blinded test set of more than 1,200 CNVs from a large cohort of individuals. The orthologous mouse phenotypes were found to be one of the most important classifiers. Given that the genomic and phenotypic coverage of the current mouse dataset are limited, the contribution to classification can only be expected to increase as data flows from the IMPC efforts together with further hypothesis driven research. This study provides an extremely useful insight into the kind of power we should expect when there is data on the complete mouse phenome.
Similar approaches to determining the pathogenicity of components of CNVs have been previously reported. Shaikh et al [Shaikh et al., 2011] used mouse phenotypic annotation of orthologs contained within the CNVs of 87 patients clinically diagnosed with a range of developmental delay (DD) syndromes. They then identified phenotypic classes of genes enriched in these DD-associated CNVs, and investigated whether the mouse orthologs of genes contained within the CNVs were significantly enriched in any of 147 nervous system phenotypic terms selected from the MP ontology. Four mouse phenotypes were identified that were each significantly enriched among all DD-associated CNVRs. Such genes are therefore strong candidates for causative mutations whose copy number change underlies the DD disorder for individual patients. This represented an 86% increase over the number expected by chance, and is an excellent example of the use of mouse phenotypes to prioritise causative disease candidates.
The use of orthology in mapping between human and mouse phenotypes has proved to be extremely robust and is widely used as the basis of, or as an important contributor to, establishing gene function or involvement in disease in humans. An important caveat however is that there is some evidence for functional divergence of orthologs. How important this is on a genomic scale remains to be seen – a question that can only be even approached when we have the genome-wide survey of phenotypes promised by the IMPC. A survey by Liao and Zhang [Liao and Zhang, 2008] suggested that >20% of essential human genes are non-essential in the mouse. Whilst essentiality is something of an acid test, the problem with attempting to make this comparison is confounded by the differing effects of modifiers and background on lethality and that most of the mouse data is based on a range of inbred lines, many of which show differences in the essentiality of specific mutant alleles themselves [Yoshiki and Moriwaki, 2006]. Comparing this rather heterogeneous data with known confounding factors with human clinical data it is rather surprising that we find a concordance as high as 80%, suggesting that the true figure may be much higher. Patterns of gene expression are also well-conserved; this was indicated by the study of Liao and Zhang and consistent with the global study of mouse and man carried out by Zheng-Bradley et al who found that global patterns of tissue-specific expression of orthologous genes are conserved in human and mouse and that expression of groups of orthologous genes co-varies in the two species [Zheng-Bradley et al., 2010].
Phenotype-based candidate gene discovery
The ability to establish candidate genes for a phenotype, for example looking for genes for rare diseases or prioritizing candidates for pathogenicity using only phenotypic data, has been hampered by the problem of comparing different phenotypes in different organisms, both because of lack of common semantics but also because of intrinsic differences between the way often fundamentally common phenotypes can be expressed in the different organisms. These issues are discussed in [Schofield et al. 2010] and [Schofield et al 2011a]. Two approaches have been taken to this; the first is to use a bridging terminology such a UMLS or MeSH. [Burgun et al. 2009] successfully used an approach involving lexical matching to map the MP ontology to UMLS permitting interrogation of OMIM and other clinical datasets using one of the more than 100 terminologies integrated into UMLS. However, more recently, an approach (termed EQ) using the matching of logical definitions of pre-composed phenotype ontologies has been developed which overcomes the intrinsic semantic problems in integrating phenotype ontologies from different species. In the EQ method, a phenotype is characterized by an affected Entity (from an anatomy or process ontology) and a Quality [from the Phenotype And Trait Ontology (PATO)) that specifies how the entity is affected [Gkoutos et al., 2004]. The affected entity can either be a biological function or process as specified in the Gene Ontology (GO), or an anatomical entity. Whilst the ontologies used to create the definitions are largely species-agnostic, such as GO, CheBI, MPATH, anatomical entities are almost exclusively specified using species-specific anatomy ontologies. In order to make mappings between these vertebrate anatomies the metazoan, species-independent UBERON ontology is used in constructing anatomically-based cross-products [Mungall et al., 2012]. This method was used by Washington et al. [2010] who annotated the phenotypes of 11 gene-linked human diseases from OMIM and computationally compared these with other ontology-based phenotype descriptions from model organisms.
Two further approaches using logical definitions of pre-composed ontologies have been recently developed. Mousefinder [Smedley et al. 2012. This issue] exploits new semantic matching software [OWLSim] to identify new candidate genes for orphan human diseases using inferred phenotype information alone. A second recent approach using EQ, PhenomeNET, integrates multiple species-specific pre-composed phenotype ontologies to generate a single cross-species phenotype ontology [Hoehndorf et al., 2011]. Efficient automated reasoning over the PhenomeNET is enabled through ontology modularization and design patterns for expressing phenotypes and their links to anatomy and physiology ontologies [Hoehndorf et al 2010, 2011a]. Once data from the model organism databases is integrated into the network this allows the direct comparison of phenotypes of multiple species, and PhenomeNET is able to perform a pairwise comparison of phenotypes using a measure of semantic similarity. PhenomeNET can rank phenotypes for diseases as well as phenotype annotations from model organism databases and can predict genes that participate in the same pathway, orthologous genes (on the basis of shared phenotypes) as well as gene–disease associations based on comparing phenotypes alone.
Prospects
The tools and approaches for mobilizing data on gene/function relationships from model organisms have only recently become available, and are already showing how useful and powerful cross-species studies can be. The model organism databases, particularly that for the mouse, MGD, have lead the way in data formalisation and integration and the human genetics community now stands to reap many benefits from that. Coding of human phenotypic data to the Human Phenotype ontology has provided a watershed in the convergent flow of data for humans and model organisms and as its use is adopted by other resources they too will yield up their data to semantic integration with all the power that brings to the study of human genetics and disease.
Both the developed and incipient resources for the mouse discussed here are landmarks in experimental biology and genetics. The availability of mutants for every coding gene in a mammal, designed in the same way and on the same genetic background is an unparalleled resource, and the prospect of systematic phenotype data for all these mutants opens up the ability to carry out computational analysis unbiased by partial phenotyping, background strain differences and the interests of the investigator. The amount of biological data which will flow from the International Mouse Phenotyping Consortium’s activities will be huge – we know nothing about nearly half of the predicted protein coding genes in the mammalian genome – the ignorome - is now very much the target of our efforts. The challenge now is for the human genetics community to take up the data, materials and computational resources offered by these projects and apply them to the understanding of disease. This is a familiar paradigm for some investigators but novel for others. One driving imperative for the work carried out on the mouse community projects over the last decade and forward into the next is that data and resources should be made as widely and freely available as possible and that the groups involved in these programmes interact with and respond to the wider scientific community. With energetic and open communications we can expect the synergy between human and mouse genetics to accelerate in an unprecedented manner as data flows, with both basic biology and medicine being the beneficiaries.
Acknowledgments
The authors would like to thank Prof. Klaus Schughart and Prof. Steve Brown for helpful comments on the manuscript. Related work in the authors’ laboratories is funded by the National Institutes of Health (R01 HG004838-02), the Commission of the European Union (EUMODIC contract number LSHG-CT-2006-037188) and the European Commission’s 7th Framework Programme, RICORDO project, grant number 248502.
References
- 1000 _genomes_consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott A. Mouse project to find each gene’s role. Nature. 2010;465:410. doi: 10.1038/465410a. [DOI] [PubMed] [Google Scholar]
- Acevedo-Arozena A, Wells S, Potter P, Kelly M, Cox RD, Brown SD. ENU mutagenesis, a way forward to understand gene function. Annu Rev Genomics Hum Genet. 2008;9:49–69. doi: 10.1146/annurev.genom.9.081307.164224. [DOI] [PubMed] [Google Scholar]
- Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM) Hum Mutat. 2011;32:564–7. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. The knockout mouse project. Nat Genet. 2004;36:921–4. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, et al. The European dimension for the mouse genome mutagenesis program. Nat Genet. 2004;36:925–7. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer WG, Shultz KL, Coombs HF, 3rd, Horton LG, Donahue LR, Rosen CJ. Multiple quantitative trait loci for cortical and trabecular bone regulation map to mid-distal mouse chromosome 4 that shares linkage homology to human chromosome 1p36. J Bone Miner Res. 2011 Sep 28; doi: 10.1002/jbmr.515. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet. 2004;36:431–2. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- Beckers J, Wurst W, de Angelis MH. Towards better mouse models: enhanced genotypes, systemic phenotyping and envirotype modelling. Nat Rev Genet. 2009;10:371–80. doi: 10.1038/nrg2578. [DOI] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Eppig JT, Kadin JA, Richardson JE. The Mouse Genome Database genotypes::phenotypes. Nucleic Acids Res. 2009;37(Database issue):D712–9. doi: 10.1093/nar/gkn886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39(Database issue):D842–8. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Lad H, Green E, Gkoutos G, Gates H, de Angelis MH. Standards of Mouse Model Phenotyping. Wiley-VCH Verlag GmbH; 2006. Eumorphia and the European Mouse Phenotyping Resource for Standardized Screens (EMPReSS) pp. 311–320. [Google Scholar]
- Brown SD, Chambon P, de Angelis MH. EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet. 2005;37:1155. doi: 10.1038/ng1105-1155. [DOI] [PubMed] [Google Scholar]
- Brown SD, Wurst W, Kuhn R, Hancock J. The Functional Annotation of Mammalian Genomes: The Challenge of Phenotyping. Annu Rev Genet. 2009;43:305–33. doi: 10.1146/annurev-genet-102108-134143. [DOI] [PubMed] [Google Scholar]
- Bult CJ. From information to understanding: the role of model organism databases in comparative and functional genomics. Anim Genet. 2006;37(Suppl 1):28–40. doi: 10.1111/j.1365-2052.2006.01475.x. [DOI] [PubMed] [Google Scholar]
- Burgun A, Mougin F, Bodenreider O. Two approaches to integrating phenotype and clinical information. AMIA Annu Symp Proc. 2009;2009:75–9. [PMC free article] [PubMed] [Google Scholar]
- Callaway E. How to build a better mouse. Nature. 2011;475:279. doi: 10.1038/475279a. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010;86:6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Finnell RH, Rossant J, Wurst W. A new partner for the international knockout mouse consortium. Cell. 2007;129:235. doi: 10.1016/j.cell.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium. The Genome Architecture of the Collaborative Cross Mouse Genetic Reference Population. Genetics. 2012;190(2):389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RD, Church CD. Mouse models and the interpretation of human GWAS in type 2 diabetes and obesity. Dis Model Mech. 2011;4:155–64. doi: 10.1242/dmm.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson M. FIMRe: Federation of International Mouse Resources: global networking of resource centers. Mamm Genome. 2006;17:363–4. doi: 10.1007/s00335-006-0001-2. [DOI] [PubMed] [Google Scholar]
- Durrant C, Tayem H, Yalcin B, Cleak J, Goodstadt L, de Villena FP, Mott R, Iraqi FA. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21:1239–48. doi: 10.1101/gr.118786.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa O, Hancock JM. A gene-phenotype network for the laboratory mouse and its implications for systematic phenotyping. PLoS One. 2011;6:e19693. doi: 10.1371/journal.pone.0019693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feero WG, Guttmacher AE, Collins FS. Genomic medicine--an updated primer. N Engl J Med. 2010;362:2001–11. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2011 update. Nucleic Acids Res. 2010;39(Database issue):D835–41. doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs H, Gailus-Durner V, Adler T, Aguilar-Pimentel JA, Becker L, Calzada-Wack J, Da Silva-Buttkus P, Neff F, Gotz A, Hans W, et al. Mouse phenotyping. Methods. 2010;53:120–35. doi: 10.1016/j.ymeth.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Fuchs H, Gailus-Durner V, Adler T, Pimentel JA, Becker L, Bolle I, Brielmeier M, Calzada-Wack J, Dalke C, Ehrhardt N, et al. The German Mouse Clinic: a platform for systemic phenotype analysis of mouse models. Curr Pharm Biotechnol. 2009;10:236–43. doi: 10.2174/138920109787315051. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Fuchs H, Adler T, Aguilar Pimentel A, Becker L, Bolle I, Calzada-Wack J, Dalke C, Ehrhardt N, Ferwagner B, et al. Systemic first-line phenotyping. Methods Mol Biol. 2009;530:463–509. doi: 10.1007/978-1-59745-471-1_25. [DOI] [PubMed] [Google Scholar]
- Gailus-Durner V, Naton B, Adler T, Afonso L, Aguilar-Pimentel J-A, Becker L. The German Mouse Clinic – Running an Open Access Platform. In: Brakebusch CTP, editor. The mouse as a model organism. Berlin: Springer Verlag; 2011. pp. 11–44. [Google Scholar]
- Gates H, Mallon AM, Brown SD. High-throughput mouse phenotyping. Methods. 2010;53:394–404. doi: 10.1016/j.ymeth.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Gkoutos GV, Green ECJ, Mallon A-M, Hancock JM, Davidson D. Building mouse phenotype ontologies. Pac Symp Biocomputing. 2004;9:178–189. doi: 10.1142/9789812704856_0018. [DOI] [PubMed] [Google Scholar]
- Groth P, Kalev I, Kirov I, Traikov B, Leser U, Weiss B. Phenoclustering: online mining of cross-species phenotypes. Bioinformatics. 2011;26:1924–5. doi: 10.1093/bioinformatics/btq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth P, Pavlova N, Kalev I, Tonov S, Georgiev G, Pohlenz HD, Weiss B. PhenomicDB: a new cross-species genotype/phenotype resource. Nucleic Acids Res. 2007;35(Database issue):D696–9. doi: 10.1093/nar/gkl662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–9. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehir-Kwa JY, Wieskamp N, Webber C, Pfundt R, Brunner HG, Gilissen C, de Vries BB, Ponting CP, Veltman JA. Accurate distinction of pathogenic from benign CNVs in mental retardation. PLoS Comput Biol. 2011;6:e1000752. doi: 10.1371/journal.pcbi.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, Dumontier M, Oellrich A, Wimalaratne S, Rebholz-Schuhmann D, Schofield P, Gkoutos GV. A common layer of interoperability for biomedical ontologies based on OWL EL. Bioinformatics. 2011;27:1001–1008. doi: 10.1093/bioinformatics/btr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, Schofield PN, Gkoutos GV. PhenomeNET: a whole-phenome approach to disease gene discovery. Nucleic Acids Res. 2011;39:e119. doi: 10.1093/nar/gkr538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, Dumontier M, Oellrich A, Rebholz-Schuhmann D, Schofield PN, Gkoutos GV. Interoperability between biomedical ontologies through relation expansion, upper-level ontologies and automatic reasoning. PLoS One. 2011a;6:e22006. doi: 10.1371/journal.pone.0022006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2010;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–22. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ. Removing the cloak of invisibility: phenotyping the mouse. Dis Model Mech. 2008;1:109–12. doi: 10.1242/dmm.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsnelson A. Breaking the silence. Nat Med. 2011;17:1536–1538. doi: 10.1038/nm1211-1536. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsios GD, Tangri N, Castaldi PJ, Ioannidis JP. Laboratory mouse models for the human genome-wide associations. PLoS One. 2010;5:e13782. doi: 10.1371/journal.pone.0013782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–97. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Leduc MS, Lyons M, Darvishi K, Walsh K, Sheehan S, Amend S, Cox A, Orho-Melander M, Kathiresan S, Paigen B, et al. The mouse QTL map helps interpret human genome-wide association studies for HDL cholesterol. J Lipid Res. 2011;52:1139–49. doi: 10.1194/jlr.M009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proceedings of the National Academy of Sciences. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–8. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon AM, Blake A, Hancock JM. EuroPhenome and EMPReSS: online mouse phenotyping resource. Nucleic Acids Res. 2008;36(Database issue):D715–8. doi: 10.1093/nar/gkm728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandillo S, Tucci V, Holter SM, Meziane H, Banchaabouchi MA, Kallnik M, Lad HV, Nolan PM, Ouagazzal AM, Coghill EL, et al. Reliability, robustness, and reproducibility in mouse behavioral phenotyping: a cross-laboratory study. Physiol Genomics. 2008;34:243–55. doi: 10.1152/physiolgenomics.90207.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- Mathes WF, Kelly SA, Pomp D. Advances in comparative genetics: influence of genetics on obesity. Br J Nutr. 2011;106(Suppl 1):S1–S10. doi: 10.1017/S0007114511001905. [DOI] [PubMed] [Google Scholar]
- McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107:6544–9. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, et al. EuroPhenome: A repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkp1007. Available online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Torniai C, Gkoutos GV, Lewis SE, Haendel MA. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012 Jan 31;13:R5. doi: 10.1186/gb-2012-13-1-r5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe RJ, Bergstrom RA, Zheng QY, Libby B, Smith R, John SW, Schimenti KJ, Browning VL, Schimenti JC. Mouse mutants from chemically mutagenized embryonic stem cells. Nat Genet. 2000;24:318–21. doi: 10.1038/73563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 2010;7:893–5. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oti M, Huynen MA, Brunner HG. The biological coherence of human phenome databases. Am J Hum Genet. 2009;85:801–8. doi: 10.1016/j.ajhg.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, Cirulli ET, Fellay J, Dickson SP, Gumbs CE, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Philip VM, Sokoloff G, Ackert-Bicknell CL, Striz M, Branstetter L, Beckmann MA, Spence JS, Jackson BL, Galloway LD, Barker P, et al. Genetic analysis in the Collaborative Cross breeding population. Genome Res. 2011;21:1223–38. doi: 10.1101/gr.113886.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;16(Spec No. 2):R168–73. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- Poot M, Badea A, Williams RW, Kas MJ. Identifying human disease genes through cross-species gene mapping of evolutionary conserved processes. PLoS One. 2011;6:e18612. doi: 10.1371/journal.pone.0018612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S, et al. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39(Database issue):D849–55. doi: 10.1093/nar/gkq879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Kohler S, Bauer S, Seelow D, Horn D, Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83:610–5. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Krawitz P, Mundlos S. Strategies for exome and genome sequence data analysis in disease-gene discovery projects. Clin Genet. 2011;80:127–32. doi: 10.1111/j.1399-0004.2011.01713.x. [DOI] [PubMed] [Google Scholar]
- Rosenthal N, Brown S. The mouse ascending: perspectives for human-disease models. Nat Cell Biol. 2007;9:993–9. doi: 10.1038/ncb437. [DOI] [PubMed] [Google Scholar]
- Sardana D, Vasa S, Vepachedu N, Chen J, Gudivada RC, Aronow BJ, Jegga AG. PhenoHM: human-mouse comparative phenome-genome server. Nucleic Acids Res. 2010;38(Web Server issue):W165–74. doi: 10.1093/nar/gkq472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebelle L, Wolf C, Stribl C, Javaheri T, Schnutgen F, Ettinger A, Ivics Z, Hansen J, Ruiz P, von Melchner H, et al. Efficient conditional and promoter-specific in vivo expression of cDNAs of choice by taking advantage of recombinase-mediated cassette exchange using FlEx gene traps. Nucleic Acids Res. 2010;38:e106. doi: 10.1093/nar/gkq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, Ehrmann F, Ruiz-Noppinger P, von Melchner H. High throughput gene trapping and postinsertional modifications of gene trap alleles. Methods. 2011;53:347–55. doi: 10.1016/j.ymeth.2010.12.037. [DOI] [PubMed] [Google Scholar]
- Schofield PN, Gkoutos GV, Gruenberger M, Sundberg JP, Hancock JM. Phenotype ontologies for mouse and man: bridging the semantic gap. Dis Model Mech. 2010;3(5–6):281–9. doi: 10.1242/dmm.002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PN, Vogel P, Gkoutos GV, Sundberg JP. Exploring the elephant: histopathology in high-throughput phenotyping of mutant mice. Dis Model Mech. 2011;5:19–25. doi: 10.1242/dmm.008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield PN, Sundberg JP, Hoehndorf R, Gkoutos GV. New approaches to the representation and analysis of phenotype knowledge in human diseases and their animal models. Brief Funct Genomics. 2011a;10:258–65. doi: 10.1093/bfgp/elr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Haldeman-Englert C, Geiger EA, Ponting CP, Webber C. Genes and biological processes commonly disrupted in rare and heterogeneous developmental delay syndromes. Hum Mol Genet. 2011;20:880–93. doi: 10.1093/hmg/ddq527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Salimova E, Rosenthal N. Cre recombinase resources for conditional mouse mutagenesis. Methods. 2011;53:411–6. doi: 10.1016/j.ymeth.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Smith CL, Eppig JT. The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip Rev Syst Biol Med. 2009;1:390–9. doi: 10.1002/wsbm.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg J, Berndt A, Sundberg B, Silva KA, Kennedy V, Bronson R, Yuan R, Paigen B, Harrison D, Schofield PN. The mouse as a model for understanding chronic diseases of aging: the histopathologic basis of aging in inbred mice. Pathobiology of Aging & Age-related Diseases. 2011;1:71719. doi: 10.3402/pba.v1i0.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson KL, Gatti DM, Valdar W, Welsh CE, Cheng R, Chesler EJ, Palmer AA, McMillan L, Churchill GA. High-resolution genetic mapping using the mouse diversity outbred population. Genetics. 2012;190:437–47. doi: 10.1534/genetics.111.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Miller DR, Churchill GA, de Villena FP. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. Ilar J. 2011;52:24–31. doi: 10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Churchill GA. Ten Years of the Collaborative Cross. G3: Genes|Genomes|Genetics. 2012;2:153–156. doi: 10.1534/g3.111.001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–9. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77:1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7:e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weinreich SS, Mangon R, Sikkens JJ, Teeuw ME, Cornel MC. Orphanet: a European database for rare diseases. Ned Tijdschr Geneeskd. 2008;152:518–9. [PubMed] [Google Scholar]
- Wilkinson P, Sengerova J, Matteoni R, Chen CK, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, et al. EMMA--mouse mutant resources for the international scientific community. Nucleic Acids Res. 2011;38 (Database issue):D570–6. doi: 10.1093/nar/gkp799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiki A, Moriwaki K. Mouse phenome research: implications of genetic background. Ilar J. 2006;47:94–102. doi: 10.1093/ilar.47.2.94. [DOI] [PubMed] [Google Scholar]
- Zhang Y, De S, Garner JR, Smith K, Wang SA, Becker KG. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med Genomics. 2010;3:1. doi: 10.1186/1755-8794-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng-Bradley X, Rung J, Parkinson H, Brazma A. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 2010;11:R124. doi: 10.1186/gb-2010-11-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouberakis M, Chandras C, Swertz M, Smedley D, Gruenberger M, Bard J, Schughart K, Rosenthal N, Hancock JM, Schofield PN, et al. Mouse Resource Browser--a database of mouse databases. Database (Oxford) 2010;2010:baq010. doi: 10.1093/database/baq010. [DOI] [PMC free article] [PubMed] [Google Scholar]