Abstract
The present study reports the large-scale discovery of genome-wide single-nucleotide polymorphisms (SNPs) in chickpea, identified mainly through the next generation sequencing of two genotypes, i.e. Cicer arietinum ICC4958 and its wild progenitor C. reticulatum PI489777, parents of an inter-specific reference mapping population of chickpea. Development and validation of a high-throughput SNP genotyping assay based on Illumina's GoldenGate Genotyping Technology and its application in building a high-resolution genetic linkage map of chickpea is described for the first time. In this study, 1022 SNPs were identified, of which 768 high-confidence SNPs were selected for designing the custom Oligo Pool All (CpOPA-I) for genotyping. Of these, 697 SNPs could be successfully used for genotyping, demonstrating a high success rate of 90.75%. Genotyping data of the 697 SNPs were compiled along with those of 368 co-dominant markers mapped in an earlier study, and a saturated genetic linkage map of chickpea was constructed. One thousand and sixty-three markers were mapped onto eight linkage groups spanning 1808.7 cM (centiMorgans) with an average inter-marker distance of 1.70 cM, thereby representing one of the most advanced maps of chickpea. The map was used for the synteny analysis of chickpea, which revealed a higher degree of synteny with the phylogenetically close Medicago than with soybean. The first set of validated SNPs and map resources developed in this study will not only facilitate QTL mapping, genome-wide association analysis and comparative mapping in legumes but also help anchor scaffolds arising out of the whole-genome sequencing of chickpea.
Keywords: chickpea, SNP, linkage map, genotyping, NGS
1. Introduction
Discovery of the DNA sequence polymorphism is a prerequisite to generating genetic markers for various applications of modern genomics research such as map-based cloning, marker-assisted breeding, association mapping and understanding genome structure and function. Among the sequence polymorphisms, single-nucleotide polymorphisms (SNPs) represent the most abundant type of variation present in DNA. SNPs are mostly biallelic,1 co-dominantly inherited, sequence tagged and occur at high density within genomes.2 They are thus amenable to the development of genetic molecular markers at low cost, which can provide sufficiently dense genome coverage for the dissection of complex traits. SNPs are known to occur at high frequencies of ∼1 per 500–1000 bp in humans3,4 and in plant genomes also, where their frequency appears to vary significantly, e.g. 1 SNP per 16 bp in eucalyptus,5 1 per 107 bp in radish,6 1 per 147 bp in rice,7 1 per 200 bp in maize,8 1 per 370 bp in soybean genes9 and 1 per 500 bp in Arabidopsis.10
Recently, technological advancements have accelerated the genome-wide SNP discovery not only in model species, but also in crop plants. Initial efforts relied on the discovery of large numbers of SNPs from EST databases such as in grapevine,11 spruce,12 cowpea,13 pea,14 wheat15 and pine.16 However, more recently, with the development of the next generation sequencing (NGS) platforms which enable the sequencing of millions of bases at deep coverage, thousands of SNPs have been identified in many species such as maize,17 soybean,18 Medicago,19 Eucalyptus,20 rice21 and sunflower.22 To utilize the millions of available SNPs, various high-throughput SNP genotyping platforms were simultaneously developed such as the GoldenGate Genotyping Technology (GGGT; Illumina, San Diego, CA, USA),23 BeadChip-based Infinium assay (Illumina),24,25 SNPStream (Beckman Coulter, USA),26 GeneChip (Affymetrix, USA)27,28 and competitive allele-specific PCR, KASPar (KBio science, UK)29 that allow large-scale genotyping of SNPs in parallel in a large set of individuals.30 These approaches vary in terms of sensitivity, reproducibility, accuracy, capability of multiplexing and throughput. Among them, one of the most versatile SNP genotyping platforms is the Illumina GGGT, which is capable of multiplexing from 96 to 1536 SNPs per assay in a single reaction over a 3-day period.23 This technology has persistently been reported as highly reliable, with high SNP conversion rates and has fostered genetic research in several major as well as in orphan plants, especially self-pollinating crops that have been plagued by a narrow genetic base with nucleotide diversities from 0.2 to 0.5%,13,31–33 as well as the cross-pollinated species with higher sequence diversities of ≥2%.12,34,35 High-throughput SNP genotyping has enabled various applications in plant genomics especially genome-wide association studies and linkage disequilibrium studies,10,36–38 synteny-based comparative genomics13,39 and high-resolution genetic mapping.40,41 A high-density genetic linkage map is one of the most important genomic tools to accelerate marker-assisted breeding. Recent studies have reported the successful utilization of high-throughput GGGT for SNP genotyping to build genetic linkage maps in legumes such as cowpea,13 soybean18,31 and pea,14 and even in plants with highly repetitive or polyploid genomes like barley,32 pine,34 grass41 and maize.42 Production of high-density SNP-based maps have facilitated the fine mapping and cloning of agronomically important genes and also anchoring and orienting the scaffolds generated by whole genome sequence assembly data.18,41,43
Chickpea (C. arietinum L.), a diploid (2n = 2x = 16), annual, self-pollinated crop, with a genome size of 740 Mb,44 represents the world's third most important legume crop that is mainly grown in the arid and semi-arid regions of Asia and Africa. It serves as a key source of protein in human nutrition and also plays an important role in the maintenance of soil fertility owing to its ability to fix atmospheric nitrogen. Due to its economic importance, there has been a recent spurt in chickpea genomics research and a large number of genomic resources, such as molecular markers and linkage maps,45–48 ESTs49–52 and NGS-based transcriptomes,53–55 have become available. Currently, there are ∼2000 co-dominant molecular markers available, which include ∼980 genomic simple sequence repeats (gSSRs),45,46,56–62 and EST-derived markers including 361 EST-SSRs, 238 Intron-Targeted Primers (ITPs), 109 Expressed Sequence Tag Polymorphisms (ESTPs) and 294 Cleaved Amplified Polymorphic Sites/SNPs.45,47–49,51,63 Using these co-dominant markers, linkage maps that have been generated define ∼500 mapped positions.45–48,58,64 Moreover, from the recent chickpea transcriptome sequencing, even though ∼5000 molecular markers have been reported,54,55 the validation and genotyping of these molecular markers is yet to be undertaken. Despite this increased availability of genomic resources, the large-scale discovery and utilization of SNPs, which serve as the most potential markers for providing sufficiently dense genetic maps, has not been carried out in chickpea. Therefore, in the present study, the large-scale identification of genome-wide SNPs was undertaken from the NGS of two parents of a mapping population (C. arietinum cv. ICC4958 and C. reticulatum PI489777). Conversion of SNPs into successful genotyping assays based on the Illumina GoldenGate technology was demonstrated for the first time in chickpea. Moreover, the SNP markers were successfully mapped in the backdrop of previously reported co-dominant markers to generate one of the most comprehensive and dense genetic maps of chickpea.
2. Materials and methods
2.1. Plant material and DNA isolation
For SNP discovery, the two genotypes C. arietinum ICC4958 (fusarium wilt resistant and drought tolerant) and C. reticulatum PI489777 (wild annual species, fusarium wilt susceptible), which are parents of a mapping population, were sequenced. Nuclear DNA from these two genotypes was isolated using the protocol of Malmberg et al.65 and used for sequencing. For SNP genotyping and linkage map generation, the internationally accepted reference mapping population consisting of 129 RILs (recombinant inbred lines) arising from an inter-specific cross between the two genotypes mentioned above was utilized. Genomic DNA was isolated from fresh young leaves of the two mapping parents and the 129 RILs using the GenElute™ plant genomic DNA Miniprep kit (Sigma). The DNA quality was checked by electrophoresis on 0.8% agarose gels. For SNP genotyping, the DNA was quantified using Quant-iT™ Pico Green® dsDNA Kit (Invitrogen) and the fluorescence was measured with the Microtiter plate reader (Varioscan from Thermo Scientific). Samples were adjusted to 50 ng/μl using Tris-EDTA buffer. The GoldenGate genotyping assay was performed using 250 ng of DNA from each of the RILs.
2.2. NGS and SNP discovery
SNP discovery was based on the high-throughput next generation whole genome sequencing of the two genotypes, i.e. C. arietinum cv. ICC4958 and its wild relative C. reticulatum cv. PI489777. Sequencing of C. arietinum ICC4958 was carried out primarily by the 454/Roche GS FLX Titanium platform, for which DNA sample preparation and construction of whole-genome shotgun (WGS) and matepair libraries were performed as described by the manufacturer,66 with some modifications. Briefly, the nuclear DNA was nebulized and size-selected for a 300–900-bp fragment size using Agencourt AMPure SPRI beads and ligated with specific adapters A and B at each end, which resulted in generating a single-stranded or a double-stranded library. One single-stranded library using the standard library preparation kit and two double-stranded libraries using a rapid library preparation kit with an average insert size of 705 bases were used for sequencing. The libraries were amplified by emPCR.66 DNA molecules that contain a single A and B adapters at each end were enriched with capture beads coated with capture DNA. Sequencing was performed using GS FLX Titanium Sequencing kits following the manufacturer's protocol (Roche Applied Sciences, Mannheim, Germany). The generated Roche 454 reads were submitted to the Short Read Archive (SRA) database of NCBI (www.ncbi.nlm.nih.gov/sra) and assigned the accession number SRA053228.1. These reads were assembled using the de novo assembly tool Newbler (GS de novo assembler, Roche Applied Sciences) version 2.3 to obtain the reference sequence. Further, two genomic DNA libraries of 3 kb average insert size, of both the parents (C. arietinum ICC4958 and C. reticulatum PI489777) were also prepared by the SOLiD Opti Mate-paired library kit following the manufacturer's protocol and sequenced using the SOLiD 4.0 platform (Applied Biosystems, Foster City, CA, USA). These reads were assigned the accession number SRA053197.5 by the SRA of NCBI. SNPs were identified by aligning and mapping the SOLiD reads of ICC4958 and PI489777 onto the Roche assembly using the tool HAPS (http://solidsoftwaretools.com/gf/project/haps/). For the selection of SNPs the following criteria were used: (i) non-reference alleles with a minimum throughput of 5X and score 0.6 were selected, (ii) the non-reference alleles with any variant (SNPs or indels) within 60 bases upstream or downstream of the consensus reference sequences were discarded and (iii) one SNP from each contig was chosen to ensure genome-wide distribution.
The second approach for SNP discovery was amplicon resequencing. This involved the identification of SNPs from chickpea ESTs generated earlier in the laboratory and is described in Choudhary et al.48 A set of 222 EST-derived primers were used to PCR amplify and resequence genomic DNA of the two parental lines (C. arietinum cv. ICC4958 and C. reticulatum cv. PI489777) of the reference mapping population.
2.3. Development of the GoldenGate SNP genotyping assays
SNP genotyping was carried out using the Illumina GGGT. For developing the GGGT assay, the identified SNPs along with the 60 bp sequence flanking it on either side were submitted to Illumina for processing by Illumina's Array Design Tool (ADT) in order to obtain a designability rank score for each SNP ranging from 0 to 1. This score serves as a reliability metric for testing whether or not a particular SNP will convert into a successful working GoldenGate assay where SNPs with the scores of 0.6 or higher possess a high probability of converting into a successful genotyping assay. Thus, SNPs having the highest ADT scores were selected for designing the custom Oligo Pool All (CpOPA) containing the allele-specific and locus-specific oligos for use in the Illumina GoldenGate assay and the first chickpea 768-OPA was generated and designated CpOPA-I. The list of SNPs and their flanking regions that constituted CpOPA-I are presented in Supplementary Table S1. Genotyping of SNPs was performed using Illumina's BeadArray Express Reader according to the standard manufacturer's protocol,23,31 with 250 ng (50 ng/μl) of genomic DNA from each individual RIL of the mapping population. The automatic allele calling for each locus was inferred with the GenomeStudio Software V2011.1 (Illumina). The fluorescence images of array matrix carrying Cy3- and Cy5-labelled beads were generated with the two-channel scanner. A genotype that is homozygous for one SNP allele will display a signal in either the Cy3 or the Cy5 channel, whereas a genotype that is heterozygous will display a signal in both channels. The intensity data were loaded in GenomeStudio, and cluster positions were assigned to all genotypes at each SNP and these were checked manually for errors and rescored while designating homozygous and heterozygous clusters. Further, the call rates and the GenTrain and GenCall score generated using the GenomeStudio software were analysed for SNP reliability. The GenTrain and GenCall50 score of ≥0.4 and the call rate of ≥95% were used as the Illumina recommended thresholds for declaring a reliable SNP.
2.4. Construction of the genetic linkage map
The inter-specific reference mapping population consisting of 129 RILs described above was used for genotyping and map generation. For linkage analysis, the GoldenGate genotyping data of the successful SNPs were utilized. In addition to this, the genotyping data of 368 polymorphic markers (52 EST-SSRs, 51 ITPs, 25 ESTPs, 2 MtESTs, 130 gSSRs and 108 STMS; Table 1) recently mapped by Choudhary et al.48 using the same reference mapping population were also combined with the SNP genotyping data for anchoring and integrating the previously mapped co-dominant markers. This complete marker data set was employed for linkage analysis using Joinmap ver. 4.0.67 A χ2 test (P < 0.05) was performed for the conformity to the expected Mendelian segregation ratio of 1:1. Markers showing segregation distortion were also integrated into the map. Markers were ordered with the regression mapping algorithm and were classified into LGs using the grouping module at LOD thresholds of 8.0–10.0 at an increment of 0.5. Linkage groups were determined at LOD 9.5 with a recombination frequency smaller than 0.49 and a maximum threshold value of 5 for the jump. The best marker order was determined using the ‘Ripple’ function (value of 1). Recombination frequencies were converted to map distances in centiMorgans (cM) using the Kosambi mapping function.68
Table 1.
Markers utilized for construction of the inter-specific linkage map of chickpea (C. arietinum ICC4958 × C. reticulatum PI489777)
| Markers analysed | Polymorphic markers used for mapping (%) | Markers mapped | |
|---|---|---|---|
| SNPs (from this study) | 768 | 697 (90.75) | 696 |
| EST-SSRs48 | 185 | 52 (28.1) | 51 |
| ITPs48 | 151 | 51 (33.8) | 51 |
| ESTPs48 | 109 | 25 (22.9) | 25 |
| MtESTs90 | 15 | 2 (13.3) | 2 |
| gSSRs48 | 310 | 130 (41.9) | 130 |
| STMS markers48,58 | 108 | 108 | 108 |
| Total | 1646 | 1065 (64.7) | 1063 |
2.5. Synteny
Synteny analysis of chickpea was carried out using genomic (gSSRs and SNPs) and genic markers (EST-SSRs, ITPs and ESTPs) from the current chickpea map. Marker sequences were aligned against the chromosome-based assembly of Medicago HapMap Mt 3.5 database [Medicago truncatula HapMap Project 2010 (http://www.medicagohapmap.org/downloads.php)] as well as with Glycine max, JGI Glyma1 [Soybean Genome Project (http://www.phytozome.net/soybean)] using local Blastn with an E-value of <1e−05. Synteny was visualized using MapChart 2.2 (Voorrips 2002). All the Blastn individual hits corresponding to markers from the chickpea LGs (CaLG1-8) were parsed into the Microsoft Excel and the whole genome dot-blots were developed by using the physical location (Mb) of M. truncatula and G. max sequences and the genetic location (cM) of chickpea sequences.
3. Results
3.1. SNP identification
SNPs were identified from the whole genome sequencing of the two parents of the chickpea reference mapping population namely C. arietinum ICC4958 and its wild progenitor C. reticulatum PI489777. Nuclear genomes of the parents were sequenced to various depths using the NGS platforms, i.e. the Roche 454 GSFLX Titanium and the Applied Biosystem's SOLiD platforms and compared for SNP identification. WGS libraries (three) of C. arietinum cv. ICC4958 nuclear DNA were used for 15 runs generating a total of 20.50 million filtered reads with 7.674 Gb high-quality (Phred score 20) bases. All the filtered reads generated by the 454 GS FLX platform were assembled by the de novo assembly tool Newbler (GS de novo assembler, Roche Applied Sciences) version 2.3 to obtain the primary assembly. The assembly parameters used were seed step 12, minimum overlap 40, minimum overlap identity 95% and seed length 16, with default quality filter, adapter and primer trimming of the input reads. The assembly yielded a total length of 350 724 940 bases in 160 883 large contigs (≥500 bases) of average size 2180 bp and the N50 value of 3541 bp with 98.66% of the bases having quality of Q40 or more. Of these, 1000 largest contigs ranging in size from 36 to 13 kb were chosen as the reference sequence for drawing out the single-nucleotide changes. Further, 3 kb paired end libraries of both the parents (C. arietinum ICC4958 and C. reticulatum PI489777) were sequenced using the SOLiD (Applied Biosystems) platform to generate 28 Gb (37.8X) and 12 Gb (16.2X) high-quality paired short reads (50 bases), respectively. SNPs in C. reticulatum PI489777 were identified using the SOLiD reads. First, the SOLiD reads of ICC4958 were aligned on the previously described ICC4958 reference sequence by the Bioscope module of the tool HAPS (http://solidsoftwaretools.com/gf/project/haps/). The mapping output file of only the properly paired reads was then utilized by the SST scaffolding module of HAPS to produce a SOLiD read-based consensus sequence of ICC4958. Next, the SOLiD reads of C. reticulatum PI489777 were mapped onto the consensus sequence for SNP identification. Stringent criteria were used for SNP calling and only those SNPs were selected that had identical SNPs of 5X coverage and 0.6 confidence score and whose flanking regions were suitable for designing allele-specific and locus-specific primers for the Illumina GoldenGate Assay. These included only those SNPs which did not have other SNPs or indels in the 60-bp regions flanking it. Hence, a set of 920 candidate SNP loci were selected from the same number of contigs and were designated as CaSNPs. Additionally, by amplicon resequencing of 222 chickpea EST loci from C. arietinum and C. reticulatum, 102 EST-derived SNPs (designated as ESNPs) were identified for genotyping as described in Choudhary et al.48 Thus, from both the approaches, a total of 1022 SNPs (920 CaSNPs + 102 ESNPs) were identified for genotyping using the GoldenGate assay. All the 1022 SNPs and their flanking sequences are available in Supplementary Table S1. Moreover, the 1022 identified SNPs were classified as transitions (C/T and G/A) or transversions (C/G, T/A, A/C and G/T) based on nucleotide substitutions. In this analysis, higher rates of transition (almost double) in comparison with the rate of transversions were found, since 650 (63.65%) transitions were found in comparison with 372 (36.4%) transversions. Among the transitions, the [C/T] type was slightly more prevalent (334, 51.4%) than [G/A] that accounted for 316 (48.6%) of single base changes, whereas among transversions, T/A scored relatively higher (123) in comparison with other transversions.
3.2. SNP genotyping
The 1022 SNPs were submitted to Illumina for the Oligo Pool All (OPA) design for use in the GoldenGate assay. Each of the individual SNPs was assigned an ADT designability score by Illumina, ranging from 0 to 1, where a score <0.4 predicted a low success rate, 0.4 to 0.6 a moderate success rate and >0.6 a high success rate for the conversion of an SNP into a successful GoldenGate assay. In all, 944 SNPs (851 CaSNPs and 93 ESNPs) were assigned designability scores ≥0.6, thereby resulting in the possibility of conversion of 92.37% of the predicted SNPs into successful GoldenGate assays. However, for convenience of customizing the array design, the 768 SNPs with the highest scores (≥0.76) were selected for inclusion in the first chickpea 768-OPA, named as CpOPA-I (Supplementary Table 1). This CpOPA-I assay was utilized for genotyping of the 129 RIL individuals along with the parental lines of the reference mapping population.
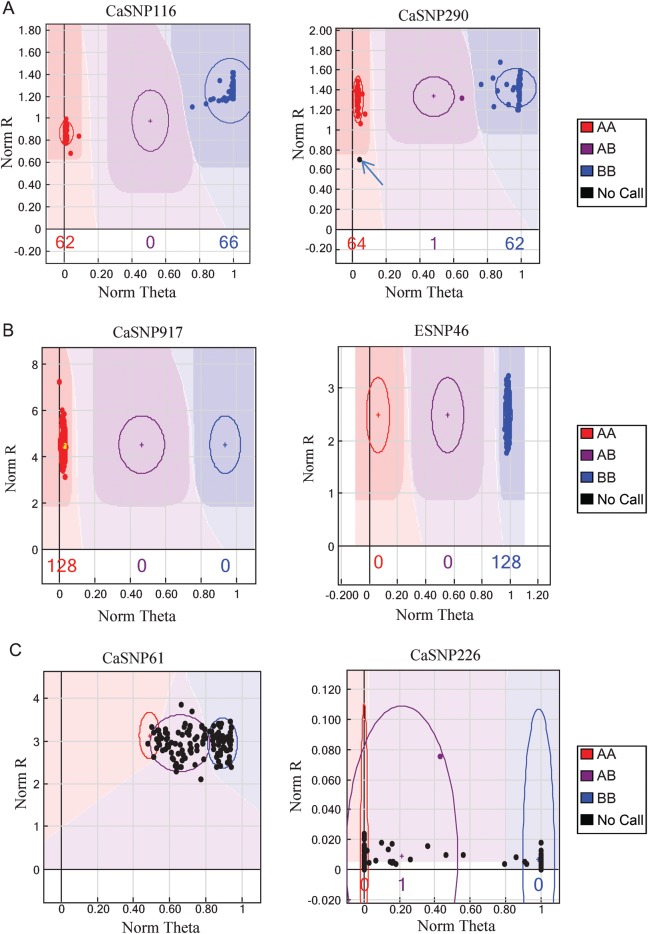
The genotyping data of the 768 SNPs across the 129 RILs were analysed using the GenomeStudio software (Illumina), which clusters and calls the data automatically, allowing visualization of the data directly for downstream analysis. For each SNP, the genotyping data representation included three main clusters corresponding to AA homozygote, AB heterozygote and BB homozygote. As in the present study, an F9-F10 RIL mapping population was used, which is expected to contribute very few heterozygotes, most of the SNP markers produced two main clusters representing the two homozygous genotypes, with sometimes a small additional cluster corresponding to heterozygous genotypes (Fig. 1A). A few data points were sometimes ambiguously located outside these clusters (indicated by arrows in Fig. 1A) and represented those for which no calls were generated and were therefore scored as missing data. In our genotyping data set, the average level of heterozygosity was 6.5%, expected for a RIL population, whereas the level of missing data per marker averaged at 5.6%.
Figure 1.
Representative clustering patterns generated by the Illumina GoldenGate SNP Genotyping assay. For a given SNP marker, genotypes are called for each sample (dots) by their normalized signal intensity (Norm R, y-axis), i.e. sum of intensities of two fluorescent signals, and allele frequency (Norm theta, x-axis) relative to a cluster position (shaded area). The data point colour codes represent: red, AA (homozygous); blue, BB (homozygous); purple, AB (heterozygous); black, no call (missing data). (A) High-quality SNPs (e.g. CaSNP116 and CaSNP290) showing well-separated clusters of homozygous alleles (red and blue) and heterozygotes (purple). Some data points located between or in the border of these clusters (marked by an arrow) are unsuccessfully genotyped samples for which no calls were generated and considered as missing data. (B) SNPs which were considered as false or monomorphic (failed to detect an SNP in the parents and the mapping population) that grouped into a single cluster (e.g. CaSNP917 and ESNP46). (C) Technically unsatisfactory SNPs (e.g. CaSNP61 and CaSNP226) represented by insufficient allele cluster separation.
In the present study, of the 768 assays, 724 SNPs generated clearly defined, well-separated clusters (as in Fig. 1A), whereas the remaining 44 SNPs did not produce clear clustering patterns and were excluded from the linkage analysis. These consisted of 28 false or monomorphic SNPs (failed to detect an SNP in the parents and the mapping population) that grouped into a single cluster (Fig. 1B), and 16 SNPs that were classified as technically unsuccessful, since they had the GenTrain and GenCall50 score of <0.40, ‘no-call’ frequencies >5% and were represented by insufficient allele cluster separation (Fig. 1C). Further, of the 724 SNP assays that showed clear clustering, 27 were found to be heterozygous in at least one of the parents and hence were excluded from linkage analysis. Therefore in all, 697 SNPs (645 CaSNPs and 52 ESNPs) out of 768 SNPs were found to be successful, demonstrating a success rate of 90.75% for the Illumina GGGT in chickpea.
3.3. SNP mapping
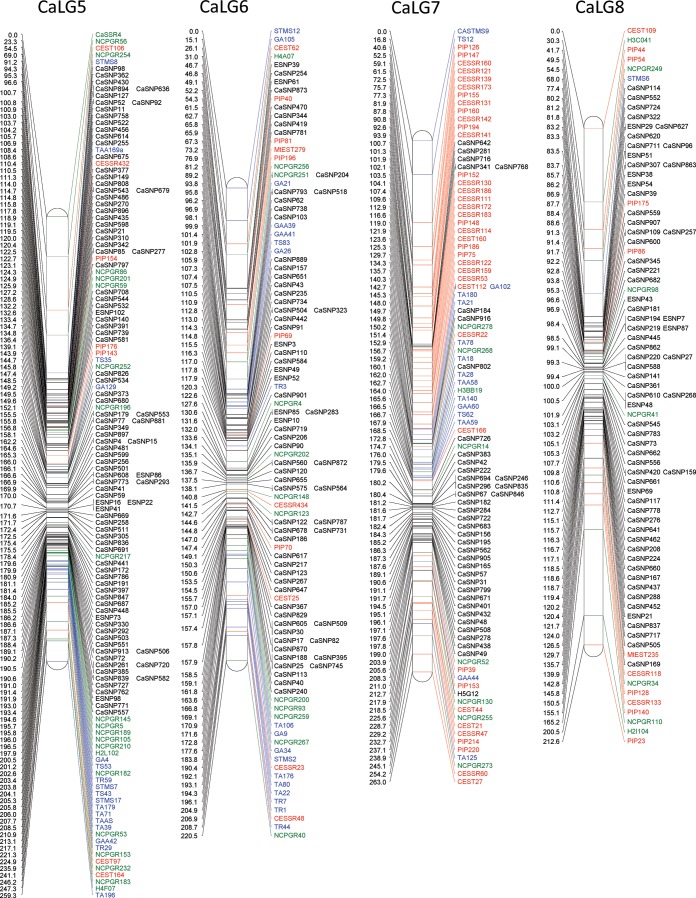
For construction of a dense genetic map of chickpea, the genotyping data of 697 polymorphic SNPs obtained from 129 RILs of the reference inter-specific mapping population (C. arietinum ICC4958 x C. reticulatum PI489777) were utilized. Further, to generate an integrated and advanced genetic map of chickpea, the genotyping data of the 697 SNPs were compiled along with the genotyping data of 368 co-dominant markers published previously by our group.48 These markers included 238 gSSRs (genomic SSRs), 52 EST-SSRs, 51 ITPs (Intron Targeted Polymorphism), 25 ESTPs and 2 MtESTs (Table 1), which served as a framework for anchoring the SNP loci. Hence, data of 1065 markers (697 SNPs and 368 previously mapped co-dominant markers) were compiled together for linkage analysis and map generation using JoinMap ver. 4.0.67 The resulting integrated linkage map of chickpea defined map positions of 1063 markers distributed over 8 linkage groups (Fig. 2). The current map spanned 1808.7 cM with an average inter-marker distance of 1.7 cM. The map contained an average of 1.43 map positions per Mb of genome (1063 map positions/740 Mb) and had one marker per 696 kb (740 Mb/1063) when considering the chickpea genome to be 740 Mb.44 The LGs were numbered (1 to 8) based on the common marker positions shared between corresponding LGs from previous studies.48,58 The genetic length of the LGs ranged from 186.8 cM (LG2) to 269.5 cM (LG1) (Table 2). LG3 was the most saturated, having 205 markers with an average marker density of 0.95 cM, whereas LG8 had the least number of markers (only 84) (Table 2). On an average, one linkage group contained 132.9 markers that spanned an average of 226 cM. All categories of markers including genic, genomic and SNPs were found on each of the LGs. However, the number of SNP markers in each of the LGs was maximum and varied in the range 65–75% except in LG7, which had only 38% SNP markers. The markers were unevenly and non-randomly distributed in the LGs and 25 clusters of markers (≥5 markers within 1 cM distance) were identified of which 20 clusters comprise only SNP markers, whereas a single cluster on LG3 contained only STMS markers. Moreover, nine gaps ranging from 20 to 35 cM were observed in four LGs (LG1, LG5, LG7 and LG8) located near the distal ends. The χ2 test performed resulted in segregation distortion for 42% (448) of the marker loci. However, 446 of these markers were finally integrated into the map so that the loss of genetic information related to these markers was minimized. Moreover, the distorted markers were found to be widely distributed throughout all the LGs even though the ratios varied from one LG to another. For example in LG1 and LG4, <25% markers showed distortion, whereas in others >30% of the mapped loci were distorted. The overall segregation distortion of 42% observed in this study was similar to the 41.3 and 38% reported earlier for the same mapping population.48,58
Figure 2.
Inter-specific linkage map of chickpea. The inter-specific linkage map of chickpea based on RILs of C. arietinum (ICC4958) × C. reticulatum (PI489777) was generated using JoinMap version 4.0. The name of the linkage groups is mentioned at the top of each LG. Distances of the loci (cM) are shown to the left and the names of loci are shown to the right side of the linkage groups. SNP markers are represented in black, while previously published markers are shown in colour: red, genic markers; green, genomic markers; blue, STMS markers of Winter et al.58 used for anchoring.
Table 2.
Distribution of markers on the eight linkage groups
| LGs | Length (cM) | Number of mapped markers | Average marker density (cM) | SNP markers (% of total) | Markers showing distortion (%) | Other markers |
|---|---|---|---|---|---|---|
| 1 | 269.5 | 119 | 2.26 | 89 (74.8) | 20.2 | 30 |
| 2 | 186.8 | 138 | 1.35 | 87 (63.0) | 56.5 | 51 |
| 3 | 196.6 | 205 | 0.95 | 134 (65.4) | 45.4 | 71 |
| 4 | 200.4 | 168 | 1.19 | 120 (71.4) | 9.5 | 48 |
| 5 | 259.3 | 141 | 1.83 | 96 (68.0) | 82.3 | 45 |
| 6 | 220.5 | 108 | 2.04 | 67 (62.0) | 50.9 | 41 |
| 7 | 263.0 | 100 | 2.63 | 38 (38.0) | 38.0 | 62 |
| 8 | 212.6 | 84 | 2.53 | 65 (77.4) | 30.9 | 19 |
| Total | 1808.7 | 1063 | 1.70 | 696 (65.48) | 42.0 | 367 |
3.4. Synteny between chickpea and other legumes
To identify the syntenic relationships between chickpea and other legume genomes, namely M. truncatula and G. max, the sequence information available for 917 of 1063 mapped loci on the current linkage map of chickpea was utilized (Table 3). These markers included 696 SNPs, 51 EST-SSRs, 94 gSSRs, 51 ITPs and 25 ESTPs for which locus sequences were available. The details about the markers on each of the chickpea linkage groups (CaLGs) that showed hits with chromosomes of G. max and M. truncatula are described in Table 3.
Table 3.
Details of CaLGs, number of markers and synteny with M. truncatula and G. max chromosomes
| Chickpea LGs | Markers | Markers selected for blast | Mt chromosome (chickpea orthologs) | Gm chromosome (chickpea orthologs) |
|---|---|---|---|---|
| CaLG1 | 119 | 107 | Mt2(35) Mt4(3) Mt1(2) Mt5(2) Mt7(2) Mt3(1) Mt6(1) Mt8(1) | Gm15(19) Gm13(17) Gm9(4) Gm8(3) Gm17(3) Gm3(2) Gm10(2) Gm11(2) Gm20(2) Gm1(1) Gm4(1) Gm5(1) Gm6(1) Gm7(1) Gm14(1) Gm19(1) |
| CaLG2 | 138 | 115 | Mt4(19) Mt5(9) Mt3(5) Mt6(4) Mt2(2) Mt7(2) Mt8(2) | Gm12(10) Gm11(7) Gm6(4) Gm9(4) Gm15(4) Gm18(4) Gm1(3) Gm5(3) Gm13(3) Gm20(3) Gm4(2) Gm7(2) Gm10(2) Gm14(2) Gm19(2) Gm3(1) |
| CaLG3 | 205 | 179 | Mt7(35) Mt5(23) Mt3(7) Mt2(6) Mt6(6) Mt8(4) Mt4(2) Mt1(3) | Gm19(29) Gm3(26) Gm1(12) Gm9(8) Gm10(7) Gm6(2) Gm7(6) Gm11(6) Gm13(5) Gm2(4) Gm5(4) Gm18(4) Gm20(4) Gm4(3) Gm6(2) Gm8(2) Gm12(2) Gm15(2) Gm16(2) Gm14(1) Gm17(1) |
| CaLG4 | 168 | 152 | Mt1(31) Mt5(5) Mt3(3) Mt4(3) Mt7(2) Mt6(1) | Gm10(18) Gm20(11) Gm17(9) Gm1(7) Gm14(7) Gm8(5) Gm11(5) Gm12(5) Gm3(4) Gm9(4) Gm18(4) Gm5(3) Gm6(2) Gm7(2) Gm13(2) Gm15(2) Gm2(1) Gm4(1) Gm19(1) |
| CaLG5 | 141 | 119 | Mt3(34) Mt1(5) Mt7(5) Mt2(4) Mt5(4) Mt4(2) Mt6(1) Mt8(1) | Gm6(19) Gm4(18) Gm20(7) Gm14(6) Gm18(6) Gm13(5) Gm11(3)Gm1(2) Gm2(2) Gm8(2) Gm17(2)Gm3(1) Gm5(1) Gm7(1) Gm19(1) |
| CaLG6 | 108 | 86 | Mt8(15) Mt5(7) Mt7(5) Mt2(4) Mt3(4) Mt4(3) Mt6(3) Mt1(2) | Gm8(11) Gm5(9) Gm13(4) Gm18(4) Gm20(4) Gm6(3) Gm7(3) Gm11(3) Gm1(1) Gm2(1) Gm3(1) Gm10(1) Gm12(1) Gm14(1) Gm16(1) Gm17(1) Gm19(1) |
| CaLG7 | 100 | 81 | Mt8(13) Mt3(6) Mt2(5) Mt5(4) Mt7(4) Mt1(3) Mt4(3) Mt6(1) | Gm7(6) Gm14(6) Gm7(6) Gm14(6) Gm5(4) Gm6(4) Gm16(4) Gm17(4) Gm1(3) Gm2(3) Gm8(3) Gm11(3) Gm13(3) Gm18(3) Gm4(2) Gm9(2) Gm19(2) Gm12(1) Gm15(1) Gm20(1) |
| CaLG8 | 84 | 78 | Mt4(22) Mt2(4) Mt6(4) Mt1(3) Mt3(3) Mt7(3) Mt8(3) Mt5(2) | Gm17(20) Gm7(8) Gm13(8) Gm5(6) Gm6(6) Gm4(5) Gm9(4) Gm15(4) Gm10(3) Gm14(3) Gm18(3) Gm19(3) Gm3(2) Gm11(2) Gm12(2) Gm16(2) Gm20(2) Gm8(1) |
| Total | 1063 | 917 | 404 | 609 |
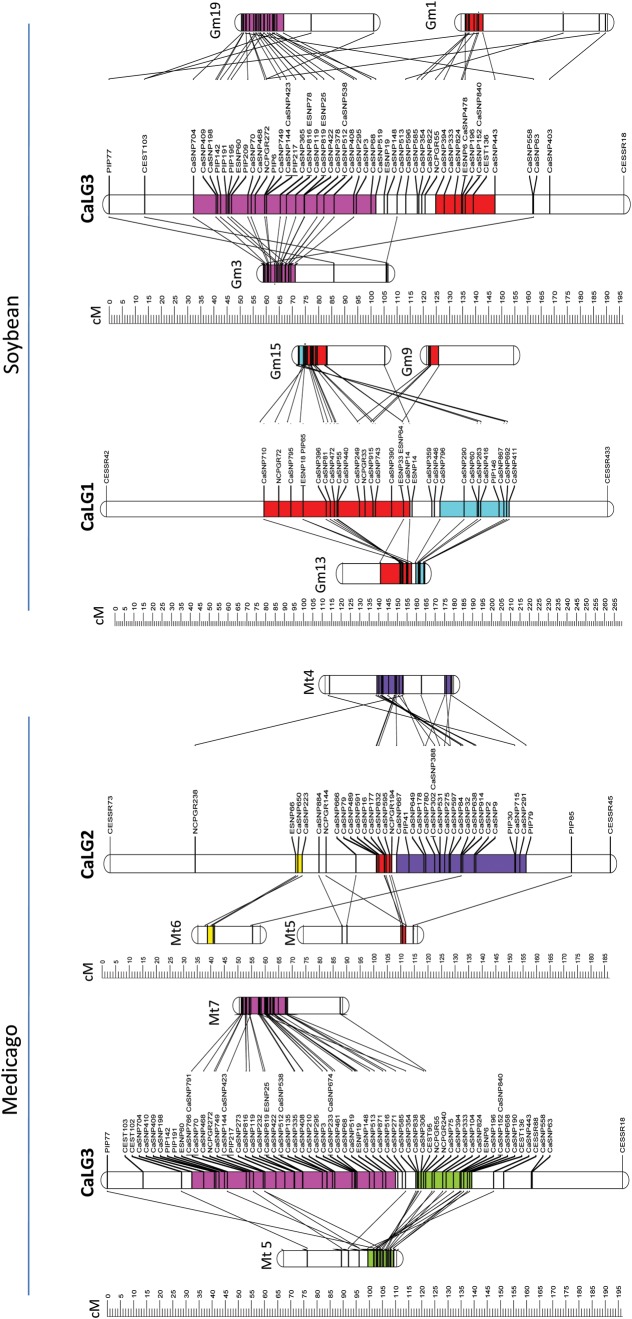
Chickpea-Medicago synteny. The chickpea-Medicago synteny was observed for 315 (34.35%) of 917 markers for which 404 significant matches were obtained on different chromosomes of Medicago. Synteny blocks were observed between chickpea and most Medicago chromosomes except Mt6 (Supplementary Fig. S1), indicating an evolutionary relationship between the two species. Many of the CaLGs were found to have syntenic blocks with more than one LG of Medicago. CaLG3 showed maximum synteny, since 68 markers showed 86 hits with Medicago chromosomes (Table 3), majority of which (35 and 23) were located on Mt7 and Mt5, respectively (Fig. 3). Similarly, synteny blocks from CaLG2 were observed on Mt4, Mt5 and Mt6 (Fig. 3). Moreover, CaLG1, CaLG4 and CaLG5 showed distinct synteny with Mt2, Mt1 and Mt3 chromosomes, respectively (Supplementary Figs. S1 and S2). Additionally, CaLG6 and CaLG7 were seen to be highly syntenic with Mt8, whereas CaLG8 was found to be syntenic with Mt4 (Supplementary Figs. S1 and S2). The number of markers that found hits in Medicago in comparison with soybean was more; however, very limited collinearity was observed between the chickpea-Medicago syntenic blocks.
Figure 3.
Syntenic relationships of chickpea LGs: CaLG2 with Mt4, Mt5 and Mt6; CaLG3 with Mt5 and Mt7; CaLG1 with Gm9, Gm13 and Gm15; CaLG3 with Gm1, Gm3 and Gm19. Coloured regions represent syntenic blocks observed in chromosomes of Medicago and soybean.
Chickpea-soybean synteny. In chickpea-soybean synteny analysis, 272 (30%) markers found a larger number of hits (609) in the soybean genome (Table 3) in comparison with Medicago. However, regions with macrosynteny, microsynteny and collinearity were observed. All CaLGs showed syntenic blocks with Gm chromosomes except CaLG7 (Supplementary Fig. S1). In the case of soybean also (as with Medicago), CaLG3 (54 markers) found maximum hits (129) in the soybean genome, of which majority were found on Gm19, Gm3 and Gm1 (Table 3; Supplementary Fig. S3). All CaLGs (except CaLG7) showed multiple synteny blocks with the Gm chromosomes (Supplementary Fig. S1). CaLG5 showed clear correspondence with regions in Gm4 and Gm6 in which the marker order was almost collinear. High collinearity was also observed between CaLG1 with Gm13, Gm15 and Gm9 (Fig. 3), followed by LG4 with Gm17. Similarly, collinear regions were also identified between CaLG6 and Gm5 and Gm8 (Supplementary Fig. S1).
4. Discussion
The present study of the chickpea genome reports the large-scale identification of SNPs from the NGS of two contrasting genotypes, which were parents of an RIL mapping population. The development and validation of the high-throughput GoldenGate SNP genotyping assay was demonstrated for the first time in chickpea with a very high success rate of 90.75% (since 697 of 768 SNPs were genotyped). Further, these SNPs were assigned map locations in the backdrop of other co-dominant markers to generate one of the most dense genetic linkage maps of chickpea, thereby clearly establishing the quality of the data and the utility of this approach. This study served to expand the repertoire of genomic resources in chickpea by providing the first set of 696 validated and mapped SNP markers, which are the most preferred next generation markers for molecular breeding, association studies and comparative genomics. Recently, there has been a spurt in the large-scale discovery of SNPs in plants, especially after the public availability of EST databases and more recently with the continuously expanding NGS of genomes and transcriptomes. SNP identification in plants range from a few thousand such as 1114 in wheat,15 2700 in Arabidopsis,69 5000 in tomato,40 6626 in pine,16 10 000 in cowpea,13 9194–36 000 in maize,17,42 22 000 in barley,32 25 000–39 022 in soybean,18,70 40 000 in Medicago19 and 67 051 in rice,71 to millions in Arabidopsis (2.5 million),72 rice (2.4–6.5 million),7,73 Medicago (3 million)38 and soybean (6.5 million).74 The thousands of SNPs generated are being extensively used for various applications such as the analysis of genetic diversity,22,74 map construction13,15,32,40 and orienting the scaffolds in whole genome sequencing projects.18,43
SNP discovery from de novo sequencing of genomes is a challenge especially in the absence of reference genomes. In the case of chickpea, since no reference genome was available, in the present study, SNPs were deduced by sequence comparisons between chickpea and its wild relative, C. reticulatum, with the aim of producing a highly useful set of molecular markers, which would be directly used for constructing a dense linkage map of chickpea. Consensus calling was done by de novo assembly of the Roche/454 long reads of C. arietinum ICC4958 to generate the reference sequence, onto which the ICC4958 SOLiD short reads were mapped to generate a consensus sequence. Next, SNP calling was done by mapping the SOLiD reads of the contrasting genotype, C. reticulatum PI489777, onto the consensus sequence. Moreover, in order to custom design a genotyping array consisting of SNPs well-distributed throughout the genome, a predetermined number of SNPs were selected from the largest sized contigs. This array proved useful for establishing the efficacy of the SNP genotyping strategy for high-resolution mapping, which is mandatory for MAS, association analysis and anchoring the chickpea genome.
Many factors may be responsible for the overall success of SNP genotyping. Firstly, SNP prediction from NGS data should be based on stringent parameters. When calling variants (especially from short reads), the quantity of reads and the accuracy of mapping the reads should be precise. This was ensured in our data since we used stringent criteria for SNP calling such as base quality ≥20, minimum read depth of 5X in both genotypes and frequency score >0.6. Secondly, the nucleotides flanking the query SNP also influence the rate of success of SNP development.16,23 In our study also, care was taken to select only those SNPs that had no variations in the 60 bases flanking it. Moreover, the Illumina ADT which provides designability scores, checks for the presence of repetitive and palindromic sequences, GC content and sequence polymorphisms in the region flanking the query SNP. High designability scores were obtained for the 920 predicted chickpea SNPs of which 851 (92.5%) had an ADT designability score ≥0.6 indicating a high success rate for the conversion of an NGS-derived SNP into a GGGT assay. Even after combining with the 102 ESNPs, designability scores ≥0.6 were obtained for 92.37% of the predicted SNPs. From these, since only 768 SNPs had to be selected for the OPA design, those having scores ≥0.76 (much higher than the threshold values) were selected for the 768-CpOPA-I. Inclusion of SNPs with such high designability scores accounted for the overall success of the genotyping assay as has also been demonstrated in pine.16 Moreover, 48 CaSNPs predicted from the NGS data were experimentally validated by resequencing the alleles of the two parental varieties and a success rate of 87.5% was obtained (data not shown).
In the present study, we also demonstrated that the high-throughput GoldenGate technology could be successfully used for genotyping predicted SNPs by conversion into working assays. This high-throughput platform of Illumina, which ensures a high level of multiplexing and reliability, has been suitably used in plants,12,13,16,32,33,75,76 where large numbers of SNPs need to be surveyed. In the present chickpea OPA, the success rate of 90.75% was obtained, which was comparable with the success rates of 89–92% reported in the previous studies in other crops such as barley,75 soybean,31 cowpea,13 maize33 and eucalyptus,20 and was higher than those reported in spruce12 and pine.16,34 The SNP success rate may be assessed in various ways and different SNP reliability thresholds may be used. The SNP success rate of 90.7% reported for chickpea was based on the percentage of successfully genotyped SNPs. However, if the criteria of the call rate (>95%) were used, the success rate of 99.8% was achieved. The success rate was 93.0% when considering the GenTrain and GenCall50 score of ≥0.4 as thresholds, as described in Grattapaglia et al.20
We present here the first SNP-based linkage map of chickpea constructed using 697 SNPs in combination with 368 previously mapped co-dominant markers including a large number of SSRs (Fig. 2). In the past few years, SSR markers have been the most preferred markers for constructing linkage maps due to their hypervariability, co-dominant nature and reproducibility, and SSR-based maps have been constructed in many plants such as wheat,77 barley,78 tobacco,79 tomato80 and Catharanthus,81 and legumes like chickpea46,48 and soybean.82 However, SSRs were found to have limitations, since they were not sufficiently dense to provide the genomic coverage necessary for the dissection of complex traits. Moreover, automation in high-throughput genotyping of SSRs was not very common. Therefore, SNPs are now proposed as the most useful markers, especially for the construction of highly saturated maps, which may have very wide and novel applications in plant genomics. Hence, modern sequencing technologies are being used to generate thousands to millions of SNPs in many plant species as mentioned previously. However, when it comes to the validation of SNPs and their use in genotyping and map generation, there are only a few examples of SNP-based high-density maps. The map of chickpea generated in this study contained 697 SNPs—much higher than the 357 in pine,16 480 in wheat15 and 558 in grass,41 but lower than the 793 in tomato,40 928 in cowpea,13 1397 in soybean31 and 2943 in barley.32 In addition to the SNPs, 368 previously mapped co-dominant markers were positioned in this chickpea map thereby generating one of the most advanced integrated maps of chickpea comprising of 1063 markers with an average marker density of 1.7 cM. This map was more saturated in comparison to other published maps of chickpea such as those of Winter et al.58 (2077.9 cM: 303 markers: AMD of 6.8 cM), Nayak et al.45 (2602.1 cM: 521 markers: AMD of 4.99 cM), Gujaria et al.47 (766.56 cM: 300 markers: AMD of 2.552 cM) and Choudhary et al.48 (1497.7 cM; 406 markers; AMD of 3.68). A distinguishing feature of this map was that it was constructed based on a single reference population (not on integration of maps from different populations), utilizing actual segregation data of each of the genomic and genic markers listed in Table 1. Consensus maps are generated in order to increase the number of mapped markers, but may compromise on the accuracy of the marker order. Hence our map may be considered as more accurate regarding the marker order on the eight linkage groups. This was confirmed as considerable conservation of marker distribution existed between the LGs of the current map and our previous map48 as well as the anchor map of Winter et al.58 Moreover, this map was a major advancement over previous maps based on co-dominant markers,45,47,48 as the mapped markers were more than doubled in the current map. Additionally, nine genic markers (four EST-SSRs, two ITPs and three ESTPs), which remained unmapped in our previous inter-specific map, were also mapped onto LG2, LG3, LG5, LG6, LG7 and LG8 in the current map. The average marker density in each of the LGs in the current map was also found to be increased (0.95–2.65) in comparison with the density reported earlier (1.77–8.01).48 Therefore, it was envisaged that this highly advanced map would not only be useful in chickpea breeding but would particularly aid and complement the efforts of orienting scaffolds of chickpea whole genome sequence being generated during its de novo sequencing at the institute. High-resolution genetic maps are critical for the assembly of sequence scaffolds into pseudomolecules corresponding to chromosomes of organisms and have been used for anchoring and orienting scaffolds in the soybean,18 watermelon,43 grape,83 cucumber84 and apple85 whole genome sequencing.
Genomic synteny can facilitate the transfer of genetic information especially between orphan and model crops. In legumes, map-based synteny has been shown in several studies such as cowpea,13 Medicago,86 peanut,87 common bean88 and asparagus bean.89 However, due to the lack of genomic resources, information on genome organization and comparative analysis was limited in chickpea. In this study, comparative analysis of chickpea using genic and genomic marker sequences with the model legume Medicago and soybean was done, which revealed higher levels of synteny with Medicago in comparison with soybean. This result indicated that the degree of synteny declines with increasing phylogenetic distance, as chickpea and Medicago both belong to the Galegoid clade, whereas soybean belongs to the Phaseoloid clade. Chromosomal rearrangements seemed to have occurred since most of the chickpea LGs were chimeric to two or more Gm chromosomal segments. Moreover, some segments of chickpea such as those on CaLG3 were found to have blocks on two Gm chromosomes (Gm3 and Gm19), thereby indicating the phenomenon of genome duplication.
In conclusion, the present study illustrates the application of NGS technology for the large-scale identification of SNPs from species without reference genomes such as chickpea. It also demonstrates that the high-throughput genotyping technologies such as the Illumina GoldenGate assay can be successfully used for genotyping and subsequently mapping the SNPs and integrating them with other co-dominant markers for the construction of a dense genetic map of chickpea. The availability of such large numbers of SNP markers, advanced genotyping technologies and the high-density marker map would serve as a foundation not only for orienting and anchoring of scaffolds arising out of whole genome sequencing projects but also for association mapping and synteny-based genomics in chickpea and other legumes.
Supplementary Data
Supplementary data are available online at www.dnaresearch.oxfordjournals.org.
Funding
We gratefully acknowledge the funding support by the Department of Biotechnology (DBT), Government of India, through their research grant (BT/PR12919/AGR/02/676/2009) for this work.
Supplementary Material
Acknowledgements
We are thankful to Dr Fred Muehlbauer, Washington State University, USA, for providing the inter-specific chickpea mapping population and genotypic data of published STMS markers.
Footnotes
Edited by Satoshi Tabata
References
- 1.Krawczak M. Informativity assessment for biallelic single nucleotide polymorphisms. Electrophoresis. 1999;20:1676–81. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1676::AID-ELPS1676>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 2.Xing C., Schumacher F.R., Xing G., Lu Q., Wang T., Elston R.C. Comparison of microsatellites, single-nucleotide polymorphisms (SNPs) and composite markers derived from SNPs in linkage analysis. BMC Genet. 2005;6:S29. doi: 10.1186/1471-2156-6-S1-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper D.N., Smith B.A., Cooke H.J., Niemann S., Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genome. Hum. Genet. 1985;69:201–5. doi: 10.1007/BF00293024. [DOI] [PubMed] [Google Scholar]
- 4.Wang D.G., Fan J.-B., Siao C.-J., et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–82. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 5.Kulheim C., Yeoh S.H., Maintz J., Foley W.J., Moran G.F. Comparative SNP diversity among four Eucalyptus species for genes from secondary metabolite biosynthetic pathways. BMC Genomics. 2009;24:452. doi: 10.1186/1471-2164-10-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F., Hasegawa Y., Saito M., et al. Extensive chromosome homoeology among Brassiceae species were revealed by comparative genetic mapping with high-density EST-based SNP markers in radish (Raphanus sativus L.) DNA Res. 2011;18:401–11. doi: 10.1093/dnares/dsr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subbaiyan G.K., Waters D.L.E., Katiyar S.K., Sadananda A.R., Vaddadi S., Henry R.J. Genome-wide DNA polymorphisms in elite indica rice inbreds discovered by whole-genome sequencing. Plant Biotech. J. 2012:1–12. doi: 10.1111/j.1467-7652.2011.00676.x. doi:10.1111/j.1467-7652.2011.00676.x. [DOI] [PubMed] [Google Scholar]
- 8.Gore M.A., Wright M.H., Ersoz E.S., et al. Large-scale discovery of gene-enriched SNPs. Plant Genome. 2009;2:121–33. [Google Scholar]
- 9.Choi I.-Y., Hyten D.L., Matukumalli L.K., et al. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics. 2007;176:685–96. doi: 10.1534/genetics.107.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atwell S., Huang Y.S., Vilhjalmsson B.J., et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–31. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troggio M., Malacarne G., Coppola G., et al. A dense single-nucleotide polymorphism-based genetic linkage map of grapevine (Vitis vinifera L.) anchoring Pinot Noir bacterial artificial chromosome contigs. Genetics. 2007;176:2637–50. doi: 10.1534/genetics.106.067462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavy N., Pelgas B., Beauseigle S.P., et al. Enhancing genetic mapping of complex genomes through the design of highly-multiplexed SNP arrays: application to the large and unsequenced genomes of white spruce and black spruce. BMC Genomics. 2008;9:21. doi: 10.1186/1471-2164-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muchero W., Diop N.N., Bhat P.R., et al. A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proc. Natl Acad. Sci. USA. 2009;106:18159–64. doi: 10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deulvot C., Charrel H., Marty A., et al. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics. 2010;11:468. doi: 10.1186/1471-2164-11-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen A.M., Barker G.L.A., Berry S.T., et al. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.) Plant Biotech. J. 2011;9:1086–99. doi: 10.1111/j.1467-7652.2011.00628.x. [DOI] [PubMed] [Google Scholar]
- 16.Chancerel E., Lepoittevin C., Le Provost G., et al. Development and implementation of a highly multiplexed SNP array for genetic mapping in maritime pine and comparative mapping with loblolly pine. BMC Genomics. 2011;12:368. doi: 10.1186/1471-2164-12-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbazuk W.B., Emrich S.J., Chen H.D., Li L., Schnable P.S. SNP discovery via 454 transcriptome sequencing. Plant Journal. 2007;51:910–8. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyten D.L., Cannon S.B., Song Q., et al. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics. 2010;11:38. doi: 10.1186/1471-2164-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Y., Kang Y., Torres-Jerez I., et al. Genome-wide SNP discovery in tetraploid alfalfa using 454 sequencing and high resolution melting analysis. BMC Genomics. 2011;12:350. doi: 10.1186/1471-2164-12-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grattapaglia D., Silva-Junior O.B., Kirst M., Lima B.M., Faria D.A., Pappas G.J. High-throughput SNP genotyping in the highly heterozygous genome of Eucalyptus: assay success, polymorphism and transferability across species. BMC Plant Biol. 2011;11:65. doi: 10.1186/1471-2229-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arai-Kichise Y., Shiwa Y., Nagasaki H., et al. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol. 2011;52:274–82. doi: 10.1093/pcp/pcr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachlava E., Taylor C.A., Tang S., et al. SNP discovery and development of a high-density genotyping array for sunflower. PLoS One. 2012;7:e29814. doi: 10.1371/journal.pone.0029814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J.B., Oliphant A., Shen R., et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 24.Oliphant A., Barker D.L., Stuelpnagel J.R., Chee M.S. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;32:S56. [PubMed] [Google Scholar]
- 25.Fan J.B., Chee M.S., Gunderson K.L. Highly parallel genomic assays. Nat. Rev. Genet. 2006;7:632–44. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 26.Bell P.A., Chaturvedi S., Gelfand C.A., et al. SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques. 2002;32:S70–7. [PubMed] [Google Scholar]
- 27.Matsuzaki H., Loi H., Dong S., et al. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–25. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzaki H., Dong S., Loi H., et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods. 2004;1:109–11. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 29.Nijman I.J., Kuipers S., Verheul M., Guryev V., Cuppen E. A genome-wide SNP panel for mapping and association studies in the rat. BMC Genomics. 2008;9:95. doi: 10.1186/1471-2164-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syvanen A.C. Toward genome-wide SNP genotyping. Nat. Rev. Genet. 2005;37:S5–10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 31.Hyten D.L., Song Q., Choi I.Y., Yoon M.S., Cregan P.B. High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor. Appl. Genet. 2008;116:945–52. doi: 10.1007/s00122-008-0726-2. [DOI] [PubMed] [Google Scholar]
- 32.Close T.J., Bhat P.R., Lonardi S., et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J.B., Yang X.H., Shah T., et al. High-throughput SNP genotyping with the GoldenGate assay in maize. Mol. Breed. 2010;25:441–51. [Google Scholar]
- 34.Eckert A., Pande B., Ersoz E., et al. High-throughput genotyping and mapping of single nucleotide polymorphisms in loblolly pine (Pinus taeda L.) Tree Genet. Genomes. 2009;5:225–34. [Google Scholar]
- 35.Myles S., Chia J.M., Hurwitz B., et al. Rapid genomic characterization of the genus Vitis. PLoS One. 2010;5:e8219. doi: 10.1371/journal.pone.0008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X., Wei X., Sang T., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–7. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 37.Tian F., Bradbury P.J., Brown P.J., et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011;43:159–62. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 38.Branca A., Paape T.D., Zhou P., et al. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc. Natl Acad. Sci. USA. 2011;108:E864–70. doi: 10.1073/pnas.1104032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo M.C., Deal K.R., Akhunov E.D., et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl Acad. Sci., USA. 2009;106:15780–5. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirasawa K., Isobe S., Hirakawa H., et al. SNP discovery and linkage map construction in cultivated tomato. DNA Res. 2010;17:381–91. doi: 10.1093/dnares/dsq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huo N., Garvin D.F., You F.M., et al. Comparison of a high-density genetic linkage map to genome features in the model grass Brachypodium distachyon. Theor. Appl. Genet. 2011;123:455–64. doi: 10.1007/s00122-011-1598-4. [DOI] [PubMed] [Google Scholar]
- 42.Jones E., Chu W.C., Ayele M., et al. Development of single nucleotide polymorphism (SNP) markers for use in commercial maize (Zea mays L.) germplasm. Mol. Breed. 2009;24:165–76. [Google Scholar]
- 43.Ren Y., Zhao H., Kou Q., et al. A high resolution genetic map anchoring scaffolds of the sequenced watermelon genome. PLoS One. 2012;7:e29453. doi: 10.1371/journal.pone.0029453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arumuganathan K., Earle E.D. Nuclear DNA content of some important plant species. Plant Mol. Rep. 1991;9:208–18. [Google Scholar]
- 45.Nayak S.N., Zhu H., Varghese N., et al. Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theor. Appl. Genet. 2010;120:1415–41. doi: 10.1007/s00122-010-1265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaur R., Sethy N.K., Choudhary S., Shokeen B., Gupta V., Bhatia S. Advancing the STMS genomic resources for defining new locations on the intraspecific genetic linkage map of chickpea (Cicer arietinum L.) BMC Genomics. 2011;12:117. doi: 10.1186/1471-2164-12-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gujaria N., Kumar A., Dauthal P., et al. Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.) Theor. Appl. Genet. 2011;122:1577–89. doi: 10.1007/s00122-011-1556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Choudhary S., Gaur R., Gupta S., Bhatia S. EST-derived genic molecular markers: development and utilization for generating an advanced transcript map of chickpea. Theor. Appl. Genet. 2012;124:1449–62. doi: 10.1007/s00122-012-1800-3. [DOI] [PubMed] [Google Scholar]
- 49.Buhariwalla H.K., Jayashree B., Eshwar K., Crouch J.H. Development of ESTs from chickpea roots and their use in diversity analysis of the Cicer genus. BMC Plant Biol. 2005;5:16. doi: 10.1186/1471-2229-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coram T., Pang E. Isolation and analysis of candidate Ascochyta blight defense genes in chickpea, Part I. Generation and analysis of an expressed sequence tag (EST) library. Physiol. Mol. Plant. Pathol. 2005;66:192–200. [Google Scholar]
- 51.Varshney R.K., Hiremath P.J., Lekha P., et al. A comprehensive resource of drought and salinity responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.) BMC Genomics. 2009;15:523. doi: 10.1186/1471-2164-10-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashraf N., Ghai D., Barman P., et al. Comparative analysis of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics. 2009;10:415. doi: 10.1186/1471-2164-10-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garg R., Patel R.K., Tyagi A.K., Jain M. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 2011;18:53–63. doi: 10.1093/dnares/dsq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garg R., Patel R.K., Jhanwar S., et al. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 2011;156:1661–78. doi: 10.1104/pp.111.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiremath P.J., Farmer A., Cannon S.B., et al. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotech. J. 2011;9:922–31. doi: 10.1111/j.1467-7652.2011.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hüttel B., Winter P., Weising K., Choumane W., Weigand F., Kahl G. Sequence-tagged microsatellite markers for chickpea (Cicer arietinum L.) Genome. 1999;42:210–7. [PubMed] [Google Scholar]
- 57.Winter P., Pfaff T., Udupa S.M., et al. Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol. Gen. Genet. 1999;262:90–101. doi: 10.1007/s004380051063. [DOI] [PubMed] [Google Scholar]
- 58.Winter P., Benko-Iseppon A.M., Hüttel B., et al. A linkage map of chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum times C. reticulatum cross: localization of resistance genes for Fusarium wilt races 4 and 5. Theor. Appl. Genet. 2000;101:1155–63. [Google Scholar]
- 59.Lichtenzveig J., Scheuring C., Dodge J., Abbo S., Zhang H.B. Construction of BAC and BIBAC libraries and their applications for generation of SSR markers for genome analysis of chickpea, Cicer arietinum L. Theor. Appl. Genet. 2005;110:492–510. doi: 10.1007/s00122-004-1857-8. [DOI] [PubMed] [Google Scholar]
- 60.Sethy N.K., Shokeen B., Bhatia S. Isolation and characterization of sequence tagged microsatellite sites markers in chickpea (Cicer arietinum L.) Mol. Ecol. Notes. 2003;3:428–30. [Google Scholar]
- 61.Sethy N.K., Shokeen B., Edwards K.J., Bhatia S. Development of microsatellite markers and analysis of intraspecific genetic variability in chickpea (Cicer arietinum L.) Theor. Appl. Genet. 2006;112:1416–28. doi: 10.1007/s00122-006-0243-0. [DOI] [PubMed] [Google Scholar]
- 62.Choudhary S., Sethy N.K., Shokeen B., Bhatia S. Development of sequence-tagged microsatellites site markers for chickpea (Cicer arietinum L.) Mol. Ecol. Notes. 2006;6:93–5. [Google Scholar]
- 63.Choudhary S., Sethy N.K., Shokeen B., Bhatia S. Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor. Appl. Genet. 2009;118:591–608. doi: 10.1007/s00122-008-0923-z. [DOI] [PubMed] [Google Scholar]
- 64.Millan T., Winter P., Jüngling R., et al. A consensus genetic map of chickpea (Cicer arietinum L.) based on 10 mapping populations. Euphytica. 2010;175:175–89. [Google Scholar]
- 65.Malmberg R., Messing J., Sussex I. Molecular Biology of Plants: A Laboratory Course Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1985. [Google Scholar]
- 66.Margulies M., Egholm M., Altman W.E., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Ooijen J. 2006. JoinMap version 4.0: Software for the calculation of genetic linkage maps in experimental population. Kyazma B.V., Wageningen, Netherlands. [Google Scholar]
- 68.Kosambi D. The estimation of map distances from recombination values. Ann. Eugenics. 1994;12:172–5. [Google Scholar]
- 69.Zeller G., Clark R.M., Schneeberger K., Bohlen A., Weigel D., Rätsch G. Detecting polymorphic regions in the Arabidopsis thaliana genome with resequencing microarrays. Genome Res. 2008;18:918–929. doi: 10.1101/gr.070169.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu X., Ren C., Joshi T., Vuong T., Xu D., Nguyen H.T. SNP discovery by high-throughput sequencing in soybean. BMC Genomics. 2010;11:469. doi: 10.1186/1471-2164-11-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamamoto T., Nagasaki H., Yonemaru J., et al. Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics. 2010;11:267. doi: 10.1186/1471-2164-11-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang X., Borevitz J.O. Global analysis of allele-specific expression in Arabidopsis thaliana. Genetics. 2009;182:943–54. doi: 10.1534/genetics.109.103499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu X., Liu X., Ge S., et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–11. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 74.Lam H.M., Xu X., Liu X., et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010;42:1053–9. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 75.Rostoks N., Ramsay L., MacKenzie K., et al. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc. Natl Acad. Sci. USA. 2006;103:18656–61. doi: 10.1073/pnas.0606133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sato K., Takeda K. An application of high-throughput SNP genotyping for barley genome mapping and characterization of recombinant chromosome substitution lines. Theor. Appl. Genet. 2009;119:613–9. doi: 10.1007/s00122-009-1071-9. [DOI] [PubMed] [Google Scholar]
- 77.Torado A., Koike M., Mochida K., Ogihara Y. SSR-based linkage map with new markers using an intraspecific population of common wheat. Theor. Appl. Genet. 2006;112:1042–51. doi: 10.1007/s00122-006-0206-5. [DOI] [PubMed] [Google Scholar]
- 78.Varshney R.K., Marcel T.C., Ramsay L., et al. A high density barley microsatellite consensus map with 775 SSR loci. Theor. Appl. Genet. 2007;114:1091–113. doi: 10.1007/s00122-007-0503-7. [DOI] [PubMed] [Google Scholar]
- 79.Bindler G., Plieske J., Bakaher N., et al. A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theor. Appl. Genet. 2011;123:219–30. doi: 10.1007/s00122-011-1578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shirasawa K., Asamizu E., Fukuoka H., et al. An interspecific linkage map of SSR and intronic polymorphism markers in tomato. Theor. Appl. Genet. 2010;121:731–9. doi: 10.1007/s00122-010-1344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shokeen B., Choudhary S., Sethy N.K., Bhatia S. Development of SSR and gene-targeted markers for construction of a framework linkage map of Catharanthus roseus. Ann. Bot. (Lond.) 2011;108:321–36. doi: 10.1093/aob/mcr162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hwang T., Sayama T., Takahashi M., et al. High-density integrated linkage map based on SSR markers in soybean. DNA Res. 2009;16:213–25. doi: 10.1093/dnares/dsp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jaillon O., Aury J.M., Noel B., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–7. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 84.Huang S., Li R., Zhang Z., et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–81. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 85.Velasco R., Zharkikh A., Affourtit J., et al. The genome of the domesticated apple (Malus x domestica Borkh.) Nat. Genet. 2010;42:833–9. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- 86.Choi H.K., Mun J.H., Kim D.J., et al. Estimating genome conservation between crop and model legume species. Proc. Natl Acad. Sci. USA. 2004;101:15289–94. doi: 10.1073/pnas.0402251101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertioli D.J., Moretzsohn M.C., Madsen L.H., et al. An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes. BMC Genomics. 2009;10:45. doi: 10.1186/1471-2164-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Galeano C.H., Fernandez A.C., Franco-Herrera N., et al. Saturation of an intra-gene pool linkage map: towards a unified consensus linkage map for fine mapping and synteny analysis in common bean. PLoS One. 2011;6:e28135. doi: 10.1371/journal.pone.0028135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu P., Wu X., Wang B., et al. A SNP and SSR based genetic map of Asparagus bean (Vigna unguiculata ssp. sesquipedalis) and comparison with the broader species. PLoS One. 2011;6:e15952. doi: 10.1371/journal.pone.0015952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gutierrez M.V., Vaz-Patto M.C., Huguet T., Cubero J.I., Moreno M.T., Torres A.M. Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theor. Appl. Genet. 2005;110:1210–7. doi: 10.1007/s00122-005-1951-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.