Abstract
High-throughput next-generation sequencing provides a revolutionary platform to unravel the precise DNA aberrations concealed within subgroups of tumour cells. However, in many instances, the limited number of cells makes the application of this technology in tumour heterogeneity studies a challenge. In order to address these limitations, we present a novel methodology to partner laser capture microdissection (LCM) with sequencing platforms, through a whole-genome amplification (WGA) protocol performed in situ directly on LCM engrafted cells. We further adapted current Illumina mate pair (MP) sequencing protocols to the input of WGA DNA and used this technology to investigate large genomic rearrangements in adjacent Gleason Pattern 3 and 4 prostate tumours separately collected by LCM. Sequencing data predicted genome coverage and depths similar to unamplified genomic DNA, with limited repetition and bias predicted in WGA protocols. Mapping algorithms developed in our laboratory predicted high-confidence rearrangements and selected events each demonstrated the predicted fusion junctions upon validation. Rearrangements were additionally confirmed in unamplified tissue and evaluated in adjacent benign-appearing tissues. A detailed understanding of gene fusions that characterize cancer will be critical in the development of biomarkers to predict the clinical outcome. The described methodology provides a mechanism of efficiently defining these events in limited pure populations of tumour tissue, aiding in the derivation of genomic aberrations that initiate cancer and drive cancer progression.
Keywords: mate pair sequencing; whole-genome amplified, laser capture microdissection; prostrate cancer
1. Introduction
In order to better stratify cancer patients for the best treatment or no treatment (active surveillance), characterization of the tumour will be required that can define the molecular changes associated with indolent or aggressive behaviour. A large effort in cancer research is aimed at identifying these cancer biomarkers that will augment the prognostic value of contemporary clinical and pathological features in stratifying risk of progression. Single-nucleotide variations (SNVs) affecting the gene expression of key regulatory genes provide both mechanism and markers for the progression of cancer. However, the vast numbers of genes affected by SNVs and the unknown impact of specific mutations on protein expression and function make the derivation of driver and passenger mutations difficult. Larger genomic rearrangements, such as amplifications, deletions and translocations, predict more distinct impacts on gene expression. The recent evaluation of the prevalence and reoccurrence of large genomic rearrangements and translocations in solid tumours has highlighted their value in defining cancer progression and prognosis.1 Significantly, clinical tests aimed at recurrent translocations/gene fusions involving Bcl-Abl, TMPRSS2-ERG, c-myc and bcl2 already impact clinical decisions.1–4
Massively parallel, next-generation DNA sequencing technologies represent a quantum advance in the ability to understand cancer at the genetic level. However, in order to accurately characterize genomic aberrations that define a tumour population, it is critical to obtain pure genetic material from the tumour with no or minimal contamination of surrounding cells. Tumours exist as outgrowths or repopulations of normal tissues with genetically modified cells, and as a result, malignant cells are often embedded and intermixed with normal cells making the sampling of pure tumour cells difficult. Specifically, the bulk sampling of a tumour mass is inevitably contaminated with varying degrees of non-neoplastic stromal, inflammatory, and epithelial cells, which complicate the genetic interrogation of the tumour cells. Laser capture microdissection (LCM) is a powerful tool to efficiently and accurately extract pure populations of tumour cells from clinical specimens, yet provides a limited number of cells and genetic material for analysis. The mate pair (MP) next-generation sequencing (NGS) protocol developed for the evaluation is capable of evaluating large genomic rearrangements using only a single lane of the Illumina GAIIx sequencer.5,6 However, this protocol currently requires DNA quantities far greater than those that can be yielded from LCM extracted tissues (5–10 μg), making the application of LCM to this NGS platform challenging.5
In this study, we report a novel methodology that enables the efficient and reproducible whole-genome amplification (WGA) of DNA from small LCM collected clinical specimens for application in the MP NGS protocol. We demonstrate the use of this technique in evaluating two pathologically distinct adjacent tumour grades (Gleason patterns) in a patient with prostate cancer.
2. Materials and Methods
2.1. Tissue selection and LCM
H&E stained sections of prostate cancer tissues were reviewed by a urologic pathologist (J.C.C.) and graded for tumour content. Frozen prostate tissue sections were cut into 10 μm size and stained with Cresyl Violet (LCM Staining Kit, Ambion; AM1935). GP3 and GP4 tumour regions were separately excised by LCM using the Arcturus PixCell II microscope and CapSure Macro LCM caps (Arcturus; LCM 0211). LCM of adjacent histologically normal (aN) peripheral zone prostatic epithelial cells (in the same tissue section as GP3 and GP4) and distal normal (dN) peripheral zone prostatic epithelial cells (separate benign tissue block from the opposite side of the prostate gland from cancer) were used as controls.
2.2. Direct in situ DNA extraction and amplification
LCM cap polymer regions containing engrafted cells were carefully excised and applied directly to WGA using a modified Repli-g protocol as follows (Qiagen, CA, USA). A modified Repli-g D2 buffer was first prepared (2.5 μl phosphate buffered saline, 3 μl Repli-g buffer (DLB) and 1 μl ditiothreitol per sample) and 6.5 μl was added to the LCM engrafted cells in 0.2 μl capped tubes ensuring complete immersion of the polymer fragments. Samples were incubated for 10 min on ice. Reactions were subsequently neutralized upon addition of 3.5 μl of Repli-g stop solution, prior to adding 40 μl of the standard Repli-g mini kit master mix and incubating for 16 h at 30°C. Final heating to 65°C for 3 min inactivated the DNA polymerase. WGA was performed on at least four independent reactions from parallel frozen sections and equal volumes pooled to minimize the amplification bias. For smaller lesions excised LCM regions from multiple sections can be applied to a single WGA reaction. WGA DNA (1 μl) was visualized on 1% agarose gels and quantitated using the Quant-iT PicoGreen dsDNA reagent (Invitrogen, P7581). Qualitative multiplex polymerase chain reaction (PCR) was performed according to the Sigma-Aldrich protocol and PCR products resolved on 4% agarose gels (Sigma-Aldrich; P0982).
2.3. Illumina MP Library Production
A modified Illumina MP library protocol was used. Briefly 10 μg of WGA DNA was fragmented to 3–5 kb using the Covaris E210 (Duty Cycle 20%, Intensity 1, 1000 cycles burst for 600 s.). End repair was extended to 30 min prior to the biotin end-labelling step and size selection on a 1% agarose gel. The biotin-labelled blunt-end fragments were then circularized and endonuclease treated, before the remaining DNA circles were fragmented again to 350–650 bp on the Covaris E210. The biotinylated terminal fragments were then immobilized on M-280 streptavidin beads (Dynal) and assembled into conventional Illumina adapter flanked paired-end (PE) libraries. Libraries were amplified (18 cycles) using conventional Illumina PCR adapter primer pairs (PE 1.0 and PE 2.0), purified and analyzed on an Agilent Bioanalyzer DNA 1000 chip. The library was loaded onto one lane of an Illumina flow cell at a concentration of 9 pM generating an average of 215 000 clusters/tile using the Illumina cluster station and PE cluster kit v4. The flow cell was sequenced as a 76 × 2 PE read on an Illumina GAIIx using SBS sequencing kit v4 and SCS v2.5. Base calling was performed using Illumina Pipeline v1.5.
2.4. Bioinformatics protocols
Bioinformatics protocols to rapidly and efficiently process NGS MP data using a 32-bit binary indexing of the Hg19 reference genome, to which consecutive 32-bit binary sequences from associated MP reads are aligned, have been previously published from our laboratory.6 This algorithm has subsequently been further optimized and is presented in depth in a separate manuscript.7 The algorithm maps both MP reads successively in the whole genome, thus minimizes miss mapping and reduces false positives. The algorithm aggressively tries to find mapping possibilities where the two reads map <15 kb apart allowing up to 10 mismatches. If mapping gave multiple good possibilities then the one with the lowest cumulative mismatch count was sent to the output. MPs mapping >15 kb apart or in different chromosomes were defined as large genomic rearrangements and selected for further analysis. When two reads map to the same chromosome more than 20 kb apart, they are defined as intra-chromosomal rearrangement (r) events. When they map to different chromosomes, they are defined as inter-chromosomal translocation (t) events. Filters, based on homology scores calculated during mapping, are applied to eliminate false positives. Fragments that represent the same event are identified as associate fragments and are clustered together. A mask table was created to further eliminate false-positive events that pass these filters defined from recurrent events in normal tissues or through BLAST-like alignment tool mapping of multiple aligning regions. After the removal of replicate read pairs, coverage calculations indicated the expected number of MPs covering a breakpoint. Replication was calculated for each chromosome and the replicate read pairs were removed. Bridged coverage was calculated as the sum of the fragment lengths (distance between read 1 and read 2) of correctly mapping MP and PE reads, divided by the mappable chromosome size. Base coverage was calculated from the total number of mappable reads, multiplied by the read length and divided by the mappable chromosome size. The coverage was calculated per chromosome and the average was found using chromosomes 1–22. For allelic coverage, the values must be divided by 2. A procedure generated count plots of the number of mapped read pairs in non-overlapping equal-length windows. The algorithm normalized each sample by calculating the window size such that windows in parts of the genome without deletions or amplifications contained an average of 300 mapped read pairs.
2.5. Validation of genomic rearrangements
MP sequence reads were mapped to the human genome, and primers spanning the fusion junctions were used in validation PCRs (50 μl, 35 cycles), on WGA tumour or normal DNA with human genomic DNA (gDNA) used as a control (G304A; Promega, WI, USA). Unique PCR products yielded from the WGA DNA, absent from control, were isolated by gel extraction (Minelute; Qiagen 28004) and Sanger sequenced. For secondary validation of genomic rearrangements on unamplified GP3 and GP4 DNA extracts, LCM was performed on fresh tissue sections and extracted by Arcturus Pico Pure DNA extraction reagents (20 μl/lesion) after incubation at 65°C for 4 h. PCRs (25 μl) were run in parallel to WGA G3/G4 DNA (100 ng, 35 cycles) but using 5 μl of unamplified material (40 cycles).
3. Results
3.1. Isolation of prostate GP3 and GP4 DNA
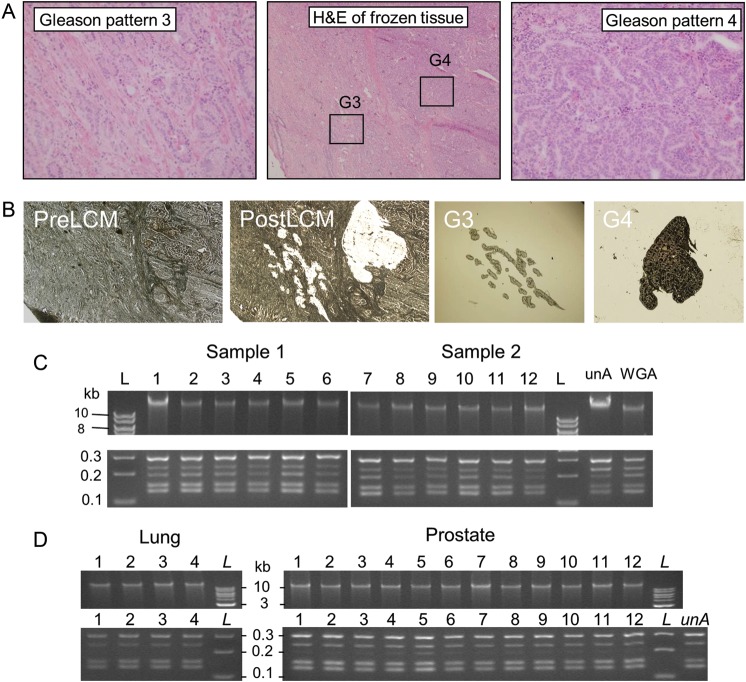
A prostate cancer case containing two regions of adjacent but distinct GP3 and GP4 tumours (Fig. 1a, central panel) was selected for the study. For the GP3 tumour (Fig. 1a, left panel), the infiltrative glands are well formed, each with a discernible round to oval shape. The GP4 tumour (Fig. 1a, right panel) shows a loss of distinct glandular differentiation in comparison with GP3, and cells are arranged in cribriform structures as well as sheets of tumour cells. LCM was used to isolate cells specific to the GP3 and GP4 populations minimizing the contamination of adjacent cells. Figure 1b demonstrates the power of the LCM technique, showing the section images before and after the laser-mediated extraction, as well as the cellular population of the GP3 and GP4 isolated on the LCM caps.
Figure 1.
LCM and WGA of prostate cancers. (a) H&E stained frozen section of prostate tissues. The central panel highlights the adjacent G3 and G4 cancers, with the individual G3 and G4 regions magnified further in the left and right panels, respectively. (b) LCM images before (PreLCM) and after (PostLCM) capture and the G3 and G4 cancer cells engrafted on the LCM caps. Products of direct in situ WGA of LCM-isolated cells (c) from two adjacent prostate lesions from six parallel tissue sections (1–6) and (d) from LCM of additional prostate (12 cases) and lung (4 cases) tissues [upper panels, 1% agarose gels, 1 kb ladders (L)]. Multiplex PCR from the corresponding tissues (lower panels, 4% agarose gels). The corresponding multiplex PCR products are shown in the lower panels (4% agarose gel with 100 bp ladders). Controls utilized gDNA (10 ng) in the WGA reaction or unamplified gDNA (unA) in the multiplex PCR.
To obtain enough gDNA to enable high-throughput MP sequencing of the specific GP of prostate cancer, WGA of the isolated DNA was required. Conventional techniques of DNA extraction and purification prior to WGA resulted in considerable variation and bias in the output quality of the DNA in pilot studies (data not shown). In order to improve on the quality of the sequencing data, a modified direct in situ WGA protocol was developed. This novel technique involved the application of engrafted LCM cellular material directly into WGA reactions. The chemical conditions of an initial DNA denaturing step facilitated the lysis of the engrafted LCM cells and the release of the nuclear reservoir of gDNA to the subsequent WGA reaction. Significantly, the crude cellular lysate did not result in observable inhibition of the enzymatic amplification reaction, generating DNA yields equivalent to control-purified gDNA (Fig. 1c). The reproducibility of this direct in situ WGA method is exemplified in parallel reactions on two sets of six consecutive LCM isolations of prostate tissue frozen sections (Fig. 1c, upper panel) yielding WGA DNA products of similar quantity and size on an agarose gel. Yields of amplified gDNA were typically in the range of 8–10 μg/50 μl WGA reactions. A multiplex PCR was used to assess amplification bias and quality of the WGA DNA. The banding patterns of five amplicons (132–295 bp) for the WGA DNA samples were equivalent to control WGA and unamplified DNA (Fig. 1c, lower panel). This result gave an initial positive indication of the representative genomic amplification in these WGA reactions. The reproducible amplification quality and quantity of WGA DNA yielded directly from LCM of 12 different prostate cases and 4 lung tissues are also presented in Fig. 1d.
3.2. MP sequencing and bioinformatics mapping algorithm
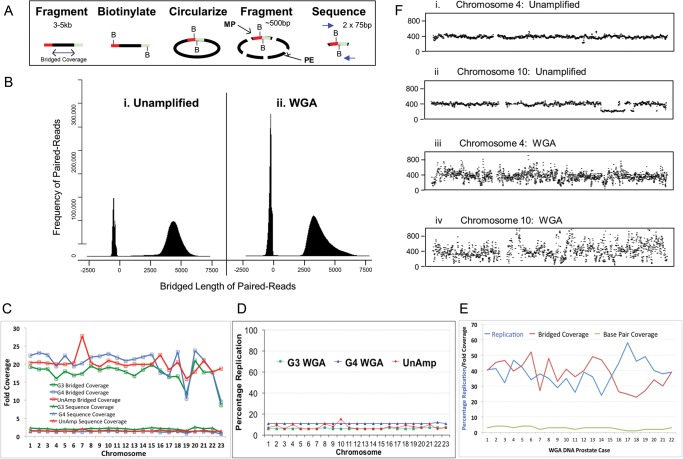
MP libraries were assembled for the prostate GP3 and GP4 WGA DNA samples using the modified MP library assembly protocol described. The conventional Illumina protocols were adapted to account for the different physical properties of the WGA DNA. The MP libraries were sequenced on single lanes of the Illumina GAIIx platform. MP sequencing of the GP3 and GP4 cancers generated 33.0 and 29.4 million mappable read pairs, respectively, consistent with results from our MP sequencing of conventional unamplified gDNA on the GAIIx sequencer, which yielded averages of 27.1 million reads. A set of algorithms was developed to detect large chromosomal aberrations with low false-positive rates. The algorithms were specifically designed to handle NGS from mate-pair protocols.7 The algorithms describe coverage in two ways: conventional base-pair coverage from the 75–100-bp reads and the theoretical bridged-coverage considering the original 3–5-kb span separating two MP reads (Fig. 2a). MP reads mapping with the expected region span allows us to infer that the correct sequence span lies between the reads. Fragment mapping profiles are presented in Fig. 2b. The predominant fragment population reflects the MP reads with 3–5 kb bridged spans. The second narrower peaks at around −300 bp consist of co-purifying PE library fragments derived from internal fragments of the circles not spanning the ligation junctions (Fig. 2a). The PE peaks map algorithmically with negative values due to the different polarity of the read pairs stemming from the circularization in the MP methodology.5 The fragment mapping profile for unamplified (Fig. 2b, i) and WGA (Fig. 2b, ii) DNA are effectively equivalent, with the WGA DNA mapping with just slightly reduced span of 2.5–5 kb indicative of the starting input of the smaller DNA fragments (Fig. 1c). Although the proportion of PE mapping fragments is still higher with the WGA DNA, additional refinements of the MP protocol could reduce this further.
Figure 2.
MP NGS of WGA DNA products. (a) A schematic of the MP library assembly, detailing the initial fragmentation of the gDNA to 3–5 kb and biotinylation of the termini of these fragments to enable isolation and sequencing of the ligated terminal regions after circularization and a second fragmentation to ∼500 bp. The bridged coverage is demonstrated as the span of the initial 3–5 kb fragments and the resulting terminal MP fusion products and conventional PE fragments depicted. (b) The bridged length of paired reads mapping to the reference genome from (i) an unamplified gDNA sample of T-cell origin and (ii) the WGA GP4 sample. (c) Bridged (open squares) and base pair coverage (open triangles) for individual chromosomes from the WGA GP3 (green) and GP4 (blue) samples and the unamplified T-cell control (red). (d) Percentage replication in the GP3 (green square), GP4 (blue triangle) and unamplified (red star) samples MP data. (e) Percentage replication (blue), together with bridged (red) and sequence (green) fold coverage for 22 additional WGA prostate cancer DNA samples following LCM isolation on the HiSeq platform. (f) Frequency coverage plots for chromosomes 4 (i and iii) and 10 (ii and iv) of unamplified DNA (i and ii) and WGA G4 DNA (iii and iv) sequencing data.
The MP bridged and base-pair coverages for the WGA DNA were also equivalent to unamplified gDNA (Fig. 2c). The average bridged coverage's of 18 and 21X were obtained for the GP3 and GP4 tumours, respectively, compared with an average of 17.5X for our unamplified DNA. The average sequence base coverages of 1–2X were observed for both DNA inputs. Base-pair and bridged coverages are presented for each chromosome in Fig. 2c. Both measures of coverage are observed to be relatively consistent across the majority of chromosomes for both the unamplified and WGA samples. The level of replication indicates how many identical MP sequences are derived from sequencing clusters. This replication of MP sequences is expected due to the PCR amplification processes involved in library assembly and is bioinformatically removed to prevent bias in library mapping. The GP3 and GP4 samples were associated with averages of 6 and 10% replication, respectively, in a range identical to an unamplified DNA sample (Fig. 2d).
While this report exemplifies a single prostate cancer case, multiple additional WGA samples have been processed using this methodology on the enhanced Illumina HiSeq platform. Figure 2e illustrates the coverage and replication from the MP sequencing of WGA DNA samples yielded from 22 additional prostate tissues. The average total number of reads increased to 76.7 million on the newer HiSeq platform, with the resultant base and bridged coverage levels averaging 3 and 39X, respectively. As expected, the mean levels of replication also increased to 37X, potentially due to sequence saturation on the flow cell. Together, these data provide us with further confidence in the reproducibility of the amplification methodology and the mapping algorithm described in this study.
Figure 2f (i and ii, respectively) shows MP count plots associated with an unamplified DNA sample on chromosomes 4 and 10. Although both chromosomes show very flat even profiles (Fig. 2f, i), chromosome 10 shows a clear drop in frequency between 98 and 112 Mb (Fig. 2f, ii), indicative of a deletion in this region. The centromeres are observed as sequence gaps due to the current lack of reference sequence for these regions due to the high levels of repetition. The equivalent WGA GP4 tumour frequency plots are shown in Fig. 2f, iii and iv. Although coverages across the chromosomes are high, the frequency profiles are much more variable than the unamplified DNA. Thus, at the fine mapping level, the amplification process results in some level of differential amplification bias across the genome. This variability in the frequency profiles does unfortunately make it difficult to confidently call copy number variations. Nevertheless, these data demonstrate that the MP sequencing from WGA DNA does result in extensive genome coverage across chromosomes.
3.3. Large genetic aberrations in the GP3 and GP4 cell populations
Table 1 lists the large genomic aberrations detected in the GP3 and GP4 of this prostate cancer by our MP sequencing. The events recorded passed bioinformatics filters in the mapping algorithm detailed in Materials and methods. Initial MP mapping predicted many more events prior to filtering, but the majority were false positives due to miss-mapping from highly repetitive or currently undefined regions within the reference human genome. The table is split into potential inter-chromosomal translocations and intra-chromosomal rearrangements (amplifications, deletions or inversions). The first significant observation is that the majority of large genomic aberrations are present in both the GP3 and GP4 cell populations of this prostate cancer, presenting strong evidence of common origin or clonality between these two Gleason patterns. However, the numbers of associated MPs varies considerably, both between the same and different chromosomal events. PCR was used to validate events in Table 1 assessing the accuracy of the algorithm used to call genomic rearrangements from WGA DNA.
Table 1.
Large genomic rearrangements in the GP3 and GP4 cancers
| Event | Loci 1 | Loci 2 | Position 1 | Position 2 | Gene 1 | Gene 2 | #MP G3 | #MP G4 |
|---|---|---|---|---|---|---|---|---|
| t(1-12)a | 1q32.3c | 12q21.33a | 213127619 | 89404575 | VASH2 | AK055062 | 11 | 21 |
| t(1-12)b | 1p36.13e | 12p11.22a | 17481805 | 30504168 | no_gene | no_gene | 0 | 3 |
| t(2-14) | 2q24.1c | 14q32.12a | 157595999 | 92919912 | no_gene | SLC24A4 | 5 | 0 |
| t(2-23) | 2q35c | Xq22.3b | 217539908 | 105359151 | IGFBP5 | no_gene | 5 | 0 |
| t(5-11) | 5p13.1c | 11q24.3b | 38567230 | 129589054 | LIFR | no_gene | 3 | 0 |
| t(7-17) | 7p21.2a | 17p13.1c | 13910005 | 7973212 | ETV1 | ALOX12B | 10 | 10 |
| t(8-11)a | 8q22.2b | 11q13.2c | 100826483 | 67861296 | VPS13B | CHKA | 4 | 3 |
| t(8-11)b | 8q22.2b | 11q24.2b | 100473404 | 125522385 | VPS13B | CHEK1 | 2 | 15 |
| t(17-19)a | 17q22b | 19q13.32a | 53592582 | 45963102 | no_gene | BCAM | 1 | 9 |
| t(7-19)b | 17q21.31c | 19q13.32a | 43268092 | 45311607 | no_gene | ERCC1 | 1 | 4 |
| r(1-1) | 1p33d | 1p34.1b | 47040276 | 46048889 | MKNK1 | NASP | 2 | 7 |
| r(2-2)a | 2q13b | 2q13b | 111600477 | 111523813 | ACOXL | ACOXL | 10 | 10 |
| r(2-2)b | 2q13d | 2q13b | 113373061 | 111874072 | no_gene | ACOXL | 6 | 4 |
| r(3-3)a | 3p22.3b | 3p22.3b | 34811976 | 34726774 | no_gene | no_gene | 12 | 7 |
| r(3-3)b | 3q24c | 3q11.2c | 145684839 | 97497658 | no_gene | ARL6 | 0 | 5 |
| r(4-4)a | 4q31.21c | 4q24a | 145217293 | 102291108 | no_gene | (PPP3CA) | 3 | 6 |
| r(4-4)b | 4q31.22a | 4q27b | 146813724 | 121967686 | ZNF827 | C4orf31 | 3 | 9 |
| r(4-4)c | 4q28.3h | 4q24a | 137887074 | 101807230 | no_gene | no_gene | 3 | 21 |
| r(4-4)d | 4q28.1c | 4q27b | 126358971 | 121952037 | FAT4 | no_gene | 3 | 20 |
| r(4-4)e | 4q31.21c | 4q24e | 146126396 | 107031959 | (OTUD4) | TBCKL | 3 | 11 |
| r(6-6) | 6q16.3d | 6q16.1e | 104492242 | 97668534 | no_gene | C6orf167 | 2 | 16 |
| r(7-7) | 7q36.2c | 7q36.2b | 154376229 | 154265528 | DPP6 | DPP6 | 1 | 5 |
| r(8-8)a | 8p23.1a | 8p23.1c | 12610164 | 10101851 | LONRF1 | MSRA | 8 | 21 |
| r(8-8)b | 8p22e | 8p23.1a | 12718032 | 12607655 | no_gene | LONRF1 | 3 | 19 |
| r(9-9) | 9q32d | 9q32d | 117100468 | 116868162 | AKNA | (KIF12) | 11 | 15 |
| r(11-11)a | 11q24.2b | 11q13.2c | 125525691 | 68317314 | CHEK1 | SAPS3 | 2 | 13 |
| r(11-11)b | 11q23.3e | 11q23.1d | 117933682 | 112273163 | no_gene | no_gene | 7 | 27 |
| r(12-12)a | 12q23.1d | 12q23.1d | 101157176 | 100665614 | ANO4 | SCYL2 | 0 | 7 |
| r(12-12)b | 12q21.1a | 12q15d | 71683589 | 71437383 | (TSPAN8) | TSPAN8 | 1 | 11 |
| r(23-23) | Xq25h | Xq25h | 128603069 | 128566668 | (SMARCA1) | SMARCA1 | 6 | 5 |
3.4. Validation of a chromosome 1–12 balanced translocation event
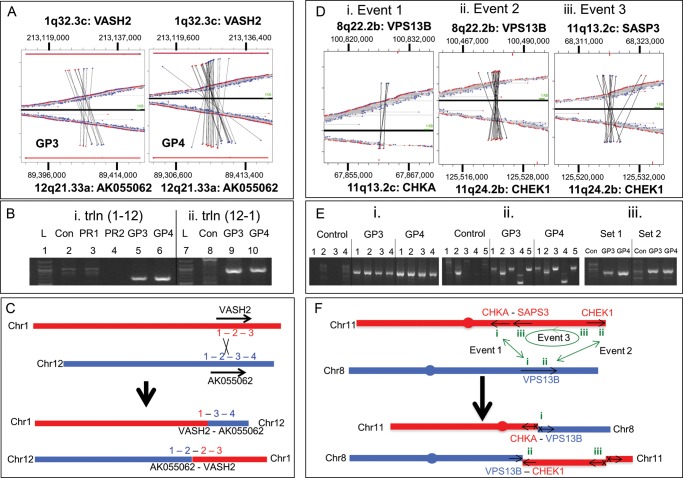
Figure 3a–c presents the validation of a balanced translocation event between 1q32.3c and 12q21.33a, involving genes VASH2 and AK055062, respectively (Table 1). The GP3 and GP4 cell populations recorded 11 and 21 MP reads, respectively (Table 1), which are mapped in Fig. 3a. The upper and lower parts of the figures refer to chromosomes 1 and 12, respectively. MP reads are illustrated as either blue or red dots, depending on the polarity of the MP sequence mapping to the reference genome, with a line linking the paired reads. Central horizontally linked red-to-blue dots represent intra-chromosomal mapping MP fragments with the concordant bridged coverage spans (3–5 kb). Vertically linked red-to-blue dots are associated with MPs specific to this translocation event between chromosomes 1 and 12 (Fig. 3a, i and ii). Two sets of red/blue-linked MP events spanning the two chromosomes are observed, red-to-blue and blue-to-red. This is indicative of a balanced translocation event between the two chromosomes, where genomic regions have been exchanged between the two chromosomes resulting in t(1-12) and t(12-1) translocation products (Fig. 3c). The regions where the red–blue and blue–red mapping events converge indicate the location of the translocation breakpoints.
Figure 3.
Validation of chromosome translocation events: mapping of two independent translocation events are depicted; a balanced translocation between chromosomes 1 and 12 (a–c) and a complex rearrangement between chromosomes 8 and 11 (e and f). (a) MP sequences mapping to chromosomes 1 and 12 for GP3 and GP4 are depicted above and below the zero axis, respectively, as red or blue dots dependent on the direction of the sequence read mapping to the reference genome. Horizontally linked red and blue dots closest to this axis depict normal mapping MP sequences to chromosome 1 or 12 alone. MP sequences linked vertically depict translocations between these two chromosomes. (b) PCR validation using primers specific to the t(1-12) event (i) for GP3 and GP4 (lanes 5 and 6) and the t(12-1) event (ii) for GP3 and GP4 (lanes 9 and 10). gDNA (lanes 2 and 8) and different prostate tumour tissues (PR1 and PR2, lanes 3 and 4) were used as controls together with 1 kb ladder (lanes 1 and 7). (c) Schematic representation of predicted VASH2 and AK055062 fusion products resulting from the translocation events described. (d) Mapping of MP sequences to three inter-linked events at genomic loci 8q22.2b, 11q13.2c and 11q24.2b. Events 1 and 2 describe two t(8-11) translocation events linking VPS13B to CHKA (i) and CHEK1 (ii), respectively. Event 3 describes an r(11-11) intra-chromosomal rearrangement linking CHEK1 with SASP3 (iii). (e) PCR validation 1% agarose gels are presented for four different primer sets (1–4) for event 1 (i), five different primers sets (1–5) for event 2 (ii) and two primer sets (1 and 2) for event 3 (iii) for the GP3 and GP4 WGA DNA and gDNA as control. (f) Schematic representation of the three fusion events on chromosome 8 (red) and 11 (blue), describing the potential products and the impact on the gene regions involved. Arrows describe the coding direction of the genes and an X represents the loss of the promoter regions.
PCR primers spanning the indicated breakpoints on chromosomes 1 and 12 were used to validate the translocation events (Fig. 3b, i and ii). Specific bands are observed for both the GP3 and GP4 samples (lanes 5, 6, 9 and 10) that are not present in the human gDNA control (lanes 2 and 8) or two additional prostate cancers from different patients (lanes 3 and 4). Sanger sequencing of excised PCR bands yielded unique fusion sites for both the t(1-12) and t(12-1) translocation events. Identical fusion junctions were observed in the GP3 and GP4 samples. The t(1-12) translocation resulted in the fusion [chr1:213 127 018] to [chr12:89 406 958]. The t(12-1) translocation mapped [chr12:89 407 157] to [chr1:213 127 057], but was more complex, involving an additional inversion of 40 nucleotides of chromosome 1 [chr1:213 127 017–213 127 056] with an overlap of four nucleotides (TAAG) at the 12–12 fusion point (Supplementary Table S1). Both fusions are intronic, located between exons 2 and 3 of VASH2 and exons 1 and 2 of AK055062 resulting in two potential fusion proteins (Fig. 3c). The consequence of these two fusion events, the translation products yielded and the impact on tumourigenesis still needs to be investigated and were beyond the scope of this current work.
3.5. Validation of chromosome 8–11 complex rearrangement
Figure 3d–f describes the mapping and validation of a more complex chromosomal rearrangement, involving two translocations from VPS13B on chromosome 8 to two distal sites on chromosome 11 involving CHKA and CHEK1. The MP mapping of these two events each predict a single junction, without associated balanced events (Fig. 3d, i and ii). While one event predicts a direct translocation between the chromosomes 8 and 11 regions with blue-to-red MPs, the second event is more complex with red-to-red MPs predicting a sequence inversion. In order to understand this complex rearrangement, a third event involving an intra-chromosomal inversion (blue-to-blue MP) of a large segment of the q-arm of chromosome 11 linking SASP2 to CHEK1 needs to be considered (Fig. 3d, iii). Considering the three events together the balanced recombination event can be predicted (Fig. 3f). While the 8-11 translocation between CHKA and VPS13B describes one half of the rearrangement, events 2 and 3 together form the balanced event. Specifically, a large fragment of the q-arm of chromosome 11 inverted, linking the CHKA adjacent gene; SASP3 to CHEK1 at the termini of chromosome 11, with the rest of the CHEK1 gene fusing with VPS13B on chromosome 8. Using primers spanning the predicted fusion sites, each event was validated by PCR generating unique bands in both the GP3 and GP4 cell populations that were not observed with control gDNA (Fig. 3e). While events 1 and 3 would result in the fusion of two gene regions with the loss of promoter sequences, event 2 results in both CHEK1 and VPS13B retaining their promoters and thus two truncated proteins may result with unknown cellular impact. Additionally, a ∼300 kb fragment spanning from CHKA to SAPS3 is unaccounted for in this description. However, the reduced coverage of MPs adjacent to the CHKA and SASP3 MP regions in Fig. 3d could indicate a deletion of these sequences.
3.6. Additional validations in the GP3 and GP4 cancers
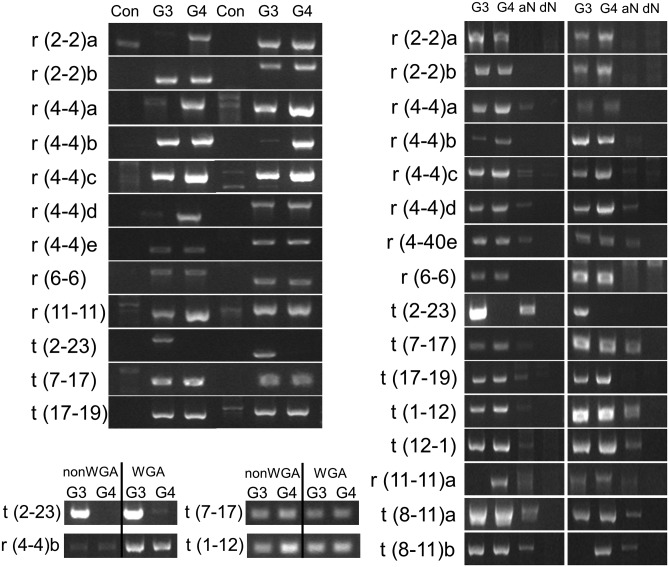
In order to further support this novel experimental methodology and bioinformatics algorithm, 12 additional events from Table 1 were selected for PCR validation. All 12 events generated unique PCR products in the prostate cancer that were absent in control gDNA (Fig. 4a). Significantly, all events but one validated in both the GP3 and GP4 samples; however, the intensity of the bands often varied between GP3 and GP4, indicating this did not result from LCM contamination. For several events, the intensity of the band was greater in the GP4 than the GP3, indicating a greater prevalence of the rearrangement in the GP4 cell population. Only the event t(2-23) validated predominantly in GP3 with no or potential very low levels observed in the GP4 (Fig. 4a–c). While it is difficult to confidently define the levels of each genomic aberration in these tumours considering the input of WGA DNA and the semi-quantitative PCR, the band intensities correlated to some extent with the numbers of associated MP reads (Table 1). Sanger sequencing of the validating PCR bands revealed the fusion breakpoints for each event (Supplementary Table S1). Significantly, for each PCR band, the exact fusion junction was identified in both the GP3 and GP4 samples. These data present strong evidence for the clonality of GP3 and GP4 in this prostate cancer. Additionally, the presence of the exact junction breakpoints in both GP3 and GP4 makes these fusion events unlikely to be artefacts of WGA.
Figure 4.
Additional validations in the GP3 and GP4 cancers: (a) 1% agarose gels of PCR validation for 12 additional chromosomal rearrangement events in the GP3 and GP4 cancers, involving two primer sets and using gDNA as control. (b) PCR evaluation of aN and dN tissues for 17 previously validated events in GP3 and GP4 tumours of the same patient. (c) PCR validation in amplified (WGA) and unamplified (nonWGA) GP3 and GP4 tissues for four validated events.
3.7. Evaluation of genomic rearrangements in histologically normal adjacent tissues
Adjacent peripheral gland normal (aN) from the same sections as the GP3 and GP4 cancers was isolated by LCM and WGA. Additional, distal peripheral normal (dN) from the opposite side of the patient's prostate and in a different frozen tissue block was similarly isolated. Fusion-specific PCRs evaluated 16 validated events in the aN and dN samples. Surprisingly, the majority of events were present in aN, although the intensity of the bands were consistently reduced (Fig. 4b). All the events associated with the previously discussed 1–12 and 8–11 rearrangements validated in the aN tissue, as did a group of five rearrangements within a 50-Mb region of chromosome 4, suggesting potential early events in cancer progression. Just r(2-2)a, r(2-2)b and r(6-6) yielded no equivalent bands in aN. Sanger sequencing of 12 aN bands of sufficient intensity to sequence without further amplification, confirmed the events (data not shown). The absence of any of these events in the dN, while predicting a potential local field effect phenomenon for this patient, also further eliminated the possibility of WGA artefacts. Although LCM contamination cannot be excluded in these analyses, the range of different banding patterns observed between the GP3, GP4 and aN tissues, specifically the contrasting t(2-23) GP3-specific event, minimizes these concerns.
3.8. Validation in non-amplified tissues
To further confirm that these rearrangements were not artefacts of the WGA process, additional validations on unamplified tissues were performed. Further LCM of the same GP3 and GP4 cell populations was performed and DNA extracted in small volumes of extraction buffer. Due to the limited quantities of DNA extracted, only a limited number of previously optimized validation PCRs could be performed. Figure 4c demonstrates that identical bands were reproduced in the unamplified tissues as were observed in the WGA samples, further indicating the reliability of the methodology.
4. Discussion
High-throughput NGS provides a powerful tool to investigate the molecular changes associated with cancer progression. To confidently identify these changes, it is vital to have methodologies to determine genetic changes specific to tumour populations. Prostate cancer like many cancers is heterogeneous with various degrees of differentiation that are assessed in the Gleason scoring system.8 A critical transition for patient prognosis and for dictating therapy is the presence of GP4 or higher.9 Men who have tumours composed entirely of GP3 have an excellent outcome regardless of treatment.10 In contrast, the presence of GP4 in addition to GP3 has a significant negative impact on survival and raises the possibility that GP3 alone is molecularly different from GP3 associated with GP4. Therefore, to understand prostate cancer progression, stratify patients by risk of progression and to provide an optimal care, it is necessary to understand the molecular changes in GP3 and GP4, and those that drive progression to GP4. The methodology described enables the accurate collection of pure populations of clinical specimens and the subsequent direct WGA of the gDNA.
The reporting of the prevalence and major role of the E-twenty six (ETS)/TMPRSS2 family of genomic rearrangements in prostate cancer has dramatically changed the perception of solid tumour biology.1 Elucidation of genomic rearrangements present in solid tumours provides a scaffold to study disease heterogeneity, as well as the relationships between distinct cell populations such as clonality and cancer progression. The NGS MP protocol provides a method to specifically interrogate a gDNA sample for large rearrangements and was thus modified for the input WGA DNA.5 The application of this methodology to WGA DNA yielded MP sequencing of similar quality to unamplified gDNA. Specifically, MP sequencing coverage and replication levels for WGA DNA were equivalent to unamplified DNA. However, frequency plots did indicate more focal variation in the WGA MP profiles compared with unamplified DNA, indicating some bias in the regional amplification, which made copy number variation calling problematic with WGA DNA. We are currently investigating complex bioinformatics algorithms aimed at compensating for this natural frequency variation from WGA profiles to enable more confident copy variation number calling. It must also be noted that the prostate cancer case exemplified in this study was performed on the Illumina GAIIx platform and more recent sequencing of WGA MP libraries using the methodology described in this report on the newer Illumina HiSeq platform generated far greater depths of sequencing (Fig. 2f and Supplemental Table S2).
In this study, LCM was used to collect pure populations of GP3 and GP4 tumour cells, together with benign-appearing prostatic cells from the same patient. Our novel methodology for WGA and MP sequencing enabled us to accurately define large genomic aberrations using algorithms developed in our laboratory. Table 1 lists 30 high-confidence genomic rearrangements within the GP3 and GP4 tumour cell populations. The 30 events involved 15 chromosomes, with 20 intra-chromosomal rearrangements and 10 inter-chromosomal translocations. Each event selected for validation was successfully verified by PCR and the fusion junctions determined, emphasizing the validity of the technique and algorithm presented in this study. While many genomic rearrangements were predicted, conservative filtering and knowledge of recurrent false positives additionally observed in normal tissues enabled us to eliminate the majority of false positives. Significantly, the GP3 and GP4 of this patient did not contain the common ERG-TMPRSS2 fusion event,1 but the validated t(7-17) fusion site lies just 15 kb downstream of the ETS family member; ETV1 gene, which is fused at a similar 3′ distance downstream of the previously unreported ETS family fusion partner ALOX12B. Though no fusion product is predicted, this case was independently demonstrated to have elevated ETV1 expression (data not shown).
The presence of identical rearrangements within the adjacent Gleason patterns provides evidence of the common origin of GP3 and GP4 in this prostate cancer, indicating the clonal expansion of GP3 with progression to GP4, a critical step in disease progression with very important clinical implications. While we cannot extrapolate this single patient observation to prostate cancer in general, this report describes powerful methodology to address this question in multiple prostate tumour samples, which is currently underway in our laboratory. Additionally, while these data may provide clues to potential GP3 to GP4 progression in the ratio of MP numbers and the intensity of PCR validation bands, care must be taken considering the input of WGA amplified DNA in the studies. More extensive and direct experimentation is required on unamplified tissues to answer these questions, which was outside the scope of this study. Our findings also suggest the presence of a significant prostate cancer field effect in this patient with 13 of 16 validated events, also present in the aN tissues but not dN prostate samples (Fig. 4b). While great care was taken in the LCM sampling, contamination cannot be ruled out. However, the varying levels of PCR banding observed for the GP3, GP4 and aN validations (Fig. 4a and b) does not reflect a level of continuity expected from a constant level of contamination in each sample.
The main difference of our procedure from previous studies relates to handling and amplification of DNA from small numbers of captured cells. To circumvent limitations related to low DNA yields and quality resulting from LCM of a few hundred cells, we developed an in situ whole-genome DNA amplification procedure to bypass the DNA extraction steps. As described, this methodology produced a good quality DNA resulting in representative MP sequencing data spanning the whole genome of tumour cells. While MP sequencing of WGA DNA did result in significant increases in uneven profiles across the genome, it still provided adequate depth of sequencing to reveal a large set of rearrangements that were validated by independent experimental methods. We conclude that the ability to genetically characterize pure populations of histologically significant lesions far outweighs these limitations introduced by WGA of the DNA.
While the successful application of WGA to the MP protocol was demonstrated in this report, special attention must be made in both library assembly and bioinformatics analysis. The reduced size of the input WGA (∼15 kb) compared with unamplified DNA (∼50 kb) required specific modifications to the Illumina MP fragmentation protocols. Over fragmentation of WGA DNA using protocols designed for normal gDNA was observed to compromise the final sequencing data, resulting in reduced MP data coverage. While the bioinformatics algorithm applied to WGA and gDNA was identical, WGA DNA was observed to result in increased background noise in the MP data. A summary of MP data from eight cases each of WGA and unamplified gDNA are presented in Supplementary Table S2. Large numbers of false-positive discordant MP reads were clearly a factor for both DNA inputs due to limitations in the Illumina MP library production protocols. The percentage of discordant MPs that passed initial filters, were as expected slightly greater with WGA DNA. This elevated false-positive fusion frequency potentially arises from artefacts of random re-priming of the 3′ termini of displaced amplification fragments to non-specific sites. While significant in the output MP data, they predominantly present as single events with no associates and the majority are removed in the final filtering steps requiring three associated MP reads to pass filter. An indication of the false positive rates can thus be reflected in the number of observed MPs that present with no associates. As expected, this population is extensive for both groups, with 83% of discordant MP having no associates for the gDNA before initial filtering. This figure was almost 10% higher with WGA DNA (91%) reflecting a larger proportion of false positives stemming from the amplification protocol. However, upon filtering the number of unique discordant MPs from the two populations is similar, reflective of the effectiveness of the bioinformatics algorithms applied to eliminate these false positives. In conclusion, while an increased number of false positives are predicted with WGA, the presence of these events as unique random events enables them to be easily eliminated upon bioinformatics filtering. However, care must always be taken in interpreting potential fusion events in MP data and PCR validation is generally required to truly validate events. Nevertheless, the robustness of the algorithm is emphasized in the level of validation observed in this study. Conversely, a significant number of genomic breakpoints may be missed due to reduced depths of sequencing or underrepresentation in the final library preparations. However, defining the level of false-negative calls in the data is far more complex and would require absolute knowledge of all the translocations present in a case to be assessed.
The Illumina MP protocol enables the sequencing of 100 nucleotides from the termini of larger spanning (2–5 kb) DNA fragments compared with conventional PE libraries (0.2–0.5 kb). This extended DNA span increases the probability of spanning a genomic breakpoint. The ‘bridged-coverage’ of >30X with a half lane of a HiSeq2000 data is highly effective at detecting these breakpoints. However, with ‘base-coverage’ in the range of 4–5X, the ability to call SNVs is limited. The reduced size of DNA yielded from formalin fixed paraffin embedded (FFPE) tissues (generally <500 bp) also precludes its application to the MP protocol and most WGA techniques. Interrogation of large genomic rearrangements in DNA from FFPE tissues currently requires whole genome sequencing of PE libraries on several lanes of an Illumina HiSeq2000 flow cell in order to attain high enough sequencing depths to confidently call fusion breakpoints. While more robust methods are now available for WGA from FFPE tissues, these methodologies target the smaller species of DNA present in these tissues, generally <500 bp which still precludes their application to current MP protocols.
In summary, this study shows the methodology of interrogating small pure cell populations by NGS technologies using LCM coupled with WGA and a modified MP library assembly protocol. The WGA DNA generated in this study is of sufficient quality and quantity for application to other next generation protocols, including whole-genome and exome capture sequencing protocols (data not shown and S.J. Murphy et al., submitted). Sampling and sequencing of distinct tumour cell populations as well as histologically normal cell populations allows the characterization of genetic changes occurring between these cell populations providing insight into cancer development and progression. The results described for the prostate tissue exemplified in this study provide major clues to the origin of this patient's cancer. Current experimentation in our laboratory is using this methodology to sequence large panels of adjacent tumours and histologically normal tissues from a variety of cancer types to identify large genomic translocations suitable for clinical targeting.
Contributions
S.J.M., J.C.C., F.K., R.J.K. and G.V. planned the project, S.J.M., J.C.C., S.Z., R.A.S. and B.W.E. carried out the experiments. S.J.M., S.H.J. and G.V. analyzed the data and S.J.M., J.C.C. and G.V. wrote the manuscript. All authors edited and commented on the manuscript.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported in part by a Waterman Biomarker Discovery grant from the Mayo Clinic Center for Individualized Medicine.
Supplementary Material
Footnotes
Edited by Masahira Hattori
References
- 1.Rubin M.A., Maher C.A., Chinnaiyan A.M. Common gene rearrangements in prostate cancer. J. Clin. Oncol. 2012;30:608–15. doi: 10.1200/JCO.2011.35.1916. doi:10.1200/JCO.2011.38.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niitsu N., Okamoto M., Miura I., Hirano M. Clinical features and prognosis of de novo diffuse large B-cell lymphoma with t(14;18) and 8q24/c-MYC translocations. Leukemia. 2009;23:777–83. doi: 10.1038/leu.2008.344. doi:10.1038/leu.2008.344. [DOI] [PubMed] [Google Scholar]
- 3.Burke B.A., Carroll M. BCL-ABL; a multifaceted promoter of DNA mutation in chronic myelogenous leukemia. Leukemia. 2010;24:1105–12. doi: 10.1038/leu.2010.67. doi:10.1038/leu.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin P., Jetly R., Lennon P.A., et al. Translocation (18;22) (q21;q11) in B-cell lymphomas: a report of 4 cases and review of the literature. Hum. Pathol. 2008;39:1664–72. doi: 10.1016/j.humpath.2008.04.007. doi:10.1016/j.humpath.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Illumina. Mate Pair Library v2 Sample Preparation Guide for 2–5 kb Libraries. 2009. http://nextgen.mgh.harvard.edu/attachments/MatePair_v2_2-5kb_SamplePrep_Guide_15008135_A.pdf.
- 6.Feldman A.L., Dogan A., Smith D.I., et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively parallel genomic sequencing. Blood. 2011;117:915–9. doi: 10.1182/blood-2010-08-303305. doi:10.1182/blood-2010-08-303305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasmatzis G., Johnson S.H., Knudson R.A., et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peritheral T-cell lymphomas. Blood. 2012 doi: 10.1182/blood-2012-03-419937. 2012 Aug 1. [Epub ahead of print] PMID: 22855598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Epstein J.I. An update of the Gleason grading system. J. Urol. 2010;183:433–40. doi: 10.1016/j.juro.2009.10.046. doi:10.1016/j.juro.2009.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Amin A., Partin A., Epstein J.L. Gleason score 7 prostate cancer on needle biopsy: relation of primary pattern 3 or 4 to pathological stage and progression after radical prostatectomy. J. Urol. 2011;186:1286–90. doi: 10.1016/j.juro.2011.05.075. doi:10.1016/j.juro.2011.05.075. [DOI] [PubMed] [Google Scholar]
- 10.Stephenson A.J., Kattan M.W., Eastham J.A., et al. Prostate cancer-specific mortality after radical prostatectomy for patients treated in the prostate-specific antigen era. J. Clin. Oncol. 2009;27:4300–5. doi: 10.1200/JCO.2008.18.2501. doi:10.1200/JCO.2008.18.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.