Abstract
Gene targeting (GT) can introduce subtle alterations into a particular locus and represents a powerful tool for genome editing. Engineered zinc finger nucleases (ZFNs) are effective for generating minor allelic alterations. Efficient detection of such minor alterations remains one of the challenges in ZFN-mediated GT experiments. Here, we report the establishment of procedures allowing for efficient detection, quantification and enrichment of such subtle alterations. In a biallelic model, polyacrylamide gel electrophoresis (PAGE) is capable of detecting rare allelic variations in the form of DNA heteroduplexes at a high efficiency of ∼0.4% compared with ∼6.3% by the traditional T7 endonuclease I-digestion and agarose gel electrophoresis. In a multiple allelic model, PAGE could discriminate different alleles bearing addition or deletion of 1–18 bp as distinct bands that were easily quantifiable by densitometry. Furthermore, PAGE enables enrichment for rare alleles. We show for the first time that direct endogenous GT is possible in medaka by ZFN RNA injection, whereas PAGE allows for detection and cloning of ZFN-targeted alleles in adults arising from ZFN-injected medaka embryos. Therefore, PAGE is effective for detection, quantification and enrichment of multiple fine allelic differences and thus offers a versatile tool for screening targeted subtle gene alterations.
Keywords: gene targeting, heteroduplex analysis, zinc finger nucleases, transcription activator-like effector nucleases, polyacrylamide gel electrophoresis
1. Introduction
Genetic modifications at specific sites of animal genomes represent a powerful method for elucidating molecular mechanisms underlying normal and pathological processes.1 In mice, gene targeting (GT) via homologous recombination (HR) in embryonic stem (ES) cells followed by the formation of germline chimeras has been a routine tool, leading to the production of numerous knockout animals to explore the gene functions in the particular processes of normal development and disease models.1,2 This classical approach of knockout production has been limited to mouse and rat,3,4 mainly due to the availability of pluripotent ES cell lines capable of whole animal production via germline transmission.
Engineered sequence-specific endonucleases, namely zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), have recently attracted considerable attention,5 because they can mediate high efficiencies of GT and thus allow for allelic alterations directly in developing embryos without the intermediate steps such as HR in ES cells, chimera formation from targeted ES cells and germline transmission, which are required in traditional GT experiments. ZFNs have successfully led to the targeted gene disruption (GD) in zebrafish and exogenous GD in medaka.6,7 More recently, TALENs have also been utilized successfully in zebrafish somatic cell cultures and embryos.8,9 These endonucleases can induce double-strand DNA breaks to increase the HR frequency, allowing for gene knockin directly in developing embryos.10 Recent progresses include the development of engineered zinc finger nickases inducing homology-directed repair with reduced mutagenic effects,11 selection-independent generation of gene knockout mouse ES cells using ZFNs12 and monomeric site-specific nucleases for genome editing.13
One of the challenges in ZFN- or TALEN-mediated GD experiments is the ability and efficiency to detect the desirable disrupted alleles, which are rare and usually contain minor differences compared with wild-type (WT) alleles,6,8 because ZFN- or TALEN-mediated GD may occur independently and generate various alterations in different cells and embryos and the individuals derived from them may be highly heterogeneous. These minor allelic alterations may be hardly detectable under the standard agarose gel electrophoresis (AGE). Traditionally, three methods are used to detect GD lesions without any requirement for special equipment. These are phenotype selection,6,14 restriction site selection8 and electrophoresis after enzymatic mismatch cleavage.15,16 Phenotype selection is not applicable to many genes that do not show an apparent phenotype after disruption. Restriction site selection requires a specific restriction site within the region of detection: upon disruption, a gene or its fragment may lose the recognition site for a restriction enzyme, leading to a change in the restriction pattern. Enzyme mismatch cleavage is a versatile approach that is widely used in mutation and polymorphism detection,17 because it enables the detection of any allelic variations independent of the corresponding phenotype and restriction site. Allelic sequences can form homoduplex (Hm) or heteroduplex (Ht) when they are subject to repeated cycles of denaturing and annealing as in PCR. The T4 endonuclease VII and T7 endonuclease I (T7) specifically recognize and cut an Ht DNA at the site of mismatch, generating two shorter DNA fragments detectable on ordinary AGE.18 This approach makes use of T7 digestion and AGE, and is thus referred to TAGE subsequently for simplicity.
Several other approaches of detecting minor sequence differences require the use of special equipment. Capillary electrophoresis19 uses particular devices and can identify single-molecule differences.20 High-resolution melting analysis21 has recently been used to detect chemically induced point mutations in medaka22 as well as DNA methylation.23 However, specialized instruments designed to monitor fluorescence changes during heating are required.
Polyacrylamide gel electrophoresis (PAGE) provides high-resolution detection of polymorphisms24 and it has also been used to study DNA mismatch repair.25 The usefulness of PAGE for detecting allelic alterations in GD experiments remains to be determined. This study was aimed at the development of procedures for the detection of allelic alterations at high efficiency and resolution. In a biallelic model system, TAGE and PAGE were found to be able to detect minor DNA differences at an efficiency of 6 and 0.4%, respectively. In a multi-allelic model system, PAGE but not TAGE allowed for rapid detection, quantification and enrichment of multiple rare alleles. Most importantly, in a real experiment for ZFN-mediated GT through direct microinjection in medaka embryos, PAGE proved its effectiveness to screen for GT at a particular locus.
2. Materials and Methods
2.1. Fish and chemicals
Work with fish followed the guidelines on the care and use of animals for scientific purposes of the National Advisory Committee for Laboratory Animal Research in Singapore and approved by this committee (permit number 27/09). Medaka (strains af, HB32C, HdrR, i1, i3 and Orange) was maintained under an artificial photoperiod of 14-h/10-h light/darkness at 26°C as described.26 Chemicals and enzymes were from Sigma-aldrich and Promega unless otherwise indicated.
2.2. Plasmids
Plasmid DNA was extracted by using the Qiagen Maxi and Midi preparation kits (Qiagen). WT1 and D18 are 321 and 303 bp in length, respectively, which were PCR-amplified by using primers ATGGTTGAGTCCCAATCTTTTG plus TCAATATCGCTCTGAAACCCAG from two plasmids containing the medaka nanog cDNAs. pWT2 contains a 346-bp insert of the first exon of medaka gsdf gene. The insert was PCR amplified by using primers GGTTCCCAGGCCCCGGGCTAGCAG plus GAAAAGGATCGTTGCAATTCGC and cloned in pGEM-T Easy vector. pD1, pD6, pD7, pA18 and pA3D7 were similar to pWT2 except for deletions (D) or additions (A) of 1, 6, 7 and 18 bp in the insert. These deletions or additions were generated from ZFN-mediated GD in medaka embryos (see below).
2.3. Genomic DNA extraction
Caudal fin clips were collected from growing or adult fish and incubated overnight at 55°C in 1.5-ml Eppendorf tubes containing 100 μl of lysis buffer (100 mM Tris–HCl, pH 8.0, 200 mM NaCl, 0.2% SDS, 5 mM EDTA, pH 8.0 and 100 μg/ml proteinase K). Following heat-inactivation of proteinase K for 10 min at 70°C, DNA was precipitated with 2.5 volume of ethanol and dissolved in 50 μl of TE (10 mM Tris–HCl, 1 mM EDTA, pH 8.0). DNA isolation from whole fish and embryos was performed as described.27
2.4. Genomic PCR and successive PCR
Genomic PCR was run in a 20-μl volume containing 1 μl of fin clips DNA, 50 ng of medaka genomic DNA or 50 pg of linearized plasmid DNA in the absence or presence of 50 ng of medaka genomic DNA for 35 cycles (94°C for 15 s, 60°C for 15 s and 72°C for 30 s) with Takara Ex-Taq DNA polymerase and the supplied buffer.
In the case of consecutive rounds of PCR for enrichment for particular fractions, 1 μl of the gel-purified PCR products was used as template for 30 cycles of PCR at the same conditions.
2.5. Heteroduplex formation
When DNA mixtures containing two or more different sequences are used as templates for PCR, heteroduplexes are formed automatically during cycles of denaturing and re-annealing.28 However, to ensure full heteroduplex formation or to generate heteroduplexes between PCR products from two separate reactions, a denaturing and re-annealing procedure was performed. PCR products or mixture of PCR products from separate reactions were heated to 94°C for 3 min and slowly returned to room temperature to form heteroduplexes.29
2.6. T7 endonuclease I digestion and agarose gel electrophoresis
For the traditional TAGE detection method, 3 μl of PCR product was subjected to digestion with 0.3 U T7 endonuclease I (New England BioLabs) in a 5-μl volume at 37°C for 1.5 h. Then, 5 μl of digested products, together with 3 μl undigested PCR product as control, were mixed with 5× loading dye, and was loaded on the 2% (w/v) agarose gel with 0.05 μg/ml of ethidium bromide to run the electrophoresis under 130 V for 15 min.
2.7. Polyacrylamide gel electrophoresis
For our PAGE detection method, 4 μl of PCR products were separated on 8% polyacrylamide gels in 1× TBE buffer using a Mini-Protean electrophoresis unit (Bio-Rad Laboratories) at 100 V for 2 h at room temperature. Gels were submerged in 1× TBE buffer containing 0.05 µg/ml of ethidium bromide for 20 min. Gels were documented on a bioimaging system (Vilber Lourmat).
2.8. Gel recovery, cloning and sequencing
DNA bands were cut from the gel under UV light and smashed in 10–20 μl TE. After incubation overnight at room temperature, the supernatant containing DNA was used for successive PCR (see above).
TA cloning was performed with pGEM-T Easy vectors. DNA mini-preps were prepared according to the standard alkaline lysis protocol30 and used for sequencing on the 3130xl Sequencer (Applied Biosystems). Sequence alignment and analysis were done by using Vector NTI and DNAman.
2.9. ZFN-mediated GD
Expression vectors pZN1gsdf and pZN2gsdf (see Supplementary data) were supplied commercially (Toolgene, South Korea). They encode a pair of ZFNs that target the first exon of the medaka gsdf gene (unpublished data). After linearization with ApaI, both vectors were used for mRNA synthesis by using the T7 RNA polymerase (Invitrogen) as described.31 The mRNAs were mixed at 1:1 ratio and microinjected at 20–100 ng/μl into embryos of af and HdrR medaka at the 1-cell stage as described.31 The injected embryos were analysed or allowed to develop into adulthood.
2.10. Step-by-step procedure
We are interested in versatile procedures for use in most ordinary laboratories without specialized equipment. Therefore, we developed a PAGE detection procedure, as is indicated in Fig. 1A, which can be streamlined as follows:
DNA extraction: extract and purify genomic DNA from gene-disrupted individuals.
Primers design: design a pair of primers flanking the ZFN/TALEN cleavage site to yield PCR products of ∼300 bp.
Genomic PCR: perform PCR with the DNA samples, one WT sample as negative control and one mixture of mutant and WT alleles as positive control if available.
- Heteroduplex formation:
- for heterozygous founder individuals containing disrupted alleles, heteroduplexes are formed during PCR. No additional step is required;
- for progenies that might be homozygous, each PCR product should be mixed with WT PCR product with a ratio of ∼1:1, heated to 94°C for 3 min and slowly returned to room temperature to form heteroduplexes.
- PAGE detection:
- prepare 8% polyacrylamide gels with 1× TBE buffer;
- 4–6 μl of PCR product is loaded with 6× loading dye to each well of the gel. Electrophoresis is carried out at 12 V/cm for ≥2 h;
- the gels are stained with 0.05 µg/ml ethidium bromide for 20 min and documented on a bio-imaging system.
Allelic alteration identification: bands of the sample are compared with those of the negative control. The presence of different bands from the control indicates allelic alterations. If positive control shows no heteroduplex band, experiment should be redone.
Repeat Steps 3–6 to reduce or eliminate false-positive samples.
- Gel recovery, cloning and sequencing:
- bands of heteroduplexes are recovered by smashing and soaking in 10–20 μl of TE in 37°C for overnight;32
- 1 μl of the recovered DNA are used as templates for one round of successive PCR of 30 cycles;
- the products are TA cloned into pGEM-T Easy vector. At least 10 colonies are picked to do mini-preps to collect plasmid DNA using the standard alkaline–SDS procedure;
- sequencing is run on the insert of the plasmid DNA and the result is compared with the WT allele to validate allelic variations.
Figure 1.
Heteroduplex production and detection by TAGE and PAGE. (A) Flowchart showing heteroduplex production by PCR and heteroduplex detection by PAGE and TAGE. (B and C) Biallelic heteroduplex model assay. Plasmids containing a WT allele (WT1) and a mutant allele with an 18-bp deletion (D18) were mixed with indicated dilution factor for PCR. (B) TAGE detection. D18 with a dilution factor of 16 can be detected. (C) PAGE detection. D18 with a dilution factor of 256 can be detected. Size markers in base pairs are indicated to the left. Arrowheads denote a double band. Asterisks depict heteroduplexes. Hm, homoduplex; Ht, heteroduplex; PAGE, polyacrylamide gel electrophoresis; T7, T7 endonuclease I; TAGE, T7-digestion and agarose gel electrophoresis. This figure appears in colour in the online version of DNA Research.
3. Results
3.1. Rationale and experimental design
Following the procedure above, after repeated PCR cycles of denaturing and annealing, genomic DNA containing gene-targeted alleles will generate three types of PCR products, namely, wild-type (WT) homoduplex (Hm), mutant (MT) Hm and WT–MT heteroduplex (Ht). These PCR products do not show a visible difference on AGE. In TAGE, Ht DNA fragments are cut by T7 at the mismatch site and become distinguishable from the uncut Hm molecules, which can be discriminated as distinct bands on AGE.
Alternatively, on native PAGE, such PCR products can be directly separated due to the differences in configuration and length. Since Ht shift slower than Hm on PAGE, Ht bands can be found clearly on the top of Hm bands, as an unambiguous indicator of successful GD. Moreover, after PAGE, all PCR products remain intact. Accordingly, Ht DNA fragments are recoverable from the gel for cloning and sequencing, or can be further re-amplified by consecutive rounds of PCR before cloning and sequencing. Therefore, PAGE features high resolution of separation and the integrity of DNA fragments, which should permit separation and quantification of each PCR product, and more importantly, selective enrichment of Ht DNA for analysis. Its quantification ability leads to an estimation of GD proportion in a sample, providing information for deciding the sample size for screening by cloning and sequencing. Its enrichment ability reduces the WT alleles by eliminating Hm fractions, facilitating the identification of GD-containing samples via reducing the workload of cloning and sequencing in each sample.
3.2. Biallelic model
The procedures and the usefulness of TAGE and PAGE were first established and evaluated for Ht detection in a biallelic model system. To this end, two plasmids, WT1 and D18, which contain an insert of 321 and 303 bp as the WT allele and 18-bp deletion allele (thus D18) of the medaka nanog gene respectively, were linearized and mixed to form a series of dilution (D18 diluted by WT1 at a factor of 2–256). The serial dilution mixtures were used as the templates for PCR and PCR products were subjected to TAGE and PAGE detection.
On TAGE, Hm WT1 and D18 fragments remained intact and appeared as bands that were slightly different in size (Fig. 1B). When mixed at a 1:1 ratio, WT1 and D18 without T7 cleavage produced a double band and, upon T7 cleavage, formed two distinct bands with a similar intensity. Serial dilution revealed that D18 remained detectable at a dilution factor of 16, producing a sensitivity level of ∼6.3% for TAGE in Ht detection (Fig. 1B). In consistence with the principle of TAGE, T7 digestion leads to reduced band intensities. These observations reveal the feasibility and sensitivity of the established TAGE procedure for Ht detection. On the other hand, in the PAGE detection procedure, D18 remained detectable even at a dilution factor of 256 (Fig. 1C). This produces a sensitivity level of ∼0.4% for PAGE in Ht detection, which is 16-fold more efficient than TAGE.
A closer inspection revealed several advantageous features of PAGE procedure for detecting allelic modifications (Fig. 1C). WT1 and D18 Hm fragments formed distinct bands of sufficient difference in size. Moreover, Ht fragments migrated at a much lower rate than Hm fragments and formed bands with a clear distance from Hm DNAs. As illustrated in Fig. 1A, two alleles will form two different configurations of Ht fragments depending on the involvement of top or bottom strands of WT or MT alleles.33 In our biallelic model, WT1 and D18 formed two configurations, which were resolved as two distinct bands on PAGE (Fig. 1C). Consequently, PCR products from a biallelic DNA template give rise to four distinct bands, compared with two visible bands in TAGE (compare Fig. 1B and C). In addition, Ht fragments on TAGE formed a faint band that was embedded in a smear at a dilution factor of 16 (Fig. 1B), compared with clear bands with little background even at a dilution factor of 256 (Fig. 1C). Most importantly, upon increasingly serial dilution, the Hm fragment of D18 as the rare allele became invisible but contributed to Ht formation, thus increasing the detection sensitivity.
To mimic the real situation of detecting allelic alterations in a targeted genome, the serially diluted mixtures of linearized WT1 and D18 (cDNA original) were added to medaka genomic DNA and subjected to analyses by PCR and PAGE. As illustrated in Supplementary Fig. S1, addition of af genomic DNA did not interfere with the formation of Ht fragments and their detection by PAGE, because the band pattern and detectable dilution factor remained unchanged with genomic DNA (compare Fig. 1C with Supplementary Fig. S1). Taken together, in a biallelic model system, TAGE is able to detect a subtle allelic alteration at a limited resolution and sensitivity of 6.3%, whereas PAGE offers high resolution and sensitivity of 0.4% in the absence and presence of genomic DNA.
It needs to be noted that, allelic differences, either natural polymorphisms or experimentally induced alterations, will generate Ht PCR products and Ht bands on PAGE. To understand the impact of sequence polymorphisms on our PAGE procedure, we tested the PCR products of gsdf locus from two medaka inbred lines along with our gene targeted fish 4 (see below) in three lengths: 572, 372 and 305 bp. Primers used are listed in Supplementary Fig. S2. The result indicates that PCR products of af sample exhibit more natural polymorphism Ht bands (asterisks) than HdrR in size of 572 and 372 bp (Supplementary Fig. S2C). Following the PAGE recovery procedure provided above, we sequenced the asterisked Ht and discover a 10 bp deletion polymorphism in af (Supplementary Fig. S2B). By shortening the PCR product size to bypass the polymorphism site (Supplementary Fig. S2B), af shows only faint background bands. Meanwhile, fish 4 PCR products of both sizes exhibit additional bands, which are generated from GT. Therefore, a WT sample serving as a negative control is required in every batch of our PAGE detection.
3.3. Multi-allelic model system
In the practice of ZFN-mediated GT experiments, additions and deletions may occur independently at different stages of development and in different cells, leading to the production of various different MT alleles. We furthered our experiments to establish and utilize a multi-allelic model system for the detection by PAGE. To this end, a set of five different alleles were produced within exon 1 of the medaka gsdf gene via ZFN-mediated GT in medaka embryos (see below). They were WT2, A18, D7, D6 and D1 (Fig. 2A). WT2 of 320 bp in length is the WT allele of gsdf (WT1 described above is the WT allele of the medaka nanog). These allelic sequences were cloned in pGEM-T Easy, and linearized plasmids were mixed at various ratios as described in Fig. 2B. As expected, PCR produced a single band on PAGE when DNA containing a single plasmid was used as template (lanes 1–5, Fig. 2B), while allelic variations, as tiny as 1-bp difference, were clearly distinguishable on PAGE as Hm and Ht forms (lanes 6, 7, 9, 11, Fig. 2B). Notably, PAGE is capable of simultaneous detection of multiple alleles as distinct bands (lanes 8, 10, 12, 13, Fig. 2B). To confirm that the Ht bands are not from artificial non-specific PCR products, PCR products manifesting single band in lanes 1–5 of Fig. 2B were mixed at the same ratio as indicated in Fig. 2B and then were subjected to a heteroduplex formation procedure. PAGE profiles (Supplementary Fig. S3) of these mixtures show exactly the same patterns as in Fig. 2B.
Figure 2.
PAGE detection of multiple alleles. (A) Partial sequences of five different alleles of the medaka gsdf gene. Additions are shown as a loop. Deletions are depicted by gaps. Bold letters indicate the cleavage site of ZFN. (B) PAGE profiles of different mixtures of the alleles. Mix ratios of the alleles are indicated above lanes. Dashed lines delineate the same heteroduplexes cross lanes. Asterisks highlight heteroduplexes. (C) Correlation between the number of alleles and total number of different PCR products which may show different bands on PAGE. Colour background depicts the positive correlation between the concentrations of each DNA strands and the corresponding band intensities. Capital letters, upper strands; small capital case letters, lower strands; underlining, homoduplexes; Hm, homoduplex; Ht, heteroduplex. This figure appears in colour in the online version of DNA Research.
A closer inspection established a correlation between the total number of bands and the total number of alleles (Fig. 2B and C): a single allele produces one PCR product (lanes 1–5), two alleles result in four products (lanes 6, 7, 9 and 11), and three alleles give rise to nine products which form six to eight distinctive bands on PAGE depending on the relative intensity and inter-band distance (lanes 8 and 10). This correlation is formulated as follows:
| (1) |
| (2) |
where N is the number of different alleles and B is the number of Ht bands. Since values are integers in our case, for shorting the calculation, Equation (2) can be simplified with a ceiling function which leads to the smallest following integer:
| (3) |
Therefore, the total number of alleles in a sample can be easily estimated by Ht band number without solving an equation. This trait of PAGE allows us to intuitively observe the main composition of the sample.
In case of multiple alleles, the number of Hm PCR products is the same as the number of alleles (Fig. 2C, underlined), and the remainder is Ht PCR products (Fig. 2C, not underlined). Abundances of these Ht products show a positive correlation with the corresponding allelic abundance as depicted by the colour gradient. Therefore, not all Ht bands are visible, and the number of Ht bands on PAGE will indicate the minimal number of allelic alterations in a sample.
3.4. Enrichment of rare alleles
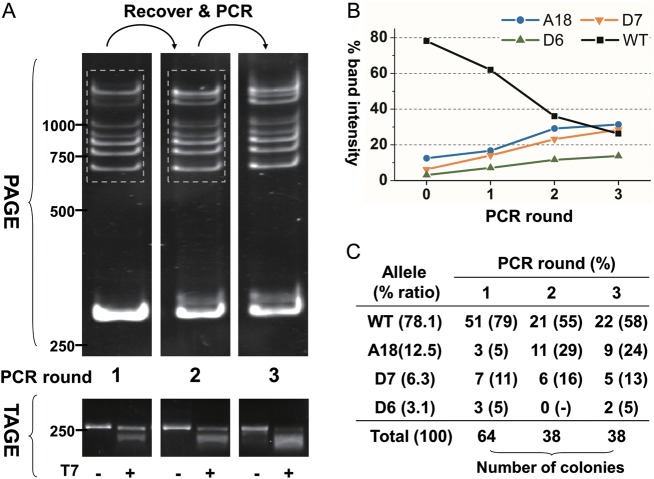
A final proof for targeted alterations in a ZFN-treated sample comes from sequence analysis. ZFN-mediated GT has been reported to occur in 1–3% cells of zebrafish embryos,34,35 where only one gene copy is targeted in most cases. Consequently, the targeted allele is usually present at ∼1% in total genomic DNA from a positive sample. Accordingly, the validation of GT is a time-consuming task in a ZFN-mediated GT experiment, because it requires preparing and sequencing several hundreds of recombinant colonies.34,35 The high resolution and band distinctness of intact PCR products on PAGE provoked us to develop a procedure of enriching for rare alleles, which is gel recovery and successive PCR (grsPCR). In grsPCR, Ht bands of first-round PCR are gel recovered, and DNA is released by diffusion from gel slices for use as template for successive rounds of PCR. To test this procedure, a DNA mixture of four alleles at defined ratio (column 1, Fig. 3C) was used for first round of PCR; Ht fractions were gel recovered collectively, and DNA released was used for second and third rounds of PCR (Fig. 3A), and PCR products were quantified by densitometry (Fig. 3B) and cloned for validation by sequencing (Fig. 3C). Upon two to three rounds of PCR, an increase in Ht fractions was indeed apparent on both PAGE and TAGE when compared with the Hm fractions (Fig. 3A). To quantify a change in intensity during grsPCR, various bands were defined and measured by densitometry (Supplementary Fig. S4A and B). According to the band patterns of this four-allelic model system as shown in Fig. 2B, major bands of the four alleles are identifiable, which permits intensity quantifications of the four alleles by combining two bands of each allele (Supplementary Fig. S4C). In this way, the intensity of dominant WT allele was found to be reduced considerably from 62.0% in the first PCR over 36.0–26.3%, while intensities of rare alleles increased after one and two rounds of grsPCR (Fig. 3B and Supplementary Fig. S4C). Cloning and sequencing led to a similar enrichment factor (Fig. 3C). After one single round of grsPCR, for example, the WT allele decreased from 79 to 55%, and rare alleles increased relatively. This verifies quantitative enrichment for rare alleles. Taken together, PAGE profile enables the quantification of each component, while grsPCR allows for the enrichment of multiple rare alleles.
Figure 3.
PAGE detection, enrichment and quantification of multiple alleles. (A) PAGE and TAGE profiles of multiple alleles after successive rounds of PCR following gel recovery (dash frame). DNA mixture containing four alleles shown in lane 12 of Fig. 2B was used as template with mix ratios indicated in (C). (B) Relative intensity of different bands of each round. Round ‘0’ represent the initial percentages of each allele. (C) The number of clones and percentages of each allele from PCR products of each round. This figure appears in colour in the online version of DNA Research.
3.5. Experimental ZFN-mediated GT system
The experiments described so far dealt with model systems by using linearized plasmids mixed at defined ratios as templates for PCR and analyses via TAGE and PAGE. We wanted to demonstrate the usefulness of PAGE in a real experiment of direct GT in embryos of a vertebrate. We chose the ZFN approach and medaka as the model organism. Medaka is a laboratory fish and a lower vertebrate model for stem cell biology27,36,37 and germ cell biology.31,38 Its genome has been sequenced (http://www.ensembl.org/index.html). There are many inbred lines/strains (http://biol1.bio.nagoya-u.ac.jp:8000/) and experimental tools including embryo microinjection.31,39,40
We are interested in the development of approaches for direct GT by using ZFNs and TALENs in medaka. By microinjection of RNAs for a pair ZFNs into embryos of af medaka, we obtained several targeted alleles at the medaka gsdf, a gene whose RNA expression is spatially and temporally correlated with early testicular differentiation.41 Four of the alleles were used in the multi-allelic model assay described above (Figs 2 and 3). More than hundred of fish derived from ZFN-injected embryos were grown into adulthood. Genomic DNA was extracted from fin clips of a subset of samples and subjected to TAGE and PAGE assays. We detected successful GT in 2 out of 48 fish examined, because the two fish (4 and 6) exhibited clear bands of Ht PCR products on PAGE (Fig. 4A), and a smear or faint band indicative of Ht products on TAGE (Fig. 4B). The band patterns of two fish samples on PAGE and TAGE were similar to that observed in model systems (Figs 1 and 2). Sequencing 72 recombinant colonies of the Ht PCR products from the third-round PCR (a total of two rounds of grsPCR) revealed three different alleles. One is the WT allele that was present in 45 clones (63%), and the remainder is targeted alleles D7 (22%) and A3D7 (15%; Supplementary Fig. S5). Interestingly, allele A3D7 has both a 3-bp addition and a 7-bp deletion, and more intriguingly, also four mismatches of 1–2 bp each at three separate positions. It follows that ZFNs can simultaneously introduce addition, deletion and mutation into one and same targeted allele. Hence, ZFN-mediated endogenous GT is possible in medaka embryos, and PAGE is effective in detecting and cloning ZFN-targeted rare alleles in fish.
Figure 4.
Detection of ZFN-mediated allelic alterations in medaka. Medaka embryos were microinjected with RNAs for a pair of ZFNs that targeted the first exon of gsdf locus, and were allowed to develop into adult animals. Genomic DNA was extracted from fin clips of eight randomly sampled adults and was subjected to PCR and PAGE detection of targeted allelic alterations. (A) PAGE profiles of PCR products, unambiguously showing the presence of heteroduplex DNA (asterisks) in two out of eight fish. (B) TAGE profile of PCR products, showing a faint band of heteroduplex DNA (asterisks) within smears in the same two fish.
4. Discussion
The introduction of engineered ZFNs6,42 and TALENs8,9 has made it possible to achieve GT directly in developing animal embryos and selection-independent GT in mouse ES cells.12 The ability to efficiently identify subtle allelic alterations is essential in these ZFN- and TALEN-mediated GT experiments. In this study, we have developed TAGE and PAGE in model systems and a real GT experiment as versatile procedures for detecting allelic variations in ordinary laboratories without specialized equipment and devices. Under our conditions, TAGE identifies the digests of mismatch-containing Ht fragment as a clear band when the rare allele is present ≥6.3% in a biallelic model system, but as a faint band within a smear in DNA containing ZFN-altered alleles. These results demonstrate the ability and limit of TAGE in allelic detection, as has well be documented.17
We showed that PAGE is of choice because of its five major advantages over TAGE. First, PAGE is straightforward and does not require any additional step of enzymatic treatment of PCR products, facilitating the cloning of wanted PCR products by gel recovery. Secondly, PAGE is capable of detecting rare alleles as low as 0.4%, sensitivity of which is 16 times higher than TAGE. Thirdly, PAGE provides an extremely high resolution, as desirable allelic alterations are present in heteroduplexes that form distinct bands with large distances from the bulk WT homoduplex, which enables the simultaneous identification and quantification of multiple different alterations. Fourthly, PAGE allows for discrimination between Ht from natural polymorphisms and those introduced by GD. Finally, on the basis of high resolution and band distinctness of intact PCR products on PAGE, we have developed a gel recovery and successive PCR procedure, which, for the first time to our knowledge, enables enrichment for rare alleles. The percentage of the rare alleles could reach 37% in our experiment (Supplementary Fig. S5); hence, only dozens of recombinant colony sequencing are needed to detect the desirable rare alleles, considerably reducing the work load compared with previous studies when hundreds of colonies were sequenced.34,35
PAGE has been used for heteroduplex analysis for years with well-established model systems.28,29,33,43 Based on these previous reports and our experience, for PAGE detection, we suggest that the design of PCR primers amplifying fragments around 300 bp, which can save the time of electrophoresis and avoid polymorphisms while containing enough sequence information. Natural polymorphisms will exhibit Ht bands, thus it is necessary to add one sample of the WT allele in every batch of our PAGE detection as negative control. Only those showing different patterns or additional bands compared with the WT sample can be deemed as positive ones. This represents one of the merits of the PAGE method because in the T7 detection method, polymorphisms cannot be distinguished from introduced mutation since they show similar shortened bands. Also the PAGE method provides a tool to survey the site of interest for natural polymorphisms, revealing the suitability of each animal population for such GT experiments.
As for the detection for progenies of GD individuals, which might contain homologous disrupted sequence, a heteroduplex formation procedure after mixing with WT PCR product is necessary to avoid omission. In addition, our protocol also suggests repeating the detection for positive samples to rule out false-positive ones.
Since PAGE is only a quick screening method, we might overlook certain one-base-deletion or point mutation in our sample. For point mutation, previous research using ‘hydrolink mutation detection enhancement gels’ cannot detect all types of the mutation via heteroduplex analysis.33 However, almost all published cases of deletion or addition occur in ZFN/TALEN-mediated GT show disruption with more than 1 bp.6–8,34,35,42 These large lesions can be detected by our PAGE method clearly.
Our first trial of ZFN-mediated GT in medaka has led to the production and identification of several different alleles from a limited number of embryos and growing animals, revealing the functionality of ZFNs used and the efficiency of this ZFN-mediated GT approach in this organism. It needs to be noted that ZFNs are capable of simultaneous introduction of addition, deletion and point mutation in a single allele, pointing the proficiency of ZFNs in producing a wide variety of targeted allelic alterations in medaka.
Although developed for analysing experimentally induced minor sequence differences, our procedures, PAGE in particular, will find a wide variety of applications in detection, quantification and enrichment of polymorphisms and alterations in basic and applied research as well as medicine.
Authors’ contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the National Research Foundation of Singapore under its Competitive Research Program (NRF-CRP7-2010-03), the Grants-in-Aid from Japan Society for the Promotion of Science (50113428) and the Biomedical Research Council of Singapore (R154-000-427-305). We acknowledge NUS scholarship to J. B. Chen, X. Zhang and T. S. Wang.
Supplementary Material
Acknowledgements
We thank Jiaorong Deng for fish breeding, Foong Choy Mei and Wong Kway Yip Veronica for laboratory management.
Footnotes
Edited by Osamu Ohara
References
- 1.Capecchi M.R. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–12. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2.Wang R.H., Sengupta K., Li C., et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–23. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins F.S., Rossant J., Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Tong C., Li P., Wu N.L., Yan Y., Ying Q.-L. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–3. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark K.J., Voytas D.F., Ekker S.C. A TALE of two nucleases: gene targeting for the masses? Zebrafish. 2011;8:147–9. doi: 10.1089/zeb.2011.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyon Y., McCammon J.M., Miller J.C., et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechol. 2008;26:702–8. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ansai S., Ochiai H., Kanie Y., et al. Targeted disruption of exogenous EGFP gene in medaka using zinc-finger nucleases. Dev. Growth Differ. 2012;54:546–56. doi: 10.1111/j.1440-169X.2012.01357.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang P., Xiao A., Zhou M., Zhu Z., Lin S., Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 9.Sander J.D., Cade L., Khayter C., et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 2011;29:697–8. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson R.D., Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez C.L., Certo M.T., Mussolino C., et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–8. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osiak A., Radecke F., Guhl E., et al. Selection-independent generation of gene knockout mouse embryonic stem cells using zinc-finger nucleases. PLoS One. 2012;6:e28911. doi: 10.1371/journal.pone.0028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleinstiver B.P., Wolfs J.M., Kolaczyk T., Roberts A.K., Hu S.X., Edgell D.R. Monomeric site-specific nucleases for genome editing. Proc. Natl Acad. Sci. USA. 2012;109:8061–6. doi: 10.1073/pnas.1117984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takasu Y., Kobayashi I., Beumer K., et al. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem. Mol. Biol. 2010;40:759–65. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–88. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeder M.L., Thibodeau-Beganny S., Osiak A., et al. Rapid ‘open-source’ engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babon J., McKenzie M., Cotton R. The use of resolvases T4 endonuclease VII and T7 endonuclease I in mutation detection. Mol. Biotechnol. 2003;23:73–81. doi: 10.1385/MB:23:1:73. [DOI] [PubMed] [Google Scholar]
- 18.Young L., Dong Q. Two-step total gene synthesis method. Nucleic Acids Res. 2004;32:e59. doi: 10.1093/nar/gnh058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlowski P., Krzyzosiak W.J. Combined SSCP/duplex analysis by capillary electrophoresis for more efficient mutation detection. Nucleic Acids Res. 2001;29:e71. doi: 10.1093/nar/29.14.e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagally E.T., Medintz I., Mathies R.A. Single-molecule DNA amplification and analysis in an integrated microfluidic device. Anal. Chem. 2001;73:565–70. doi: 10.1021/ac001026b. [DOI] [PubMed] [Google Scholar]
- 21.Wittwer C.T., Reed G.H., Gundry C.N., Vandersteen J.G., Pryor R.J. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003;49:853–60. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa T., Kamei Y., Otozai S., et al. High-resolution melting curve analysis for rapid detection of mutations in a Medaka TILLING library. BMC Mol. Biol. 2010;11:70. doi: 10.1186/1471-2199-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez C.M.R., Asenjo B.G., Lloyd A.J., Wilkinson M.J. Direct detection and quantification of methylation in nucleic acid sequences using high-resolution melting analysis. Anal. Chem. 2010;82:9100–8. doi: 10.1021/ac1024057. [DOI] [PubMed] [Google Scholar]
- 24.Walton K.E., Styer D., Gruenstein E.I. Genetic polymorphism in normal human fibroblasts as analyzed by two-dimensional polyacrylamide gel electrophoresis. J. Biol. Chem. 1979;254:7951–60. [PubMed] [Google Scholar]
- 25.Li G.M., Wang H., Romano L.J. Human MutSalpha specifically binds to DNA containing aminofluorene and acetylaminofluorene adducts. J. Biol. Chem. 1996;271:24084–8. [PubMed] [Google Scholar]
- 26.Hong N., Li Z., Hong Y. Fish stem cell cultures. Int. J. Biol. Sci. 2011;7:392–402. doi: 10.7150/ijbs.7.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Y., Winkler C., Schartl M. Production of medakafish chimeras from a stable embryonic stem cell line. Proc. Natl Acad. Sci. USA. 1998;95:3679–84. doi: 10.1073/pnas.95.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruano G., Kidd K.K. Modeling of heteroduplex formation during PCR from mixtures of DNA templates. PCR Methods Appl. 1992;2:112–6. doi: 10.1101/gr.2.2.112. [DOI] [PubMed] [Google Scholar]
- 29.Highsmith W.E., Jr., Jin Q., Nataraj A.J., et al. Use of a DNA toolbox for the characterization of mutation scanning methods. I: construction of the toolbox and evaluation of heteroduplex analysis. Electrophoresis. 1999;20:1186–94. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1186::AID-ELPS1186>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Birnboim H.C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–23. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., Hong N., Xu H., et al. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009;126:366–381. doi: 10.1016/j.mod.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D.W. Isolation of DNA fragments from polyacrylamide gels by the crush and soak method. Cold Spring Harb Protoc. 2006 doi: 10.1101/pdb.prot100479. doi:10.1101/pdb.prot2936. [DOI] [PubMed] [Google Scholar]
- 33.Nataraj A.J., Olivos-Glander I., Kusukawa N., Highsmith W.E., Jr. Single-strand conformation polymorphism and heteroduplex analysis for gel-based mutation detection. Electrophoresis. 1999;20:1177–85. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1177::AID-ELPS1177>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Foley J.E., Maeder M.L., Pearlberg J., Joung J.K., Peterson R.T., Yeh J.R.J. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat. Protoc. 2009;4:1855–68. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng X.D., Noyes M.B., Zhu L.H.J., Lawson N.D., Wolfe S.A. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong Y., Winkler C., Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes) Mech. Dev. 1996;60:33–44. doi: 10.1016/s0925-4773(96)00596-5. [DOI] [PubMed] [Google Scholar]
- 37.Yi M., Hong N., Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430–3. doi: 10.1126/science.1175151. [DOI] [PubMed] [Google Scholar]
- 38.Hong Y., Liu T., Zhao H., et al. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc. Natl Acad. Sci. USA. 2004;101:8011–6. doi: 10.1073/pnas.0308668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong N., Li M., Zeng Z., et al. Accessibility of host cell lineages to medaka stem cells depends on genetic background and irradiation of recipient embryos. Cell. Mol. Life Sci. 2010;67:1189–202. doi: 10.1007/s00018-009-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu T.M., Liu L., Wei Q.W., Hong Y.H. Sperm nuclear transfer and transgenic production in the fish Medaka. Int. J. Biol. Sci. 2011;7:469–75. doi: 10.7150/ijbs.7.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata Y., Paul-Prasanth B., Suzuki A., et al. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns. 2010;10:283–9. doi: 10.1016/j.gep.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Cui X.X., Ji D.N., Fisher D.A., Wu Y.M., Briner D.M., Weinstein E.J. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 2011;29:64–7. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 43.Rossetti S., Corra S., Biasi M.O., Turco A.E., Pignatti P.F. Comparison of heteroduplex and single-strand conformation analyses, followed by ethidium fluorescence visualization, for the detection of mutations in four human genes. Mol. Cell. Probes. 1995;9:195–200. doi: 10.1006/mcpr.1994.0028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.