Abstract
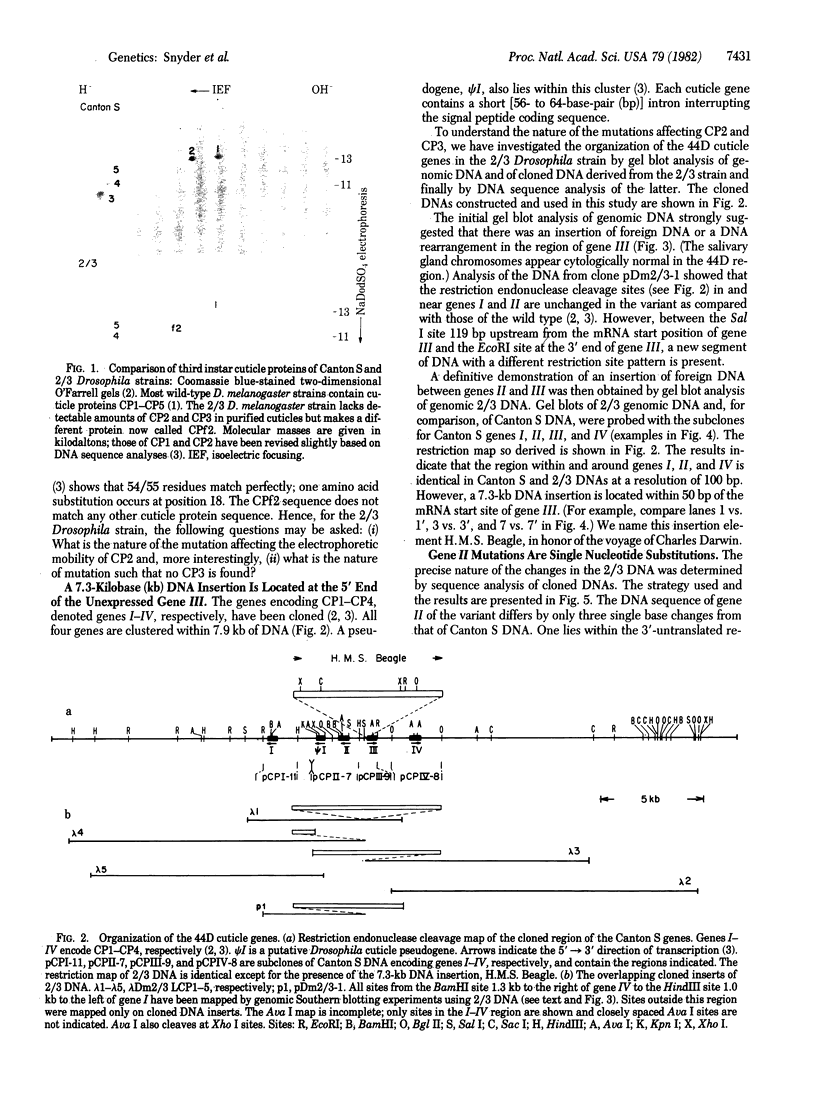
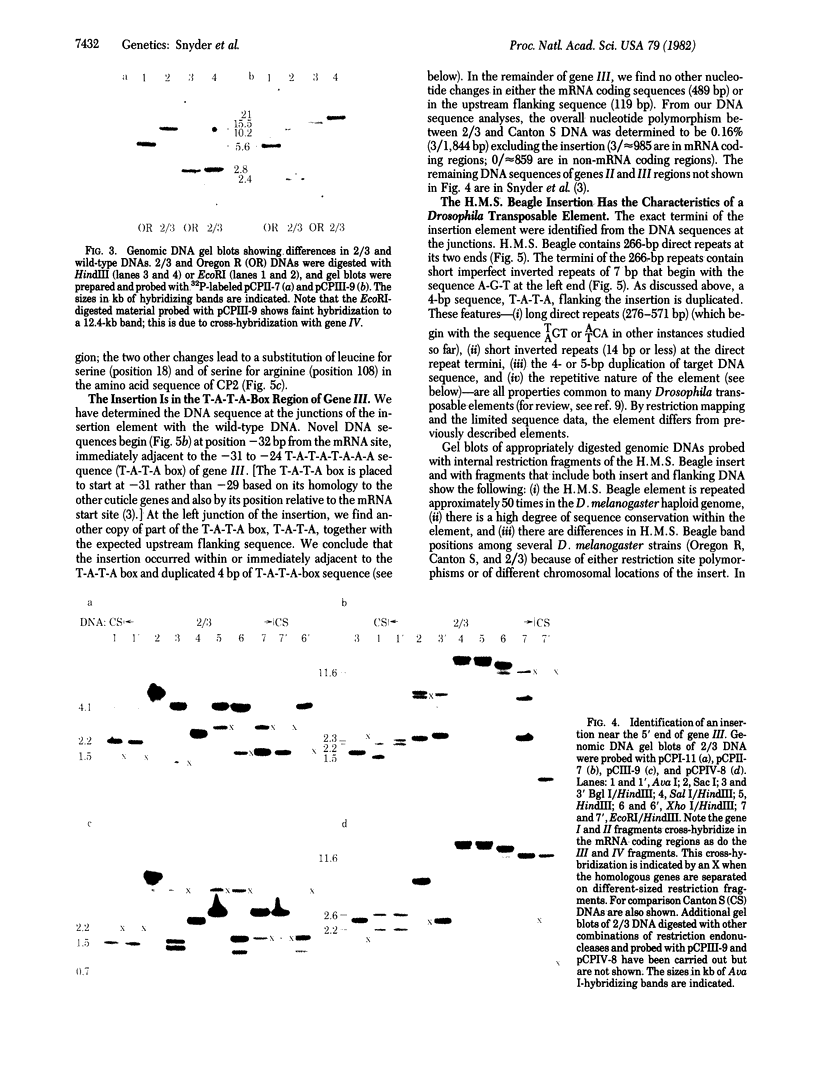
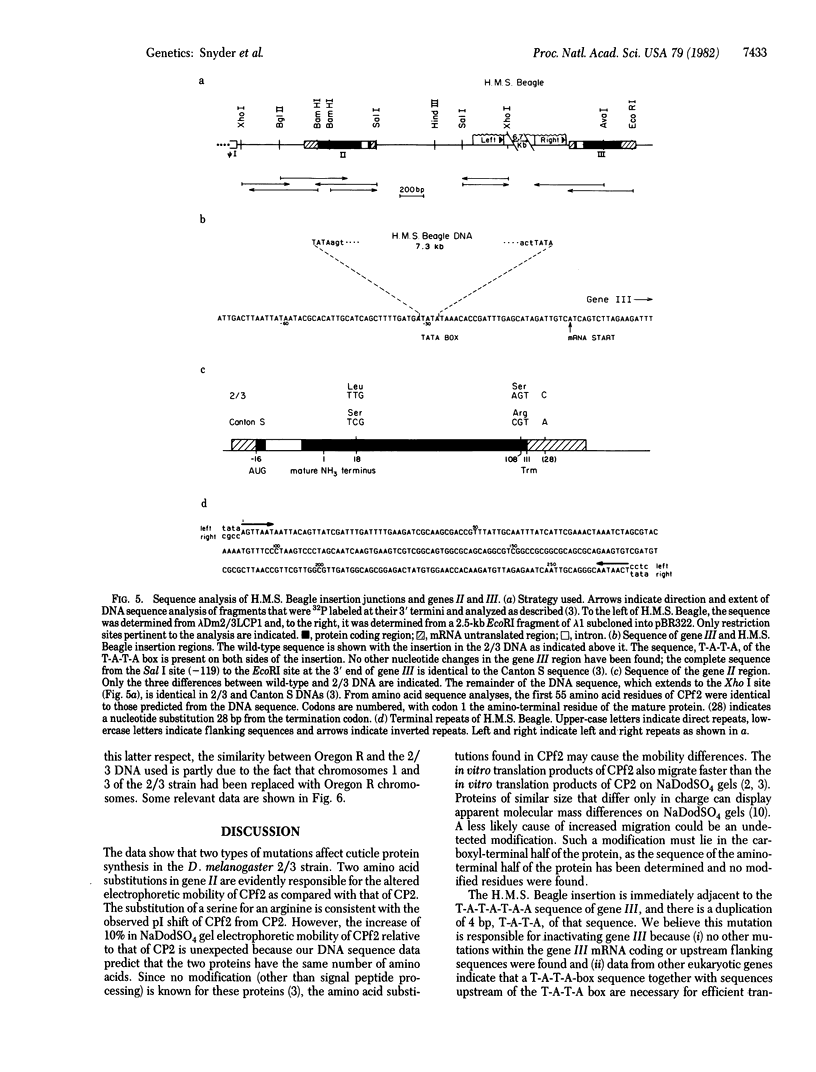
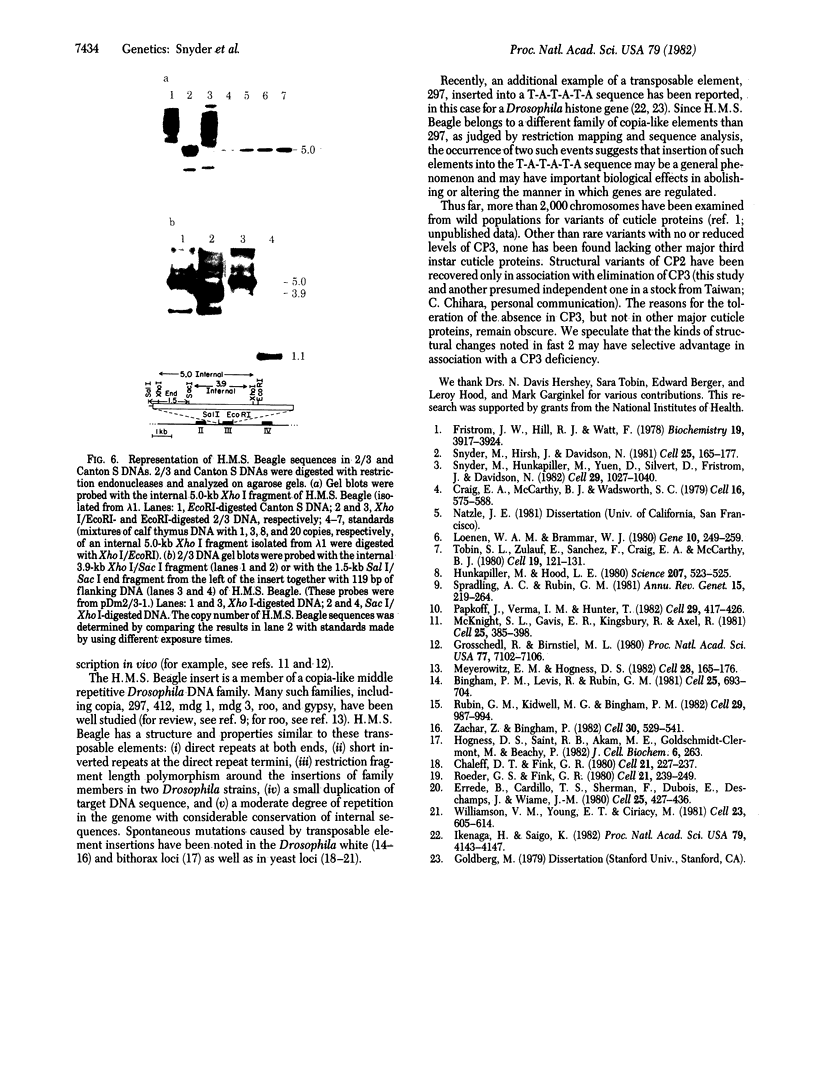
Two mutations that affect larval cuticle protein gene expression in the 2/3 variant Drosophila melanogaster strain were investigated. We demonstrate that this strain synthesizes an electrophoretic variant, fast 2 (CPf2), of wild-type cuticle protein 2(CP2). It also lacks detectable amounts of cuticle protein 3 (CP3). The other major cuticle proteins are still present. Protein and DNA sequence analyses indicate that point mutations cause two amino acid substitutions that change the electrophoretic mobility of CPf2 relative to that of CP2. The mutation abolishing the expression of CP3 was found to be a 7.3-kilobase DNA insertion located within the T-A-T-A box region of this gene, at -31 base pairs from the mRNA start site. This DNA insertion, called H.M.S. Beagle, belongs to a conserved family of repeated DNA elements that have characteristics similar to those of previously characterized Drosophila transposable elements. H.M.S. Beagle elements are repeated approximately 50 times in the haploid genome and exhibit restriction fragment-length polymorphisms around points of insertion between Canton S, Oregon R, and 2/3 Drosophila strains. Sequence analysis indicates that H.M.S. Beagle contains 266-base-pair direct repeats at its termini and is flanked by a duplication of 4 base pairs of target DNA sequence, T-A-T-A, in the CP3 gene insertion. Thus, insertion of a transposable element into the putative promoter region of the CP3 gene is evidently responsible for inactivating CP3 gene expression.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Chaleff D. T., Fink G. R. Genetic events associated with an insertion mutation in yeast. Cell. 1980 Aug;21(1):227–237. doi: 10.1016/0092-8674(80)90130-0. [DOI] [PubMed] [Google Scholar]
- Craig E. A., McCarthy B. J., Wadsworth S. C. Sequence organization of two recombinant plasmids containing genes for the major heat shock-induced protein of D. melanogaster. Cell. 1979 Mar;16(3):575–588. doi: 10.1016/0092-8674(79)90031-x. [DOI] [PubMed] [Google Scholar]
- Errede B., Cardillo T. S., Sherman F., Dubois E., Deschamps J., Wiame J. M. Mating signals control expression of mutations resulting from insertion of a transposable repetitive element adjacent to diverse yeast genes. Cell. 1980 Nov;22(2 Pt 2):427–436. doi: 10.1016/0092-8674(80)90353-0. [DOI] [PubMed] [Google Scholar]
- Fristrom J. W., Hill R. J., Watt F. The procuticle of Drosophila: heterogeneity of urea-soluble proteins. Biochemistry. 1978 Sep 19;17(19):3917–3930. doi: 10.1021/bi00612a005. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. New protein sequenator with increased sensitivity. Science. 1980 Feb 1;207(4430):523–525. doi: 10.1126/science.7352258. [DOI] [PubMed] [Google Scholar]
- Ikenaga H., Saigo K. Insertion of a movable genetic element, 297, into the T-A-T-A box for the H3 histone gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4143–4147. doi: 10.1073/pnas.79.13.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Gavis E. R., Kingsbury R., Axel R. Analysis of transcriptional regulatory signals of the HSV thymidine kinase gene: identification of an upstream control region. Cell. 1981 Aug;25(2):385–398. doi: 10.1016/0092-8674(81)90057-x. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E. M., Hogness D. S. Molecular organization of a Drosophila puff site that responds to ecdysone. Cell. 1982 Jan;28(1):165–176. doi: 10.1016/0092-8674(82)90386-5. [DOI] [PubMed] [Google Scholar]
- Papkoff J., Verma I. M., Hunter T. Detection of a transforming gene product in cells transformed by Moloney murine sarcoma virus. Cell. 1982 Jun;29(2):417–426. doi: 10.1016/0092-8674(82)90158-1. [DOI] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. DNA rearrangements associated with a transposable element in yeast. Cell. 1980 Aug;21(1):239–249. doi: 10.1016/0092-8674(80)90131-2. [DOI] [PubMed] [Google Scholar]
- Rubin G. M., Kidwell M. G., Bingham P. M. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982 Jul;29(3):987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Snyder M., Hirsh J., Davidson N. The cuticle genes of drosophila: a developmentally regulated gene cluster. Cell. 1981 Jul;25(1):165–177. doi: 10.1016/0092-8674(81)90241-5. [DOI] [PubMed] [Google Scholar]
- Snyder M., Hunkapiller M., Yuen D., Silvert D., Fristrom J., Davidson N. Cuticle protein genes of Drosophila: structure, organization and evolution of four clustered genes. Cell. 1982 Jul;29(3):1027–1040. doi: 10.1016/0092-8674(82)90466-4. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Tobin S. L., Zulauf E., Sánchez F., Craig E. A., McCarthy B. J. Multiple actin-related sequences in the Drosophila melanogaster genome. Cell. 1980 Jan;19(1):121–131. doi: 10.1016/0092-8674(80)90393-1. [DOI] [PubMed] [Google Scholar]
- Williamson V. M., Young E. T., Ciriacy M. Transposable elements associated with constitutive expression of yeast alcohol dehydrogenase II. Cell. 1981 Feb;23(2):605–614. doi: 10.1016/0092-8674(81)90156-2. [DOI] [PubMed] [Google Scholar]
- Zachar Z., Bingham P. M. Regulation of white locus expression: the structure of mutant alleles at the white locus of Drosophila melanogaster. Cell. 1982 Sep;30(2):529–541. doi: 10.1016/0092-8674(82)90250-1. [DOI] [PubMed] [Google Scholar]