Abstract
The huge amount of information that needs to be assimilated in order to keep pace with the continued advances in modern medical practice can form an insurmountable obstacle to the individual clinician. Within radiology, the recent development of quantitative imaging techniques, such as perfusion imaging, and the development of imaging-based biomarkers in modern therapeutic assessment has highlighted the need for computer systems to provide the radiological community with support for academic as well as clinical/translational applications. This article provides an overview of the underlying design and functionality of radiological decision support systems with examples tracing the development and evolution of such systems over the past 40 years. More importantly, we discuss the specific design, performance and usage characteristics that previous systems have highlighted as being necessary for clinical uptake and routine use. Additionally, we have identified particular failings in our current methodologies for data dissemination within the medical domain that must be overcome if the next generation of decision support systems is to be implemented successfully.
What is a decision support system?
Decision support systems (DSS) are a set of manual or computer-based tools that assist in some decision-making activity. In today's information-driven environment, DSS are commonly understood to be a variety of computerised information management systems, designed to help resolve complicated problems and/or questions by supporting the decision-making process. DSS are gaining increasing popularity in various domains including business, engineering, the military and medicine. These systems are especially valuable in situations where the amount of available information is prohibitive for the intuition of a human decision maker and where precision and optimal performance are of importance [1].
Medical decision support systems
The huge amount of information that needs to be assimilated in order to keep pace with continued advances in modern medical practice can form an insurmountable obstacle to the individual clinician. In medical applications decision quality is of crucial importance, whilst human decision-making performance can be suboptimal and deteriorate as the complexity of the problem increases. For these reasons, the development of medical DSS is becoming increasingly important [1] and the routine uptake of these “intelligent” systems is becoming more common [2]. One of the earliest rule-based expert systems, DENDRAL [3], was implemented in the 1960s and was designed to provide support to organic chemists. This was further developed over the early 1970s by the same team at Stanford University into arguably the first rule-based medical DSS, MYCIN [4]. This system attempted to identify bacteria causing severe infections and recommend appropriate antibiotics. From these early DSS and the subsequent development of knowledge engineering, we now have DSS based on established architectures.
Specific radiological considerations
The development of DSS has received particular attention in radiology in both diagnostic as well as service-planning roles. Recent work has highlighted the value of DSS to guide order entry on outpatient imaging tests [5]. This article will focus on the application of DSS aimed at improving clinical performance through the provision of real-time diagnostic support.
Radiological practice has seen huge technical advances from direct image visualisation on phosphor screens through to the digital radiographic techniques we now employ. More significantly, we now find ourselves entering an era of advanced complex, computationally intensive quantitative imaging techniques that are used as specific biomarkers of both disease and treatment response. Unless we see the application of DSS to facilitate the uptake of these techniques, these advances will not be fully realised in the clinical setting. Indeed, some of the authors have personal experience of failure when attempting translational development of complex techniques developed in their own academic practice [6–9].
The understanding of what functionality and architectures the next generation of DSS require will facilitate their uptake in academic as well as clinical radiological applications.
Computer-aided diagnosis vs decision support systems
What is the distinction between computer-aided detection (CAD) systems, which have seen significant development within radiological practice, and DSS systems? Although in many instances they share common underlying architectures, their objectives differ significantly. CAD systems are very successful at undertaking repetitive tasks and tasks that suffer from a lack of interobserver concordance, as well as performing well in domains where there is a lack of trained domain experts [10]. CAD systems are in effect autonomous image analysis and processing systems tasked to specific roles within radiological practice or indeed a series of complementary processes [10] with minimal initial human interaction. At a specific time in the CAD cycle the system will, for example, prompt the human user into reviewing the image through prompts highlighting specific regions of the image, whereupon the cycle restarts. More advanced systems also incorporate an element of image processing to facilitate further this methodology [11].
In comparison, DSS aim to improve human clinical performance or function as training systems through their interaction with the user. They use prompts designed to steer the operator through a diagnostic conundrum providing information in an unsolicited and structured manner. Such systems do not specifically undertake any image processing or analysis functions, rather relying on the operator passing imaging information to the DSS itself.
In future applications this distinction will undoubtedly become blurred as elements of DSS are incorporated into CAD systems and vice versa; however, this review is restricted purely to DSS applications.
Route of analysis
We begin by examining the earlier rule-based expert systems (ES) that were often designed to function in a manner more akin to a domain expert replacement rather than as a support system. We then review more recent systems based around neural networks designed to emulate human reasoning techniques. From there we examine case-based reasoning, which emulates the decision-making processes employed by physicians themselves. Finally, we discuss DSS based on Bayesian networks, which many consider to represent the “state-of-the-art” in DSS design.
Although more recent systems have evolved towards a Bayesian design, we envisage that new systems should have architectural components best suited to the volume and knowledge type available at the time of implementation. The objective is, through the analysis of the evolution of such systems and of modern DSS implementations, to elicit the specific requirements for the next generation of radiological DSS. In doing so, we hope to identify the knowledge and technological gaps within this specific and important branch of health informatics and e-medicine that require resolution if such systems are to gain the clinical uptake they deserve.
Fundamental characteristics of decision support systems
The modern DSS should aim to facilitate optimal human performance by harnessing the most efficient features of the computer system for use in conjunction with the end user's own decision-making skills and abilities. A most important feature of any DSS is the level of its performance: the speed with which the system can solve a problem and the accuracy and appropriateness of the system's result. These two facets are not mutually exclusive, as even the most accurate diagnostic information will have little value if it is available too late to be effectively used. Conversely, no matter how fast a system's problem solving, the user will not be satisfied if the result is incorrect.
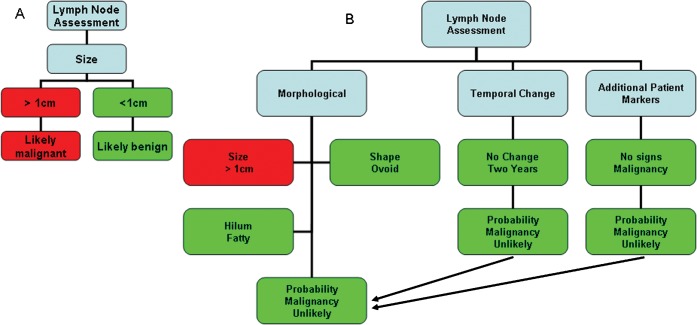
In theory, conventional computer programs always provide the same “correct” solutions, but in practice the system can give appropriate support only if the data are complete and exact. When the data are incomplete or include errors, a conventional program will provide either no solution or, more problematically, an incorrect one. In addition, deterministic systems might be based on domain-specific assumptions that oversimplify the decision-making process (Figure 1).
Figure 1.
The decision algorithms in modern decision support system (DSS) are commonly complex and designed with recognition that the available information might be incomplete or poorly defined. For instance, in (A) if a choice algorithm states “if a lymph node is >1 cm in short axis diameter then it is pathological” this decision will be subject to the same errors whether it is applied by a human or is automatically implemented. For this reason, in (B) a modern DSS would assess a lymph node as follows: “if a lymph node is >1 cm in short axis diameter and has a fatty hilum, is ovoid in shape and is unchanged over 2 years in a patient with no other tumour markers, then it is likely to be benign”.
Rule-based expert systems
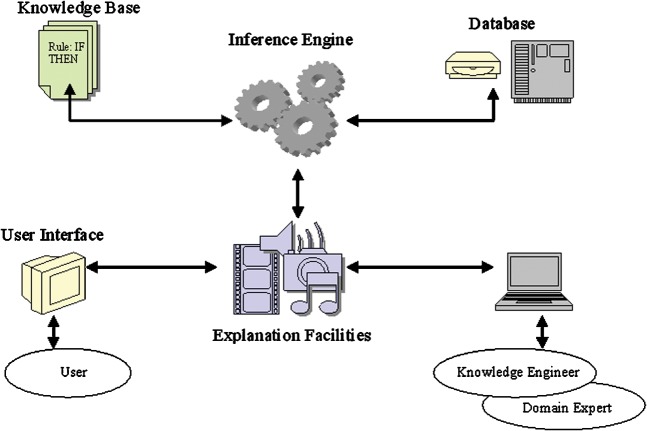
Early medical DSS attempted to simulate the judgement and behaviour of a human domain expert. A rule-based ES has five main components (Figure 2) [12]. Such systems would often attempt to replace the human user and in doing so would often perform unsatisfactorily. For any system to work effectively it must assimilate the knowledge and decision-making framework of the domain expert, as well as integrating new knowledge, and display this knowledge in a form that is easy to visualise and understand. In other words, the system should be able to explain how it has reached its particular conclusion.
Figure 2.
Diagrammatic representation of the five components of a rule-based expert system. (1) The knowledge base contains information provided by the domain expert and is used for problem solving in the form of “rules” that usually have a condition/action (if/then) structure. (2) The database contains the facts that have been provided to match against the condition part of the rules. Using the database, the system can search for the appropriate “if” statement to be satisfied before triggering the action structure. (3) The inference engine carries out this reasoning by linking the knowledge base with the facts in the database. (4) The explanation facilities enable the user to query the system as to how a particular conclusion was reached or why a specific fact was/is needed. These facilities also allow the user to interrogate the system as to the rules and knowledge stored within the database and knowledge base. (5) The user interface is not only important in facilitating accurate and easy information provision, but is invaluable in determining the accuracy and methodology of the data visualisation provided to the end user.
Rule-based expert systems in radiology
RENEX [13], an ES for the diagnosis of renal obstruction from nuclear medicine scans, has been seen to perform at a level approximating that of a domain expert and is subject to further testing [14].
The explanatory systems in this and other ES are of particular benefit as training tools by explaining the importance of findings to clinicians who do not hold the domain experience to understand the implicit knowledge inferred from the system's recommendations [15–17]. The importance of explanatory facilities in a rule-based ES was first expounded within the domain of neuroradiology by Teather et al [18] and du Boulay et al [19]. The BRAINS expert system was able to give recommendations concerning diagnosis and further imaging on the basis of statistical data with explanatory features “of benefit to less experienced radiologists” [18, 19]. This system used language-based descriptive inputs from the reporting radiologist. Teather et al [18] identified certain features that were essential in providing a clinically acceptable system, features that still hold true today:
The system must not usurp the radiologist's position and usual working practice.
The system must have an adaptable, clinically orientated user interface with help on demand.
Diagnostic advice must be given in a probabilistic form in terms of likely incidences of errors with explanation and justification of conclusions available.
The above features must be available independently of diagnostic advice.
They found that the BRAINS system demonstrated an unexpected lack of decisiveness in the diagnosis of certain pathological processes. This finding was partly related to the fact that variations in the language used for radiological descriptors meant that users had difficulty entering image description data in a reliable, consistent and repeatable manner [20].
The problem of implementing formal image description and underlying statistical granularity became even more manifest in the follow-up system that was designed to aid in MRI of the brain [21]. This difficulty was compounded by the variations in imaging parameters that introduced further statistical variation. Despite these shortcomings, both systems are good examples of the radiological reporting process driving an ES or DSS. In addition, as we shall see later, the application of standardised imaging descriptors has been seen to significantly improve the overall performance of modern DSS.
Lejbkowicz et al [17] developed Bone Browser, a diagnostic ES for bone tumour diagnosis. This system uses a rule-based ES along with a probabilistic inference engine to assess data entered by the radiologist reporting the examination. Data are entered in the form of radiological features, such as the size of the lesion or presence of soft-tissue involvement, as well as patient data such as relevant medical history or symptoms. This system performed as well as the expert; indeed it often improved the expert's performance. However, it did identify another shortcoming of ES design, namely it tended to produce differential diagnoses of twice the length! Further improvements in the system's diagnostic capacity were not thought possible owing to the disparity in imaging characteristics of the same pathological process in different parts of the skeleton and the paucity of data on certain rare conditions.
Summary: rule-based expert systems
Early implementations attempted to replace the clinician, but this has now been seen to be both an unrealistic as well as an undesirable aim. Recent systems perform well when the clinical question is well defined and relatively narrow. As the complexity of the clinical domain increases, so the ability of such systems to supply results with sufficient statistical power becomes less feasible. In general, rule-based ES do not have the ability to learn from experience. Unlike a human expert, who knows when to “break the rules”, an ES cannot easily modify its knowledge base. The inability of such systems to learn from new supplied data negates their use in settings where the acquired domain knowledge is incomplete at the time of design.
Such systems introduced the concept of using the DSS as teaching tools by offline searching and analysis of the DSS knowledge base. In this context, rule-based ES can perform poorly because, although individual rules can be relatively simple, their interactions within a larger set of rules might be opaque. Rule-based ES make it difficult to observe how individual rules serve the overall strategy [12].
BRAINS and MRI advisor [18–21] highlighted two important concepts. Firstly, DSS perform better with standardised descriptor inputs. Hence, restrictive language entry can improve DSS function, a feature that is now seen to be important in the majority of DSS implementations irrespective of domain [22]. Secondly, advanced imaging was seen to suffer from differing implementations of the same underlying technique (e.g. different scan parameters in MRI to produce generically similar T2 weighted images). This implies that the next generation of DSS needs to inform and take into account similarities and differences between generically similar imaging processes. This is of particular importance with regards to advanced imaging techniques, such as perfusion imaging [23], which have high variance in how they can be performed and interpreted.
Artificial neural networks
Neural networks are designed to model the human reasoning process and in particular its ability to process information in a parallel fashion [24]. The network is composed of a series of “nodes” interconnected by axonal equivalents known as “arcs” (Figure 3). The nodes are the sites of processing with the arcs affecting a node by positively or negatively weighting its outcome [12, 24]. This statistical weighting has the effect of influencing both the input to a node as well as its output and hence its influence on “downstream” nodes [25].
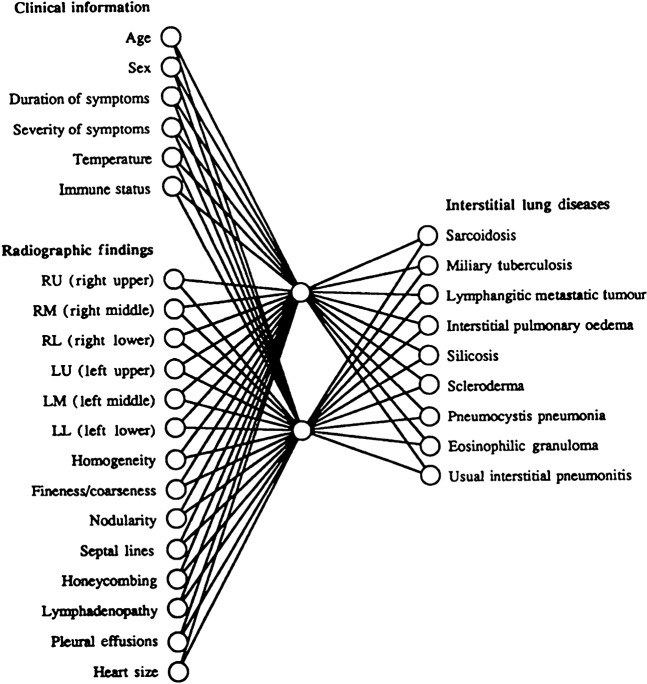
Figure 3.
Example of a typical neural network; in this case the network is used for the diagnosis of interstitial lung disease. The network shows multiple input nodes with two hidden nodes. These hidden nodes act as a form of feature analysis and detection system; they exert both positive and negative effects on the output diagnosis nodes dependent on their inputs. (Figure reproduced with permission from Asada et al [25].)
Neural networks do not use domain expertise but are “trained” using known input and resultant output variables. This establishes the “arcs” and sets the values for these internodal connections. When subsequent new cases are presented to the system it uses these trained arcs and nodes to give its output and can “learn” from new cases, which is the greatest strength of this approach [24].
Neural network decision support systems in radiology
Neuroradiology
Erol et al [26] used a neural network with input parameters of patient age, sex and data from ultrasound Doppler studies of the carotid arteries and the middle cerebral arteries to aid in the assessment of intracerebral haemodynamics following minor head injuries. They found this technique to be as effective a support aid as direct vascular studies without the comorbidity associated with angiography.
Sinha et al [27] developed a neural network DSS to aid in determining whether to subject paediatric patients to a CT scan following closed head injury. The system performed with a sensitivity of 82.2% compared with the physician's 62.2% for predicting intracranial abnormalities in closed head injury with similar specificities of 96%. Use of this DSS would have prevented six CT scans from being performed out of a total group of 382 patients, reducing radiation exposure in this very radiation-sensitive group.
Finally, Fukuda et al [28] used a neural network with input parameters of patient age and sex, as well as six morphological details of the intracranial ventricular system obtained from CT scanning, to design a neural network for the prediction of ventricular enlargement. The system gave a concordant result with four radiologists in over 90% of patients. This is a good example of a DSS using input data obtained directly from the radiologist who reported the CT scan.
Cardiovascular/thoracic radiology
Mobley et al [29] have demonstrated a neural network-based DSS that has been used to predict the necessity for cardiac catheterisation using 11 input variables in patients who presented for the first time with cardiac chest pain. The system had a sensitivity of 100% and, despite its poor specificity (26%), it is thought it could be of benefit in reducing unnecessary cardiac catheterisations [29, 30].
Swietlik et al [31] discussed the use of neural networks in both cardiac and pulmonary embolic disease estimation and found their performance similar to that of the radiologist when functioning in isolation; however, they identified improvements in performance when the systems were used as decision support aids as opposed to radiologist replacement systems.
Breast radiology
Breast radiology has seen the largest acceleration in the uptake, use and evaluation of not only neural networks but almost all forms of DSS. In 1993, Wu et al [32] identified the ability of neural networks to cope with relatively poor input data, giving relatively robust results in the output nodes. They applied this to the reading of mammograms [32] using input features extracted from mammograms by experienced radiologists. Initially, 43 features were used, but this was reduced to 14 with no degradation in performance. These features were all morphological, such as the length of spiculations or the presence of similar patterns elsewhere. The system was seen to outperform attending and resident radiologists and even experienced mammographers. Even greater performance gains were possible when the diagnosis of the decision system and of the radiologists were used in conjunction. Of great importance is the fact that the authors acknowledged that performance is determined to some extent by the ability of the radiologist reading the images to reliably extract the correct data. They felt that the “community of radiologists” can extract imaging features as reliably as an experienced mammographer, even if they might not be able to merge those features into a diagnosis without the aid of a DSS.
In 1995, Suryanarayanan et al [33] were aware of the success of neural networks in predicting malignancy using inputs in the form of imaging data from experienced mammographers. The authors took this a step further by using the Breast Imaging Recording and Data System of the American College of Radiology (BI-RADS). This had the effect of standardising the inputs between radiologists and in doing so was able to improve the specificity for biopsy of malignant lesions from 30% to 62% for radiologists alone when the sensitivity was set to 95%. They went on to analyse for intra- and interobserver variability and found no significant difference when the lexicon was used to define the data inputs to the neural network [33].
Summary: neural networks
A major advantage of neural networks is their ability to learn from new information as it is presented to the network. Although this process necessitates retraining the network, it is much easier to implement than re-writing the whole of a rule-based ES decision tree. Additionally, such updating of the underlying domain knowledge does not necessitate radiologist expert intervention, but can be achieved by the computing expert responsible for maintaining the network.
This highlights one of the major failings of neural networks; however, unlike rule-based systems, it has traditionally been thought that neural networks cannot produce meaningful explanations of how they came to their conclusion, being unable to explain their rationale in a comprehensible manner. Physicians generally will not accept and act on the advice of a decision system without knowing the basis for the system's diagnosis [18, 19, 34]. However, recent work has enabled rule generation from neural networks, so this shortcoming might be overcome in the future [35].
The advantages of implementing standardised inputs by using a restrictive data entry terminology first saw significant success in neural network implementations. As we shall see, this concept of restrictive language entry has also seen success in other DSS design architectures such as Bayesian systems.
Case-based reasoning
Case-based reasoning (CBR) is the process of solving new problems based on the solutions of similar past problems, for example a doctor who instigates a particular treatment based on his experience with previous patients who exhibited similar clinical findings or symptoms. In effect, CBR is a type of evidence-based analogy making. CBR is not only a powerful method of computer reasoning, but is a ubiquitous behaviour in everyday human problem solving. CBR can be broken down into a four-step process for the purposes of implementation [36]:
Retrieval: given a target problem, previous cases are retrieved from memory that are thought to be relevant to solving the problem. The cases themselves consist of the previous problem, how it was solved and details on how the solution was reached.
Re-use: the solution from the retrieved case needs to be mapped to the new target problem and might involve adaptation of the solution to fit the new situation.
Revision: the process by which, having mapped the, previous solution to the target situation, the new solution is tested in the real world (or a simulation) and, if necessary, revised.
Retain: the final process whereby after the solution has been successfully adapted to the target problem, the resulting experience is stored as a new case in memory.
Case-based reasoning applications in radiology
CBR has been applied in a system called ISIS (Intelligent Selection of Imaging Studies) [37]. This was a development of the ordering system protoISIS, which was trained using 200 previous radiology requests to select the appropriate ultrasound or CT imaging test based on the requesting data [37]. The designers realised that for the system to be clinically useful it had to have enhanced explanatory features and the ability to critique its user's actions and requests.
CBR has also been applied in the field of CT reporting, assessing the features of previous reports and comparing them to a current case [38]. This has enabled the extraction from store of previous images that the system thought relevant to a current case with an accuracy of up to 95.6%.
Once again, breast imaging has made significant inroads into the use of CBR. An example of this was developed by Bilska-Wolak and Floyd [39, 40], which used the BI-RADS lexicon as the basis for 1443 biopsy-proven mammographic cases. The system reduced benign breast lesion biopsies by 27%. As approximately 66–90% [39, 40] of breast biopsies are on benign lesions, this is a significant reduction. It is interesting to note that this CBR system was initially driven by 10 features identified from the BI-RADS lexicon to describe each case. These features were then reduced to a subset of the most influential features by performing an exhaustive feature search of all possible feature combinations and picking those combinations with the highest partial receiver operating characteristic (ROC) areas (area under the curve 0.9).
Summary: case-based reasoning
As described above, a requirement for any medical DSS is the explanatory features with regards to how a particular diagnosis was reached. This requires knowledge of the users' level of knowledge and an ability to find alternative explanatory mechanisms to justify themselves, much like a human expert does. In a CBR system this cannot be achieved, as by definition there will only be a single reasoning paradigm [41]. Additionally, one can say that the CBR approach will make use of the domain expert's anecdotal evidence, as its underlying operating principle has no underlying statistically relevant data. As discussed above with regards to Teather et al's general rules for DSS acceptance [18], “diagnostic advice must be given in a probabilistic form in terms of likely incidences of errors with explanation and justification of conclusions available.”
However, in situations where domain knowledge is too scarce for statistical relevance, clinicians' own performance is inherently based on anecdotal evidence, and for this reason CBR is often used where experts find it hard to articulate their thought processes when solving problems. Knowledge acquisition for say a Bayesian network (see below) would be extremely difficult in such domains, and is likely to produce incomplete or inaccurate results. Additionally, CBR allows the case base to be developed incrementally, while maintenance of the case library is relatively easy and can be carried out by domain experts with little necessity for non-domain intervention or interpretation.
Bayesian networks
Bayesian networks and probability
Bayes' theorem, developed by the Reverend Thomas Bayes, an 18th century mathematician and theologian, was first published in 1763 [42]. We are all familiar with Bayesian network DSS even if we might not realise it. The Microsoft Office (Microsoft Corporation, Redmond, WA) help wizard, most notably visualised as a “talking paper clip”, uses a Bayesian network to underpin its actions. In fact, Microsoft considers Bayesian network development to be a vital component of artificial intelligent learning support systems and one that will underpin most system development in its future systems [43].
A Bayesian network has a graphical structure similar to that of a neural network with nodes and arcs. Typically, one specific node is termed the “root” node, and in medical DSS this is often the disease process of interest (e.g. breast disease). For example, in Figure 4 we see the way in which patient age, family history (FHx) and hormone replacement therapy (HRT) affect the probability of breast disease [44].
Figure 4.
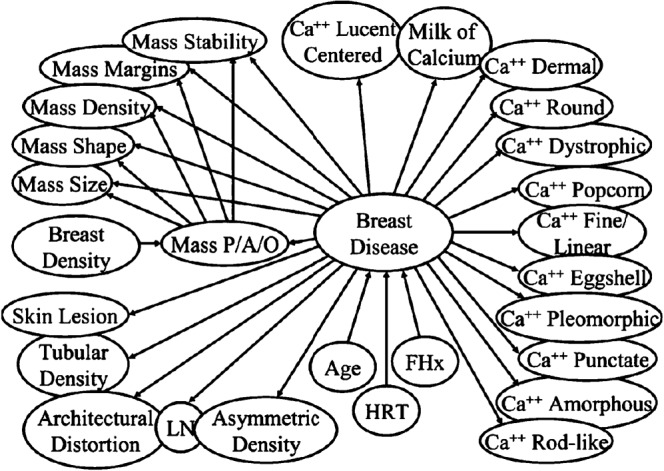
Example of a complex Bayesian network for mammography. All nodes can be thought of as representing specific variables relating to the root node such as diagnostic tests, patient demographics, clinical findings and so on. Each node is connected to the other nodes over which they exert influence by arcs that depict the fact that the nodes can affect the probability of the nodes to which they are connected. Ca++, calcifications; FHx, family history of breast cancer; HRT, hormone replacement therapy; LN, intramammary lymph node; mass P/A/O, mass present, absent or partially obscured. (Figure reproduced with permission from Burnside [44].)
Bayes' theorem
Examining Table 1, we see that the probability of a disease being present (prob dis) is calculated by F/I. The probability of a particular diagnostic test being positive (prob test) is given by H/I. In addition, Bayes' theory works on the probability of a particular test finding being positive given a particular disease state (prob test¦dis).
Table 1. A 2 × 2 contingency table illustrating the relationship between disease status and a single associated test.
| Diagnostic test |
Cell | Finding | ||||
| Negtive | Positive | |||||
| A | True negative | |||||
| Disease | Negtive | A | B | C | B | False positive |
| Positive | D | E | F | D | False negative | |
| G | H | I | E | True positive | ||
The post-test probability (which underpins Bayesian theory) is the probability of a pathology being present given that a certain test finding is positive or (prob dis¦test), which is calculated by E/H. Bayes' formula enables one to calculate the probability of a disease state on the basis of the probability of the presence of the disease, the probability of a particular test finding and the probability of the disease in the face of a particular finding using the following formula [44]:
 |
We can further examine this using the worked example of the simple Bayesian network shown in Figure 5, which relates the probabilistic relationship between subarachnoid haemorrhage and a CT scan of the brain. Examining Table 2, we see that the probability of a subarachnoid haemorrhage in a patient who has been referred to the hospital with a headache is:
Figure 5.
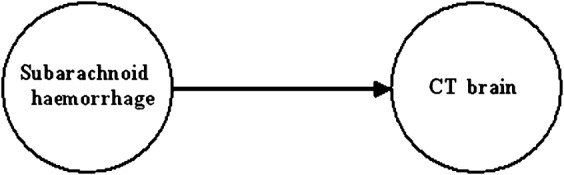
Simple Bayesian network relating subarachnoid haemorrhage to associated CT scan of the brain. In this case the parent node (as well as the primary node) is subarachnoid haemorrhage with its arc pointing to CT brain (the child node).
Table 2. A 2 × 2 contingency table relating CT scan of the brain to subarachnoid haemorrhage.
| CT brain scan |
||||
| Negative | Positive | Total | ||
| Subarachnoid haemorrhage | Negative | 1343 | 26 | 1369 |
| Positive | 3 | 102 | 105 | |
| Total | 1346 | 128 | 1474 | |
(prob dis) = f/I = 105/1474 = 7%
The probability of a positive CT scan of the brain is:
(prob test) = 128/1474 = 9%
The post-test probability of a subarachnoid haemorrhage given a positive CT scan of the brain is therefore:
(prob dis¦test) = 102/128 = 80%
Each node within a Bayesian network contains specific probabilities related to all the possible states of the node dependent on the findings in the surrounding nodes. This list of probabilities is termed a conditional probability table (CPT). In the example given above, the node “subarachnoid haemorrhage” is the parent node and has two possible states, which in this case are mutually exclusive, a haemorrhage is either present or absent. Using data from previous studies, the CPT for this node can be calculated as well as the CPT for the child node, which is the CT scan (Table 3). This table reflects not only the possible outcomes of the CT scan, but the outcomes in conjunction with the parent node.
Table 3. Conditional probability table for subarachnoid haemorrhage and CT brain nodes in Bayesian network (e.g. Figure 5).
| Subarachnoid haemorrhage | Probability (%) | Subarachnoid haemorrhage | CT brain scan | Probability (%) |
| Present | 7 | Present | Positive | 97 |
| Absent | 93 | Present | Negative | 3 |
| Absent | Positive | 2 | ||
| Absent | Negative | 98 |
One can see that as the number of nodes increases the CPT can become very large and, as each value within the contingency table needs to be populated when building the network, problems occur when data are not available for each relationship. There are several approaches to overcome this: the probabilities can be obtained from domain experts, data from the literature or from trial data. Alternatively, a large data set of cases can be used to train the probabilities.
Bayesian inference
Inference, or model evaluation, is the process by which Bayes, theorem underpins the function of a Bayesian network. Considering Figure 4 [44], one can see a complex network where each node is associated with its own CPT. To begin, the CPT reflects the prior probabilities derived by the network creators. This is often obtained from the “beliefs” of the domain experts who have been tasked with facilitating the network design. They are known as prior probabilities as their value reflects probabilities derived before any data concerning the current problem have been entered.
When data are entered into the network (e.g. information on the degree and type of calcification on a mammogram or patient demographic data), values within specific nodes are “clamped” to that specific value related to the observation made by the user. At this point the mathematical process is undertaken whereby the probabilities of all the connected nodes are recalculated to represent the new evidence available to the network. This process is referred to as inference, and following this procedure the new probabilities are referred to as posterior probabilities, as they now reflect to some degree the computed beliefs in light of new evidence. These posterior probabilities now reflect the new probabilities of all the possible outcomes in the model based on new evidence and the original probabilities within the model derived from the network creator.
Bayesian decision support systems in mammography
As discussed earlier, breast radiology has been at the forefront of DSS use for many years. One hypothesis for this is that in the multidisciplinary setting of the “breast clinic” the radiologist is often the lead clinician for both screening and diagnostic work and as such is responsible for clinically complex diagnostic and predictive tasks. Bayesian networks have been used to predict the probability of cancer using the probabilistic relationships between breast disease and mammographic findings and in so doing have been used to estimate the risk of malignancy [45, 46]. A Bayesian system has been used to improve the positive predictive value of breast lesion biopsy, a goal that was started through earlier work with neural networks [32, 33, 47]. The system performed as well as the breast radiologist in not missing a cancer (p<0.001) and improved the positive predictive value of biopsy from 21.6% to 31.2%.
The same system had its Bayesian probabilities updated with data from 92 pathology-proven biopsies and correlation to the BI-RADS descriptors of mammographic findings. This modification gave a sensitivity of 100% and specificity of 91%, with the potential to alert the radiologist to biopsy results that were discordant with mammography findings and to discover cases where biopsy sampling errors might have occurred [48, 49]. A similar system was trialled as far back as 1995 by Kahn et al [50, 51], who used input parameters including 5 patient history findings, 2 patient physical findings and 15 mammographic features that were extracted from the images by an experienced radiologist. This system was again used to offer predictive information on the probability of malignancy vs benignity of breast biopsies giving invaluable decision support.
Bayesian network summary
A potential problem highlighted by, but not specific to, Bayesian networks is related to end users making information requests in a manner not anticipated by the network's design. Such queries not covered by the network's priors illustrate a failing of the network design related to deficiencies within the domain of knowledge underpinning the system. This often relates to knowledge that exists but has not been “discovered” for representation at the time of network creation, or alternatively to knowledge that has been “guessed” by the domain expert at the time of DSS development.
This leads us to a significant problem in DSS development that relates to the quality and domain-specific volume of data used for prior belief calculation and network representation. For a clinical Bayesian network to function appropriately, the prior knowledge must be reliable otherwise the prior beliefs will distort the entire network and invalidate the results. Additionally, methodologies are needed to correlate all data within a domain and to enable network update as new knowledge becomes available from recent peer-reviewed publications, which remain the cornerstone of medical knowledge validation and dissemination.
Discussion
We have thematically examined the main techniques that have been developed and implemented to provide radiological DSS; namely, rule-based expert systems, neural networks, case-based reasoning and Bayesian networks. In doing so we have reviewed the differing approaches that can be employed to provide DSS to the clinical radiologist. Table 4 lists these differing techniques and their main features and differences.
Table 4. Comparison of artificial intelligence techniques.
| Type | Knowledge representation | Unsupervised (learns from data without teacher) | Robust (can handle unforeseen data) | Adaptive (can learn new data) | Rational (can explain reasoning) |
| Rule-based | Production of if then rules | No | No | No | Yes |
| Neural network | Graphic | Yes | No | Yes | No |
| Case-based | Graphic | No | No | No | Yes |
| Bayesian network | Cases and indexes | No | Yes | Yes | Yes |
Table modified from Kahn et al [37].
This review has identified certain aspects of DSS design that must be avoided in future applications and highlights other features that are, if not a definite requirement, certainly desirable in the next generation of DSS.
Firstly, the most successful DSS must not attempt to replace the radiologist. Rather, they should harness the excellent skills and abilities of humans in image assessment and feature extraction; these data are then passed on to the DSS. In some instances, users are not necessarily radiology domain experts, but can include students, interns and non-radiologists. Through the symbiotic interaction between user and DSS, overall performance has been seen to be elevated beyond that of even domain experts alone [27, 47].
This finding highlights a further important feature that must be implemented in the next generation of DSS, the symbiotic relationship with the user. By passing to the user requests for image feature extraction and its characteristic appearances, users are prompted to search for imaging findings that provide maximal statistical power with regards to outcome prediction for input into the support system. This directs users to refine their search patterns looking for findings of importance, even if they themselves are not immediately aware of the significance of these findings. In this way the user interface of the DSS must include a feedback mechanism giving statistical prediction on the basis of information already in the system and explaining why new information being sorted is of importance and how it will affect outcome prediction.
The underlying design of CBRs, which function without complete domain knowledge using “anectodal evidence”, is functionally successful but highlights a requirement for future DSS design such as Bayesian systems. We require new mechanisms for domain knowledge acquisition and storage, such that data from multiple sources and of different types can be assimilated to create a more complete knowledge representation within specific branches of radiology. For example, we require systems to assimilate prospective and retrospective studies, review articles, meta-analyses and case reports into single data repositories. Ideally, these repositories need to sit beneath the DSS such that, as it is updated with new knowledge, the overlying DSS will seamlessly update itself. We also propose that the next generation of DSS should be able to integrate with the world knowledge-base held in such repositories: data from local or distant centres regarding individual patients who have been run through the system would augment the world knowledge-base in real time as the systems are in use. The internet and the development of grid based computing means that change to a distributed model for data acquisition is indeed now a reality. In improving our understanding of domain-specific knowledge acquisition, assimilation, storage and searching, we will be able to overcome one major failing of DSS design: the problem of passing data from a domain expert to a knowledge engineer for creation of a DSS. With a Bayesian network without such complete knowledge discovery, the priors will be inaccurate and the network itself incomplete.
Complete knowledge discovery within a domain will also facilitate data entry into the next generation of “ideal” DSS, where the inputs are standardised and limited to those of maximal statistical outcome prediction. Indeed, the most successful systems have all incorporated standardised descriptive terms as their inputs, be they descriptors originated by the group who devised the DSS or commonly used descriptors in a certain field such as the BI-RADS lexicon. These descriptive terms are used to prompt the user into feature identification from the imaging series with the additional benefit of explanatory facilities to highlight the importance of the features being searched for. Only when the whole domain has been mapped can these terms be discovered and applied. In some respects this would represent domain-specific ontologies, where the ontologies have been reduced to a specific subset of terms with maximal statistical predictive power to a defined outcome. This is in effect a form of controlled language data entry [22, 52] implemented not only to facilitate search within the DSS data repository, but also to guide the radiologist to the imaging features statistically likely to improve the diagnostic performance of the system.
Many groups have recognised that the most efficient and successful use of a DSS is one where the system gives feedback to the user in the manner of appropriate statistics to act as a second opinion for consultation by the user. It has also been shown that for reasonable uptake and use of a system, access to the underlying statistical proofs and methodology is preferred even when the system is not being used to assess a particular case. This further enhances the value of any DSS by additionally allowing its use as an aid for training and teaching. As long as the DSS is kept up to date with the relevant medical literature, then the system can be used with training data sets to highlight the importance of specific imaging features, as well as providing the trainee with the underlying background data on the importance of the features being searched or prompted for.
The use of DSS in clinical radiological practice is set to continue and increase as we see an explosion in health informatics and e-science. However, further work is required to look at outcome indicators, determinant features and the integration of improved techniques for feature identification (e.g. formalised ontologies and natural language models) as further improvements in DSS design and implementation are sought. This will only be achievable with a move towards more quantitative knowledge discovery techniques; the development of new methods will allow for the assimilation, integration and storage of data [22] from disparate sources that can then be analysed and used to underpin DSS design, specifically Bayesian systems built upon such data repositories. These necessities are as true today as they were when first expounded by Russ Altman of Stanford University, who in 1997 stated that “There is a need to develop methods for representing biological knowledge so that computers can store, manipulate, retrieve, and make inferences about this information in standard ways” [2].
As radiologists we are moving towards quantitative imaging techniques that are difficult to apply and certainly complex to interpret; consequently, we require and should be at the forefront of DSS uptake. Radiologists have need of real-time systems to guide them through the implementation and analysis of advanced imaging techniques. Without such systems, these new methodologies will not find clinical acceptance through translational application and we will be significantly underperforming with regards to what can and should be achieved in clinical practice.
The United States National Academy of Engineering has stated that the provision of health informatics systems to provide “just in time just for me” support at the point of care is one of the great engineering challenges of the 21st century [53]. To paraphrase Altman, “the primary goals of any medical informatics project”, such as the DSS discussed here, “as for any other branch of biomedical research, are to improve the overall health of patients by combining basic scientific and engineering insights with the useful application of these insights to important problems” [2]. It is understanding these DSS, their underlying design and architecture, and how they should function and be implemented that will guide their deployment and future development, not only in radiology but in the wider realm of advanced modern clinical and academic medical practice as a whole.
References
- 1.Druzdzel MJ, Flynn RR. Decision support systems. New York: Marcel Dekker Inc, 2002 [Google Scholar]
- 2.Altman RB. Informatics in the care of patients: ten notable challenges. West J Med 1997;166:118–22 [PMC free article] [PubMed] [Google Scholar]
- 3.Lederberg J. How DENDRAL was conceived and born. Proceedings of ACM conference on history of medical informatics 5th November 1987. Bethesda, MD: ACM, 1987 [Google Scholar]
- 4.Various Rule-based expert systems: the MYCIN experiments of the Stanford heuristic programming project. Addison-Wesley, MA, 1984 [Google Scholar]
- 5.Sistrom CL, Dang PA, Weilburg JB, Dreyer KJ, Rosenthal DI, Thrall JH. Effect of computerized order entry with integrated decision support on the growth of outpatient procedure volumes: seven-year time series analysis. Radiology 2009;251:147–55 [DOI] [PubMed] [Google Scholar]
- 6.Thacker NA, Varma AR, Bathgate D, Stivaros S, Snowden JS, Neary D, et al. Dementing disorders: volumetric measurement of cerebrospinal fluid to distinguish normal from pathologic findings — feasibility study. Radiology 2002;224:278–85 [DOI] [PubMed] [Google Scholar]
- 7.Stivaros SM, Sinclair D, Bromiley PA, Kim J, Thorne J, Jackson A. Endoscopic third ventriculostomy: predicting outcome with phase-contrast MR imaging. Radiology 2009;252:825–32 [DOI] [PubMed] [Google Scholar]
- 8.Selvarajah J, Scott M, Stivaros S, Hulme S, Georgiou R, Rothwell N, et al. Potential surrogate markers of cerebral microvascular angiopathy in asymptomatic subjects at risk of stroke. Eur Radiol 2009;19:1011–18 [DOI] [PubMed] [Google Scholar]
- 9.Stivaros SM, Jackson A. Changing concepts of cerebrospinal fluid hydrodynamics: role of phase-contrast magnetic resonance imaging and implications for cerebral microvascular disease. Neurotherapeutics 2007;4:511–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summers RM. Road maps for advancement of radiologic computer-aided detection in the 21st century. Radiology 2003;229:11–13 [DOI] [PubMed] [Google Scholar]
- 11.Krupinski EA. Computer-aided detection in clinical environment: benefits and challenges for radiologists. Radiology 2004;231:7–9 [DOI] [PubMed] [Google Scholar]
- 12.Negnevitsky M. Artificial intelligence: a guide to intellient systems. 2nd ed Assison-Wesley, Ma ; 2005 [Google Scholar]
- 13.Garcia EV, Taylor A, Halkar R, Folks R, Krishnan M, Cooke CD, et al. RENEX: an expert system for the interpretation of 99mTc-MAG3 scans to detect renal obstruction. J Nucl Med 2006;47:320–9 [PubMed] [Google Scholar]
- 14.Taylor A, Manatunga A, Garcia EV. Decision support systems in diuresis renography. Semin Nucl Med 2008;38:67–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahn CE., Jr Validation, clinical trial, and evaluation of a radiology expert system. Methods Inf Med 1991;30:268–74 [PubMed] [Google Scholar]
- 16.Sotos JG. MYCIN and NEOMYCIN: two approaches to generating explanations in rule-based expert systems. Aviat Space Environ Med 1990;61:950–4 [PubMed] [Google Scholar]
- 17.Lejbkowicz I, Wiener F, Nachtigal A, Militiannu D, Kleinhaus U, Applbaum YH. Bone Browser a decision-aid for the radiological diagnosis of bone tumors. Comput Methods Programs Biomed 2002;67:137–54 [DOI] [PubMed] [Google Scholar]
- 18.Teather D, Morton BA, du Boulay GH, Wills KM, Plummer D, Innocent PR. Computer assistance for CT scan interpretation and cerebral disease diagnosis. Stat Med 1985;4:311–15 [DOI] [PubMed] [Google Scholar]
- 19.du Boulay GH, Teather D, Morton BA, Wills KM, Innocent PR, Plummer D. Brains — a computer advisor system to aid in CT scan interpretation and cerebral disease diagnosis. Neuroradiology 1987;29:196–9 [DOI] [PubMed] [Google Scholar]
- 20.Teather D, Teather BA, Wills KM, du Boulay GH, Plummer D, Isherwood I, et al. Evaluation of computer advisor in the interpretation of CT images of the head. Neuroradiology 1988;30:511–17 [DOI] [PubMed] [Google Scholar]
- 21.du Boulay GH, Woods AJ, Teather D, Teather BA, Wills KM, Plummer D. Towards an advisor for MRI. Neuroradiology 1988;30:245–51 [DOI] [PubMed] [Google Scholar]
- 22.Power R, Scott D, Evans R. What you see is what you meant: direct knowledge editing with natural language feedback. In: Prade Henri.13th European Conference on Artificial Intelligence, 1998:Brighton, UK, Proceedings John Wiley and Sons, NJ [Google Scholar]
- 23.Jackson A, O'Connor J, Thompson G, Mills S. Magnetic resonance perfusion imaging in neuro-oncology. Cancer Imaging 2008;8:186–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich E, Knight K. Artificial intelligence. London: McGraw-Hill, 1991 [Google Scholar]
- 25.Asada N, Doi K, MacMahon H, Montner SM, Giger ML, Abe C, et al. Potential usefulness of an artificial neural network for differential diagnosis of interstitial lung diseases: pilot study. Radiology 1990;177:857–60 [DOI] [PubMed] [Google Scholar]
- 26.Erol FS, Uysal H, Ergun U, Barisci N, Serhathoglu S, Hardalac F. Prediction of minor head injured patients using logistic regression and MLP neural network. J Med Syst 2005;29:205–15 [DOI] [PubMed] [Google Scholar]
- 27.Sinha M, Kennedy CS, Ramundo ML. Artificial neural network predicts CT scan abnormalities in pediatric patients with closed head injury. J Trauma 2001;50:308–12 [DOI] [PubMed] [Google Scholar]
- 28.Fukuda H, Inoue Y, Nakajima H, Usuki N, Saiwai S, Miyamoto T, et al. Potential usefulness of an artificial neural network for assessing ventricular size. Radiat Med 1995;13:23–6 [PubMed] [Google Scholar]
- 29.Mobley BA, Schechter E, Moore WE, McKee PA, Eichner JE. Neural network predictions of significant coronary artery stenosis in men. Artif Intell Med 2005;34:151–61 [DOI] [PubMed] [Google Scholar]
- 30.Mobley BA, Schechter E, Moore WE, McKee PA, Eichner JE. Predictions of coronary artery stenosis by artificial neural network. Artif Intell Med 2000;18:187–203 [DOI] [PubMed] [Google Scholar]
- 31.Swietlik D, Bandurski T, Lass P. Artificial neural networks in nuclear medicine. Nucl Med Rev Cent East Eur 2004;7:59–67 [PubMed] [Google Scholar]
- 32.Wu Y, Doi K, Metz CE, Asada N, Giger ML. Simulation studies of data classification by artificial neural networks: potential applications in medical imaging and decision making. J Digit Imaging 1993;6:117–25 [DOI] [PubMed] [Google Scholar]
- 33.Suryanarayanan S, Karellas A, Vedantham S, Glick SJ, D'Orsi CJ, Baker SP, et al. Comparison of tomosynthesis methods used with digital mammography. Acad Radiol 2000;7:1085–97 [DOI] [PubMed] [Google Scholar]
- 34.Teach RL, Shortliffe EH. An analysis of physician attitudes regarding computer-based clinical consultation systems. Comput Biomed Res 1981;14:542–58 [DOI] [PubMed] [Google Scholar]
- 35.Setiono R, Thong JYL. An approach to generate rules from neural networks for regression problems. Eur J Operational Res 2004;155:239–50 [Google Scholar]
- 36.Amodt A, Plaza E. Case-based reasoning: foundational issues, methodological variations and system approaches. Artificial Intelligence Commun 1994;7:39–52 [Google Scholar]
- 37.Kahn CE., Jr Artificial intelligence in radiology: decision support systems. Radiographics 1994;14:849–61 [DOI] [PubMed] [Google Scholar]
- 38.Tsukamoto N. Supporting system for CT diagnosis referring to previous cases. Nippon Igaku Hoshasen Gakkai Zasshi 1993;53:1415–25 [PubMed] [Google Scholar]
- 39.Baydush AH, Floyd CE., Jr Improved image quality in digital mammography with image processing. Med Phys 2000;27:1503–8 [DOI] [PubMed] [Google Scholar]
- 40.Bilska-Wolak AO, Floyd CE., Jr Development and evaluation of a case-based reasoning classifier for prediction of breast biopsy outcome with BI-RADS lexicon. Med Phys 2002;29:2090–100 [DOI] [PubMed] [Google Scholar]
- 41.Georgin E, Bordin F, McDonald PJR. Using prototypes in case based diagnosis of stream turbines. Case based reasoning: prospects for applications. IEE Colloquium 1995;1:1–4 [Google Scholar]
- 42.Stutz J, Cheeseman P. A short exposition on Bayesian inference and probability. NASA Ames Research Centre: Computational Sciences Division, Data Learning Group, Available at: http://ic.arc.nasa.gov/ic/projects/bayes–group/html/bayes–theorem–long.html 1994 [Google Scholar]
- 43.Helm L. Improbable inspiration. Los Angeles Times, 28 October 1996 [Google Scholar]
- 44.Burnside ES. Bayesian networks: computer-assisted diagnosis support in radiology. Acad Radiol 2005;12:422–30 [DOI] [PubMed] [Google Scholar]
- 45.Burnside ES, Rubin DL, Fine JP, Shachter RD, Sisney GA, Leung WK. Bayesian network to predict breast cancer risk of mammographic microcalcifications and reduce number of benign biopsy results: initial experience. Radiology 2006;240:666–73 [DOI] [PubMed] [Google Scholar]
- 46.Burnside ES, Rubin DL, Shachter RD. Using a Bayesian network to predict the probability and type of breast cancer represented by microcalcifications on mammography. Medinfo 2004;11:13–17 [PubMed] [Google Scholar]
- 47.Floyd CE, Jr, Lo JY, Yun AJ, Sullivan DC, Kornguth PJ. Prediction of breast cancer malignancy using an artificial neural network. Cancer 1994;74:2944–8 [DOI] [PubMed] [Google Scholar]
- 48.Burnside ES, Rubin DL, Shachter RD, Sohlich RE, Sickles EA. A probabilistic expert system that provides automated mammographic-histologic correlation: initial experience. AJR Am J Roentgenol 2004;182:481–8 [DOI] [PubMed] [Google Scholar]
- 49.Burnside E, Rubin D, Shachter R. A Bayesian network for mammography. Proc AMIA Symp. 2000:106–10 [PMC free article] [PubMed] [Google Scholar]
- 50.Kahn CE, Jr, Roberts LM, Shaffer KA, Haddawy P. Construction of a Bayesian network for mammographic diagnosis of breast cancer. Comput Biol Med 1997;27:19–29 [DOI] [PubMed] [Google Scholar]
- 51.Kahn CE, Jr, Roberts LM, Wang K, Jenks D, Haddawy P. Preliminary investigation of a Bayesian network for mammographic diagnosis of breast cancer. Proc Annu Symp Comput Appl Med Care. 1995:208–12 [PMC free article] [PubMed] [Google Scholar]
- 52.Tablan V, Damljanovic D, Bontcheva K. A natural language query interface to structured information. Proceedings of the 5th European semantic web conference on the semantic web: research and applications, 2008; Tenerife, Spain. Berlin: Springer-Verlag, 2008;5021:361–75 [Google Scholar]
- 53.Engineering NAo Advances in health informatics: ten great engineering challenges of the 21st centuryWashington, 2008 [Google Scholar]