Abstract
The cDNA clone ERD5 (early responsive to dehydration), isolated from 1-h-dehydrated Arabidopsis, encodes a precursor of proline (Pro) dehydrogenase (ProDH), which is a mitochondrial enzyme involved in the first step of the conversion of Pro to glutamic acid. The transcript of the erd5 (ProDH) gene was undetectable when plants were dehydrated, but large amounts of transcript accumulated when plants were subsequently rehydrated. Accumulation of the transcript was also observed in plants that had been incubated under hypoosmotic conditions in media that contained l- or d-Pro. We isolated a 1.4-kb DNA fragment of the putative promoter region of the ProDH gene. The β-glucuronidase (GUS) reporter gene driven by the 1.4-kb ProDH promoter was induced not only by rehydration but also by hypoosmolarity and l- and d-Pro at significant levels in transgenic Arabidopsis plants. The promoter of the ProDH gene directs strong GUS activity in reproductive organs such as pollen and pistils and in the seeds of the transgenic plants. GUS activity was detected in vegetative tissues such as veins of leaves and root tips when the transgenic plants were exposed to hypoosmolarity and Pro solutions. GUS activity increased during germination of the transgenic plants under hypoosmolarity. The relationship between Pro metabolism and the physiological aspects of stress response and development are discussed.
Osmotic or water stress caused by drought or high salinity is the most serious factor that limits plant growth and productivity (Boyer, 1982). These conditions induce dehydration of plant cells, which may trigger physiological, biochemical, and molecular responses (Shinozaki and Yamaguchi-Shinozaki, 1996, 1997). To counteract osmotic stress, some plants accumulate several kinds of compatible osmolytes, such as Pro, Gly betaine, and sugar alcohols, that function as osmotica and protect macromolecules such as proteins and membranes (Delauney and Verma, 1993). Among the compatible solutes, Pro is the most widely distributed osmolyte in water-stressed plants. It has been suggested to function as a nitrogen-storage compound (Ahmad and Hellebust, 1988), as an energy or reducing power sink (Walton et al., 1991), and as a radical scavenger (Smirnoff and Cumbes, 1989).
The accumulation of Pro in dehydrated plants is caused not only by the activation of Pro biosynthesis but also by the inactivation of Pro degradation; conversely, a decrease in the level of accumulated Pro in rehydrated plants is caused not only by the inhibition of Pro biosynthesis but also by the activation of Pro degradation. Genes that encode enzymes in Pro biosynthesis have been isolated from various plants, and their expression and the functions of their products have been characterized (Delauney and Verma, 1993; Yoshiba et al., 1995, 1997; Igarashi et al., 1997). However, the molecular mechanism of Pro degradation is poorly understood.
We isolated the ProDH gene (Pro dehydrogenase) from Arabidopsis (Kiyosue et al., 1996) that is identical to that of Verbruggen et al. (1996) and Peng et al. (1996). Sequence analysis of an Arabidopsis cDNA clone, ERD5 (early responsive to dehydration), isolated from plants dehydrated for 1 h revealed that it encodes a protein having homology with products of the yeast PUT1 gene and the Drosophila melanogaster sluggish-A gene (Kiyosue et al., 1996). Their gene products are precursors of ProDH proteins (Pro oxidases: EC 1.5.99.8), which are the first enzymes involved in the conversion of Pro to Glu. We show that the products of ERD5 cDNA are localized in the mitochondrial fraction. Fusion genes for ERD5 and PUT1 complemented a put1 mutant of yeast, allowing put1 to grow with Pro as the source of nitrogen. RNA gel-blot analysis demonstrated that transcripts of the ProDH gene were undetectable when plants had been dehydrated for 10 h, but that large amounts of the transcript accumulated when plants were subsequently rehydrated. Elevated levels of the transcript were also found in plants incubated in a medium that contained Pro. These results suggest that the ProDH gene is regulated at the transcriptional level by both dehydration and rehydration of plants.
To investigate the regulatory mechanism for the expression of the ProDH gene, we examined its expression by northern-blot analysis and demonstrated that the ProDH gene is up-regulated by hypoosmolarity and Pro treatment. Then, we isolated the promoter region of the ProDH gene and analyzed the regulatory mechanisms in response to environmental and developmental signals in transgenic Arabidopsis, which contained a fused gene consisting of the ProDH promoter and the coding region of the reporter gene for GUS. The roles of Pro dehydrogenase in stress response, seed and pollen development, and germination are discussed.
MATERIALS AND METHODS
Plant Materials
Plants (Arabidopsis ecotype Columbia) were grown on vermiculite beds or aseptically on GM (germination medium containing 0.09 m Suc) (Valvekens et al., 1988) containing 0.8% Bacto-agar (Difco, Detroit, MI) for 2 to 4 weeks under continuous light (3000 lux), as previously described (Kiyosue, 1993b, 1994; Nakashima et al., 1997).
Stress Treatments
Arabidopsis rosette plants were subjected to dehydration on chromatography paper (3MM, Whatman) at 60% to 70% RH and 22°C, under dim light (100 lux) (Nakashima et al., 1997). The Arabidopsis plants grown on vermiculite beds or GM agar medium were transferred to distilled water, GM liquid medium, 0.09 m or 0.26 m l- or d-Pro, respectively, and incubated for 1 to 24 h under dim light. In each case, the plants were subjected to the stress treatment for varying durations and were then immediately frozen in liquid nitrogen.
Osmolarity Measurement
The osmolarity of solutions used in northern-blot analysis was measured at 20°C by the ONE-TEN osmometer (Fiske, Norwood, MA).
Mapping of the Transcription Start Site by Primer Extension
The primer-extension experiment was performed according to the method of Yamaguchi-Shinozaki et al. (1989) using the [γ-32P]ATP-labeled oligonucleotide, which corresponds to the complementary sequence upstream of the coding region of the ProDH gene: 5′-ATAATTTCTCTTCTC-3′ (complementary to positions +66 to +80). The mRNA for the primer extension was extracted from Arabidopsis rosette plants incubated in 0.09 m l-Pro for 2 h by the guanidine thiocyanate/CsCl method, and was purified on an oligo(dT) column.
Isolation of RNA and RNA Gel-Blot Analysis
RNA was isolated from whole rosette plants as previously described (Kiyosue, 1993a). Fragments of the ERD5 cDNA were labeled by the random-primer method with [α-32P]dCTP (Amersham) using the random-primed DNA-labeling kit from Boehringer Mannheim. The labeled fragments were hybridized with RNA according to standard protocols (Kiyosue et al., 1994).
Transgenic Plants
A 1.4-kb fragment of the ProDH gene upstream sequence was prepared using double digestion with BamHI and HindIII of the pBluescript II vector (Stratagene). It was ligated into the BamHI and HindIII site of the promoterless GUS expression vector pBI101.1 (Clontech, Palo Alto, CA) and was transferred from Escherichia coli DH5α into Agrobacterium tumefaciens LBA4404 via three-way mating with an E. coli strain containing a mobilizing plasmid, pRK2013. Transformation of Arabidopsis (ecotype Wassilewskija) was performed as previously described (Valvekens et al., 1988; Benfey et al., 1989). T2 seeds of transgenic Arabidopsis were germinated on GM containing 20 μm kanamycin at 22°C. Whole plants were transferred to chromatography paper (3MM, Whatman) for dehydration stress or to Petri dishes containing distilled water, GM liquid medium, and 0.09 m l- or d-Pro and incubated under dim light (100 lux) for 24 h. To analyze GUS activity in germinating seeds, the transgenic plants were grown on sterilized filter paper soaked with distilled water or GM solution under continuous light (3000 lux).
Assay of GUS Activity and Histochemistry
GUS activity was assayed in whole-plant extracts by fluorimetric determination of the production of 4-methylumbelliferone from the glucuronide precursor using a standard protocol (Jefferson et al., 1986). Histochemical localization of GUS activity was performed by incubating whole transgenic plants in 1 mm 5-bromo-4-chloro-3-indolyl glucuronide at 37°C for 3 h to overnight, fixing, and then incubating in a 50% to 100% ethanol series (Nakashima et al., 1997).
RESULTS
Analysis of the Expression of the ProDH Gene in Arabidopsis Plants
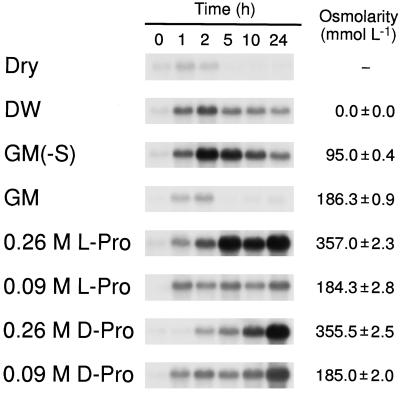
Induction of the ProDH gene in Arabidopsis plants by dehydration or transfer of plants from agar plates to various solutions was analyzed by northern-blot analysis (Fig. 1). Expression of the ProDH gene was transiently induced within 1 h after dehydration in Arabidopsis plants, as reported by Kiyosue et al. (1996). The transient expression of the gene was observed when the plants were transferred from GM agar plates to hydroponic conditions in GM liquid medium (Fig. 1). These results indicated that during the dehydration treatment the transient expression may have been caused by touching or wounding the plants when they were transferred from the GM agar plates. The gene was strongly induced within 1 h after the initiation of incubation, when the plants were transferred from the same GM agar plates to hydroponic conditions in deionized water or GM − Suc. The osmolarity of deionized water and GM − Suc was lower than that of GM (Fig. 1). Incubation in GM − Suc with 9% PEG (PEG 4000; final osmolarity about 185 mmol/L) gave a similar result to that of GM (data not shown). These results indicate that the high-level expression of the ProDH gene might be due to hypoosmolarity.
Figure 1.
Northern-blot analysis of the expression of the ProDH gene in 4-week-old Arabidopsis plants after several treatments. Each lane was loaded with 10 μg of total RNA from 3- to 4-week-old Arabidopsis plants grown in GM agar plates that were dehydrated (Dry), transferred to hydroponic conditions for 24 h in deionized water (DW) or in GM with and without Suc, with 0.9 m or 0.26 m l-Pro, and with 0.9 m or 0.26 m d-Pro. Numbers above each lane indicate the number of hours after the initiation of treatment prior to the isolation of RNA. The osmolarity of each solution is shown at the right side of the blots. Data represent means ± se (n = 3).
We previously reported that the level of ProDH mRNA in Arabidopsis plants was strongly affected by incubation with GM containing 0.26 m l- or d-Pro instead of 0.09 m Suc (Kiyosue at al., 1996). Since the osmolarity of the medium containing 0.09 m Pro (approximately 185 mmol kg−1; Fig. 1) was the same as that of the control GM solution, we examined, by northern-blot analysis, the induction of the ProDH gene by incubation with GM containing 0.09 m Pro. When the Arabidopsis plants were transferred from agar plates to hydroponic growth in the 0.09 m l-Pro or d-Pro solution, the ProDH gene was significantly induced within 1 h, although the induction level of the ProDH mRNA by 0.09 m Pro was lower than that by 0.26 m Pro (Fig. 1). Thus, we confirmed the effect of l- and d-Pro in the induction of the ProDH gene in Arabidopsis plants.
Cloning and Sequence Analysis of the Promoter Region of the Arabidopsis ProDH Gene
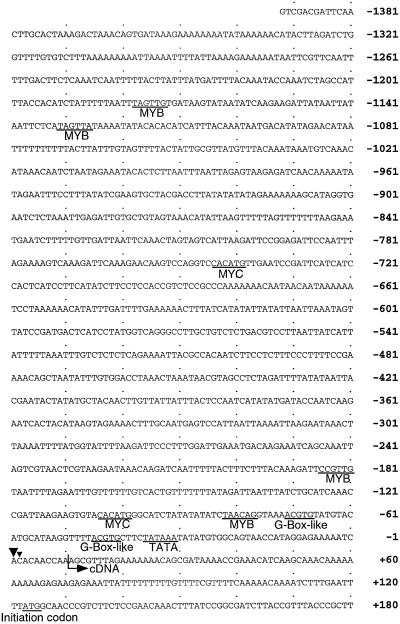
We screened a genomic library prepared from Arabidopsis plants using the insert DNA fragment of the ERD5 cDNA and isolated a 1.4-kb DNA fragment. The nucleotide sequence of this genomic DNA was determined (Fig. 2). The 5′ end of the transcript was determined by primer extension (data not shown). Two bands appeared by primer-extension assay at the +1 (adenine) and +2 (cytosine) sites. Since the band in the +1 site was thicker than that in the +2 site, we determined that the transcription start site was at the +1 site. The initiation codon is 123 nucleotides downstream from the site of initiation of transcription. A typical TATA-box sequence is located at position −37 (TATAAA). A number of sequence motifs have been identified that may have a role in the regulation of transcription of plant genes. The upstream region of the ProDH gene was searched for cis-acting elements. We found two G-box-like motifs (ACGTG at −46 and −72) similar to the G-box sequence (CACGTG; Schindler et al., 1992; Shinozaki and Yamaguchi-Shinozaki, 1997) and the ABA-responsive element sequence (PyACGTGGC; Marcotte et al., 1989; Yamaguchi-Shinozaki et al., 1989; Shinozaki and Yamaguchi-Shinozaki, 1997). Moreover, we found four putative motifs (TAACAG at −83, CCGTTG at −186, TAGTTA at −1132, and TAGTTG at −1179) that resembled a MYB recognition site (PyAACNPu; Biedenkapp et al., 1988; Nakagoshi et al., 1990; Shinozaki and Yamaguchi-Shinozaki, 1997) and two putative motifs (CACATG at −105 and −745) that resembled a MYC recognition site (CANNTG; Murre et al., 1989; Shinozaki and Yamaguchi-Shinozaki, 1997).
Figure 2.
The nucleotide sequence of the promoter region of the ProDH gene. The 5′ end of the transcript is indicated in the nucleotide sequence as position +1. A putative TATA box (TATAA), G-box-like sequence (ACGTG), MYB-like sequence (PyAACNPu), and MYC-like sequence (CANNTG) are underlined. The nucleotide sequence was analyzed with the GENETYX software system (Software Development, Tokyo, Japan).
Expression Analysis of GUS Reporter Gene Driven by the ProDH Promoter in Transgenic Arabidopsis Plants
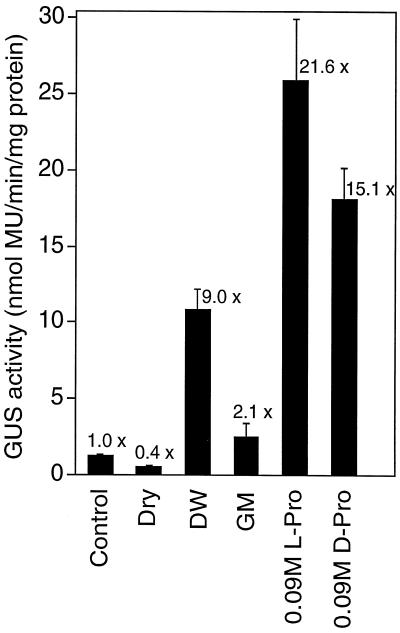
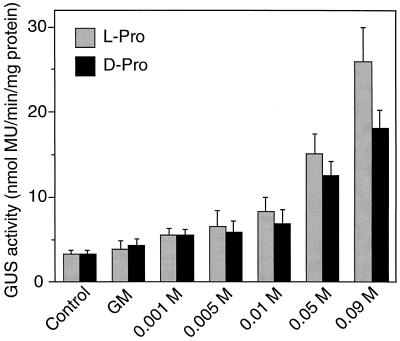
To examine whether the cis-acting elements involved in the hypoosmolarity and Pro-responsive expression of the ProDH gene are located in the isolated 1.4-kb DNA fragment, a chimeric gene consisting of the ProDH promoter region (−1393 to +122, Fig. 2) fused to the GUS reporter gene was constructed and introduced into Arabidopsis plants through A. tumefaciens. Stable transformant lines were obtained. Figure 3 presents the analysis of the expression of the GUS reporter gene in three independent, regenerated, T2 transgenic Arabidopsis plants (n = 5). GUS activity was repressed within 24 h of dehydration of Arabidopsis plants. Incubation with deionized water and treatment with 0.09 m l-Pro or d-Pro increased GUS activity, whereas GM incubation did not. Incubation with 100 μm ABA or GA3 had no effect on the expression of GUS activity (data not shown). These data suggest that the isolated 5′-upstream region of the ProDH gene contains cis-element(s) involved in the dehydration-repressive, hypoosmolarity-, and Pro-inducible expression. Dose-response analysis of GUS activity revealed that incubation in 0.001 to 0.09 m l-Pro or d-Pro induced expression of the ProDH promoter in the three independent, regenerated transgenic Arabidopsis plants (n = 5; Fig. 4).
Figure 3.
Stress-inducible GUS activity in 4-week-old T2 transgenic Arabidopsis plants, each of which carries 1.4-kbp of the ProDH promoter-GUS fusion gene. Some plants were used immediately for assay of GUS activity (Control), and the other plants were treated for 24 h by dehydration (Dry) or by hydroponic growth in distilled water (DW), GM, or GM with 0.09 m l-Pro or d-Pro. The values of GUS activity are the averages of values obtained from five plants of three independent transformant lines. Bars indicate se.
Figure 4.
Dose-response analysis of Pro-inducible GUS activity in T2 transgenic Arabidopsis plants, each of which carries the ProDH promoter-GUS fusion gene. One-month-old aseptic transgenic Arabidopsis plants were transferred to GM liquid medium with 0 (GM), 0.001, 0.005, 0.01, 0.05, or 0.09 m l-Pro or d-Pro, instead of the same molar amount of Suc, and incubated for 24 h. Some plants were used immediately for assay of GUS activity (Control). The values of GUS activity are the averages of values obtained from five plants of three independent transformant lines. Bars indicate se.
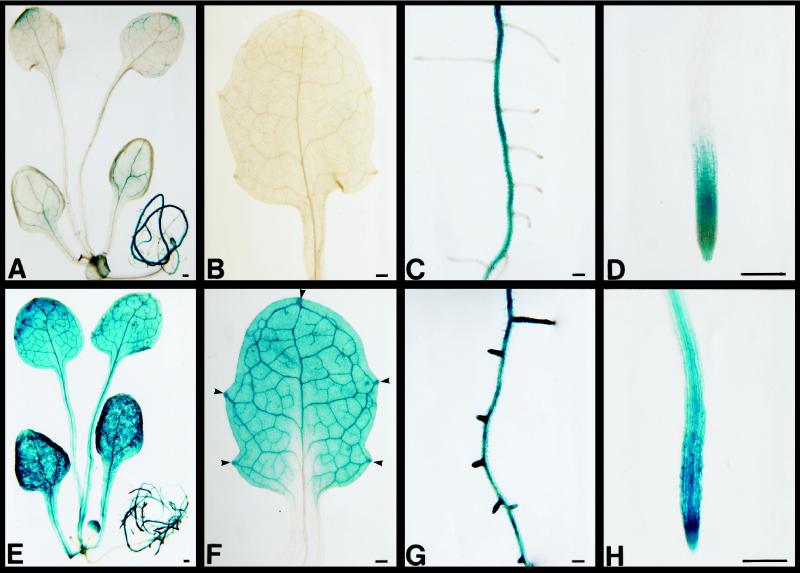
We examined the localization of GUS reporter gene expression under control of the ProDH promoter in transgenic Arabidopsis plants. Weak GUS activity was observed by histochemical analysis in unbolting, whole transgenic Arabidopsis seedlings 2 weeks after germination without any treatment (Fig. 5A). GUS activity in the leaves was relatively low (Fig. 5B). GUS expression was detected in the main roots (Fig. 5C) and some, but not all, root tips (Fig. 5D). After 0.09 m l-Pro incubation, intense GUS staining was observed in the veins and hydathodes of leaves (Fig. 5E) and in the lateral and main roots (Fig. 5G), especially in the root vascular cylinder and meristem (Fig. 5H). Examination of tissue sections of leaves revealed that the strong expression in veins was not due to a smaller cell volume than in mesophyll cells but, rather, to high expression in vascular cells (data not shown). The same expression pattern was found in transformants after 0.09 m d-Pro or water incubation (data not shown). l- and d-Pro had a stronger effect on ProDH gene expression than water in the same plant tissues.
Figure 5.
Histochemical localization of GUS activity in 10- to 14-d-old transgenic Arabidopsis plants containing the ProDH promoter-GUS fusion gene. A, Overview of GUS activity, showing weak activity in rosette plants stained for 6 h. B, Little GUS activity in leaves stained for 2 h. C, Strong GUS staining in main root stained for 6 h. D, GUS staining in some root primordia stained for 2 h. E through H, GUS staining in plants after 24 h of l-Pro treatment. E, Overview of GUS activity in roots and leaves stained for 6 h. F, Strong GUS staining in veins and hydathodes (arrowheads) of leaves stained for 2 h. G, GUS staining in root tip and veins stained for 6 h. H, Strong GUS staining in root tips stained for 2 h. Bars = 200 μm.
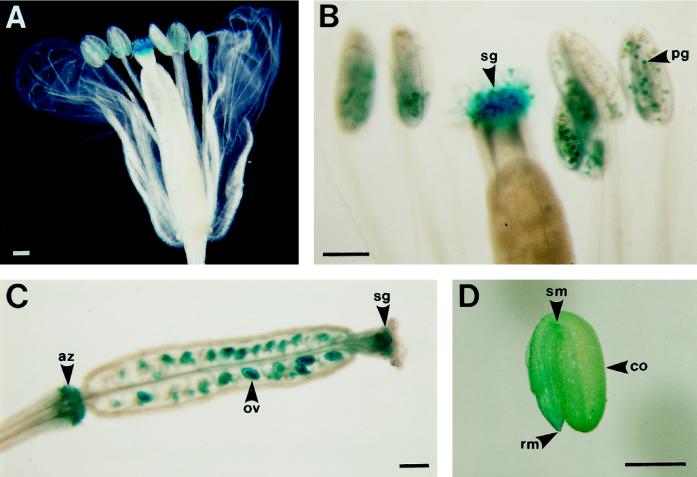
In 8-week-old transgenic Arabidopsis plants, expression of GUS was observed in the flowers (Fig. 6A) without any treatment, especially in the pollen and the stigmas with pollen (Fig. 6B), the stigma of immature siliques (shorter than 5 mm), the ovules, and the abscission zone of the petals and sepals (base of the silique; Fig. 6C), which is a vestige of the stigma. Observation by microscopy of the flower staining showed that GUS activity around the stigmas in the flowers and the tips of the siliques was due to pollen attached to the stigmas rather than to the stigmas themselves (data not shown). The stigma surface of the longer siliques (longer than 5 mm), but not their ovules, showed GUS staining. Mature siliques (longer than 7 mm) showed no GUS staining. However, high GUS activity was observed in seeds of mature siliques that had been cut to facilitate the penetration of 1 mm 5-bromo-4-chloro3-indolyl glucuronide (Fig. 6D). These results indicate that the seed coats inhibited penetration of the GUS substrate. In the seeds high GUS activity staining was observed in the tips of the cotyledons and in the shoot and root meristem.
Figure 6.
Histochemical localization of GUS activity in 3-month-old flowering transgenic Arabidopsis plants and the mature seed containing the ProDH promoter-GUS fusion gene. A, Flower stained overnight. B, Strong GUS staining in pollen grains and stigmas of flowers stained overnight. C, Silique showing strong GUS staining in the stigma, ovules, and abscission zone stained overnight. D, Longitudinal sections of the mature seed showing strong GUS staining, stained for 4 h. az, Abscission zone; co, cotyledon; ov, ovule; pg, pollen grain; rm, root meristem; sg, stigma; and sm, shoot meristem. Bars = 200 μm.
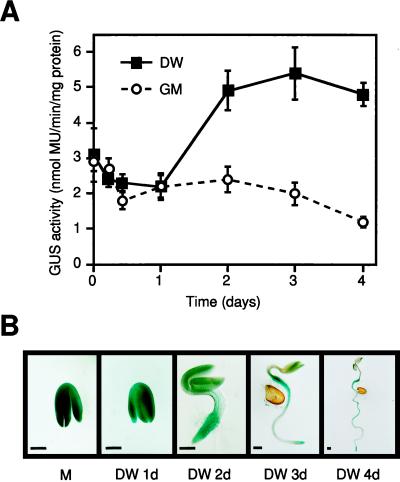
When transgenic T2 seeds were germinated on filter paper soaked with water, GUS activity increased in the germinating seeds under hypoosmolar conditions (Fig. 7). Strong GUS activity was observed mainly in the root tips and veins of germinating seeds (Fig. 7 B). However, when transgenic plants were grown on filter paper soaked with GM solution, GUS activity did not increase during germination (Fig. 7), and only weak GUS activity was observed in the root tips of germinating seeds. The germination process of the seeds in the GM solution was slower than that in water.
Figure 7.
Changes of GUS activity during germination of transgenic Arabidopsis plants containing the ProDH promoter-GUS fusion gene. A, GUS activity in germinating T2 transgenic Arabidopsis plants, each of which carries the gene. The transgenic plants were grown on sterilized filter papers soaked with distilled water (DW) or GM solution. The values of GUS activity are averages of two repeats of three independent transformant lines (n = 5–10). Bars indicate se. B, Histochemical localization of GUS activity in germinating transgenic Arabidopsis plants containing the ProDH promoter-GUS fusion gene. M, Mature seed showing strong GUS staining, stained for 6 h; DW 1d to DW 4d, germinating seeds on a filter paper soaked with distilled water at 1, 2, 3, and 4 d after planting (stained for 6 h). Bars = 200 μm.
DISCUSSION
We have shown, by northern-blot analysis, that the ProDH gene is repressed by dehydration but induced by rehydration and by l- and d-Pro treatment (Kiyosue et al., 1996). We analyzed the expression of the ProDH gene and found that it is up-regulated by hypoosmolarity and by Pro treatment (Fig. 1). The induction of ProDH gene expression under rehydration after 10 h of dehydration is caused not only by accumulated Pro but also by hypoosmolarity. Therefore, the ProDH gene is controlled by three different factors: up-regulation by hypoosmolarity and Pro and down-regulation by dehydration stress.
We then isolated a 1.4-kb promoter region of the ProDH gene and analyzed the function of the promoter using transgenic plants with the 1.4-kb ProDH promoter-GUS fusion gene. We demonstrated that GUS activity driven by the 1.4-kb promoter region was depressed by dehydration (Fig. 3). Northern-blot hybridization and promoter analysis using transgenic plants revealed that the 1.4 kb of the ProDH promoter was activated by hypoosmolarity treatment with GM − Suc solution and water (Figs. 1 and 3). Because the osmolarity of these solutions was lower than the control medium, GM, hypoosmolarity induces ProDH gene expression. Kiyosue et al. (1996) previously reported that a slight induction in ProDH gene expression was detected when the plants were incubated in GM − Suc or water for 10 h. However, analysis of the ProDH mRNA by northern-blot analysis revealed that accumulation of the ProDH mRNA began within 1 h after the transfer of the plants from the agar plates to GM − Suc or water solutions, reached a maximum after 2 h of incubation, and then decreased. Northern analysis and analysis using transgenic Arabidopsis also revealed that the promoter of the ProDH gene was up-regulated by 0.001 to 0.09 m Pro (Fig. 4). The dose dependency of the promoter was consistent with the northern-analysis results of Kiyosue et al. (1996). GUS activity was not detected in dehydrated Arabidopsis plants (Fig. 3). These results indicate that the 1.4-kb promoter region contains all of the cis-acting elements involved in up-regulation by Pro and hypoosmolarity and down-regulation by dehydration.
Kiyosue et al. (1996) reported that 0.26 m d-Pro induced more stable expression of the ProDH gene than 0.26 m l-Pro in Arabidopsis. Figure 1 also suggested that 0.09 m d-Pro treatment for 24 h results in the stronger induction of the ProDH gene expression than 0.09 m l-Pro. However, GUS activity data in Figures 3 and 4 contradict this observation, showing that l-Pro induced a higher level of the ProDH promoter activity. This contradiction may have been due to an experimental variation in the GUS activity caused by differences of insertion positions of the ProDH promoter-GUS fusion gene in transgenic lines. Alternatively, it could have been due to posttranscriptional effects in synthesizing or degradation of the GUS proteins by exogenous amino acids, especially d-Pro.
When the Arabidopsis plants were treated with water or Pro, the ProDH gene was expressed at high levels in all vegetative tissues analyzed (Fig. 5). Highest expression was found in the veins and hydathodes of leaves and root tips under water or Pro supply (Fig. 5, E–H). Histochemical analysis showed that Pro has a stronger effect on the ProDH gene expression than does water in the same plant tissues. Although Pro is not supplied from roots in natural conditions, it is reasonable to assume that excess Pro in cells would be degraded by ProDH. It is suspected that the applied Pro may be transported to almost all parts of plants by Pro transporters such as ProT1 and ProT2 (Rentsch et al., 1996), and that it triggers ProDH gene expression. When alfalfa plants are exposed to water-deficit conditions, Pro accumulates in the phloem sap (Girousse et al., 1996). Expression of the ProDH gene in veins of rehydrated plants indicated that vascular cells convert the accumulated Pro under water-deficit conditions for Glu by ProDH in rehydrated plants. Hydathodes are secretory structures that remove water from the interior of a leaf and deposit it on the leaf surface. Water is forced out of the hydathodes by hydrostatic pressure, and an osmolarity gradient may be established inside the hydathodes. The level of Pro, which is controlled by synthesis and degradation processes, may play a role in the osmolarity gradient of hydathodes. Alternatively, hydathodes may have a role in reabsorption of nutrients, including Pro from vascular bundles, and degrade the Pro by ProDH. Expression in root meristems indicates that ProDH might play a role in catabolism of Pro to amino acids, as well as nitrogen, carbon, energy, and reducing power for elongation of roots in rehydrated plants.
Under normal growth conditions, the ProDH gene was expressed at low levels in all of the vegetative tissues analyzed (Fig. 5, A–D). High levels of expression were found in reproductive tissues such as pollen grains and seeds (Fig. 6). Pro has been found to accumulate in tissues such as florets and seeds, which have a low water content, whereas tissues such as rosette leaves, which have a high water content, contain low levels of Pro (Chiang and Dandekar, 1995). In Arabidopsis reproductive tissues (florets and seeds), Pro represents 17% to 26% of total free amino acids, whereas in vegetative tissues (rosette leaves and roots), Pro contributes only 1% to 3%. Accumulation of Pro was noted in the inflorescence and siliques of Brassica napus (Flasinski and Rogozinska, 1985). In naturally desiccated tissues such as pollen, a high concentration of Pro is correlated with protection against pollen germination at unfavorable temperatures (Zhang and Croes, 1983). The elevated expression of the ProDH gene in pollen grains and seeds in Arabidopsis is consistent with an increased accumulation of Pro in these organs. Pro in pollens and seeds is considered to be dehydrogenated by ProDH to supply Glu or derived compounds and energy for their growth and development.
When transgenic plants containing the ProDH promoter-GUS fusion were germinated in water, GUS activity increased (Figs. 7 and 8). However, when transgenic plants were germinated in GM, GUS activity did not increase. These data indicate that the Pro in seeds might be oxidized to control the osmolarity of seed cells under hypoosmolarity, and used for seed germination and elongation under conditions of insufficient nutrients.
Mutant analysis indicates that the synthesis and sensitivity of GA3 are essential for seed germination and elongation growth in Arabidopsis (Koornneef and Karssen, 1994). Because the expression of the ProDH gene is not affected by GA (Kiyosue et al., 1996; and our results using the transgenic plants), ProDH gene expression during seed germination in water may be due to hypoosmolarity. Thus, ProDH might play a role in supplying nutrients and energy from accumulated Pro for development and germination of reproductive organs such as seeds.
In higher plants Pro is synthesized via two routes, from Orn and from Glu. In young Arabidopsis plantlets, the Orn pathway, together with the Glu pathway, plays an important role in Pro accumulation during osmotic stress (Roosens et al., 1998). In adult Arabidopsis plantlets, the free Pro increase is mainly due to the activity of the enzymes of the Glu pathway (Yoshiba et al., 1997; Roosens et al., 1998). Genes encoding P5CS and P5CR for the Glu pathway have been cloned in Arabidopsis (Savouré et al., 1995; Strizhov et al., 1997; Yoshiba et al., 1997). P5CS has been shown to catalyze the limiting step in Pro accumulation in response to osmotic stress in adult Arabidopsis plants (Strizhov et al., 1997; Yoshiba et al., 1997).
Recently, Hua et al. (1997) reported that the promoter of At-P5R, a gene encoding P5CR in Arabidopsis, directs strong GUS activity in root tips, the shoot meristem, guard cells, hydathodes, veins, pollen grains, ovules, and developing seeds. Among these sites, the root tips, hydathodes, veins, pollen, and seeds overlap the GUS expression sites directed by the ProDH gene, although the timing of the expression of the ProDH gene is different from that of the P5CR gene. GUS staining in the root tips, hydathodes, and veins of transformants with the P5CR promoter-GUS construct was observed under normal conditions (Hua et al., 1997), whereas GUS staining of transformants with the ProDH promoter-GUS construct was strong under high-Pro or hypoosmolar conditions (Fig. 5). Osmolarity of plant tissues may be controlled by the synthesis and degradation of Pro in these sites. During seed ripening GUS activity driven by the P5CR promoter generally decreased, and when the siliques reached 1 cm in length, no staining could be observed (Hua et al., 1997). This pattern is similar to that in the siliques of transformants containing the ProDH promoter-GUS fusion gene (Fig. 6C). Although seeds containing the P5CR promoter-GUS fusion gene were stained after they had been cut apart and treated with 1 mm 5-bromo-4-chloro-3-indolyl glucuronide, no staining was observed (Hua et al., 1997), whereas mature seeds with the ProDH promoter-GUS fusion gene were stained (Fig. 6D). Germinating seeds with the ProDH promoter-GUS fusion gene were also stained under hypoosmolarity (Fig. 7).
These data indicate that the P5CR gene for Pro synthesis is expressed during the early developmental stage of seeds and that the ProDH gene for Pro degradation is expressed during the later developmental stage and during germination of seeds. The ProDH gene was expressed in veins and root tips when plants were exposed to water or Pro, whereas the P5CR gene was expressed in these sites in the absence of stress. However, higher levels of mRNA of the Pro-transporter ProT1 were detected in flowers, and the expression was down-regulated during development in Arabidopsis (Rentsch et al., 1996). We presume that the Pro accumulated in flowers might be synthesized and/or transported at an early developmental stage and degraded to Glu by ProDH for use as a source of nutrients and energy for ripening and germination of pollen and seeds, respectively. We previously reported that P5CS is the key enzyme for Pro biosynthesis in Arabidopsis (Yoshiba et al., 1995). Histochemical analysis of the promoter of the P5CS gene will give us more information about the role of Pro in the development of plants and their acclimation to environmental stress.
This is the first report, to our knowledge, of a promoter that is negatively regulated by dehydration and up-regulated by hypoosmolarity and Pro. The isolated promoter region is also developmentally regulated, especially in reproductive organs. Detailed analysis of the promoter is required to clarify the molecular mechanism of dehydration-rehydration and Pro metabolism in plants.
ACKNOWLEDGMENTS
We thank Ms. A. Konishi and Ms. S. Yoshida (Japan International Research Center for Agricultural Sciences) for their excellent technical assistance. We also thank Dr. Y. Yoshiba (Advanced Research Laboratory, Hitachi Ltd., Japan) for helpful discussions.
Abbreviations:
- P5CR
Δ1-pyrroline-5-carboxylate reductase
- P5CS
Δ1-pyrroline-5-carboxylate synthetase
Footnotes
This research was supported in part by the Program for Promotion of Basic Research Activities for Innovative Biosciences, the Human Frontier Science Program, the Special Coordination Fund of the Science and Technology Agency, and a grant-in aid from the Ministry of Education, Science and Culture of Japan.
The accession number for the DNA sequence reported in this article is AB008810.
LITERATURE CITED
- Ahmad I, Hellebust JA. The relationship between inorganic nitrogen metabolism and proline accumulation in osmoregulatory responses of two euryhaline microalgae. Plant Physiol. 1988;88:348–354. doi: 10.1104/pp.88.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua N-H. The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 1989;8:2195–2202. doi: 10.1002/j.1460-2075.1989.tb08342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer K-H. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–837. doi: 10.1038/335835a0. [DOI] [PubMed] [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Chiang H-H, Dandekar AM. Regulation of proline accumulation in Arabidopsis thaliana (L.) Heynh during development and in response to desiccation. Plant Cell Environ. 1995;18:1280–1290. [Google Scholar]
- Delauney AJ, Verma DPS. Proline biosynthesis and osmoregulation in plants. Plant J. 1993;4:215–223. [Google Scholar]
- Flasinski S, Rogozinska J. Effect of water deficit on proline accumulation, protein and chlorophyll content during flowering and seed formation in winter rape. Acta Agrobot. 1985;38:11–21. [Google Scholar]
- Girousse C, Bournoville R, Bonnemain J-L. Water deficit-induced changes in concentrations in proline and some other amino acids in the phloem sap of alfalfa. Plant Physiol. 1996;111:109–113. doi: 10.1104/pp.111.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X-J, Van de Cotte B, Montagu MV, Verbruggen N. Developmental regulation of pyrroline-5-carboxylate reductase gene expression in Arabidopsis. Plant Physiol. 1997;114:1215–1224. doi: 10.1104/pp.114.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi Y, Yoshiba Y, Sanada Y, Yamaguchi-Shinozaki K, Wada K, Shinozaki K. Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol Biol. 1997;33:857–865. doi: 10.1023/a:1005702408601. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Burgess SM, Hirsh D. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Pro Natl Acad Sci USA. 1986;83:8447–8451. doi: 10.1073/pnas.83.22.8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of cDNA for a dehydration-inducible gene that encodes a Clp A, B-like protein in Arabidopsis thaliana L. Biochem Biophys Res Commun. 1993a;196:1214–1220. doi: 10.1006/bbrc.1993.2381. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD11 and ERD13) for dehydration-inducible genes that encode putative glutathione S-transferases in Arabidopsis thaliana L. FEBS Lett. 1993b;335:189–192. doi: 10.1016/0014-5793(93)80727-c. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Cloning of cDNA for genes that are early responsive to dehydration-stress (ERDs) in Arabidopsis thaliana L.: identification of three ERDs as HSP cognate genes. Plant Mol Biol. 1994;25:791–798. doi: 10.1007/BF00028874. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yoshiba Y, Yamaguchi-Shinozaki K, Shinozaki K. A nuclear gene encoding mitochondrial proline dehydrogenase, an enzyme involved in proline metabolism, is upregulated by proline but downregulated by dehydration in Arabidopsis. Plant Cell. 1996;8:1323–1335. doi: 10.1105/tpc.8.8.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM. Seed dormancy and germination. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334. [Google Scholar]
- Marcotte WR, Jr, Russell SH, Quatrano RS. Abscisic acid response sequences from the Em gene of wheat. Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, myoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Nakagoshi H, Nagase T, Kanei-Ishii C, Ueno Y, Ishii S. Binding of the c-myb proto-oncogene product to the simian virus 40 enhancer stimulates transcription. J Biol Chem. 1990;265:3479–3483. [PubMed] [Google Scholar]
- Nakashima K, Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Plant J. 1997;12:851–861. doi: 10.1046/j.1365-313x.1997.12040851.x. [DOI] [PubMed] [Google Scholar]
- Peng Z, Lu Q, Verma DPS. Reciprocal regulation of Δ1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol Gen Genet. 1996;253:334–341. doi: 10.1007/pl00008600. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Hirner B, Schmelzer E, Frommer WB. Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell. 1996;8:1437–1446. doi: 10.1105/tpc.8.8.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosens NHCJ, Thu TT, Iskander HM, Jacobs M. Isolation of the ornithine-δ-aminotransferase cDNA and effect of salt stress on its expression in Arabidopsis thaliana. Plant Physiol. 1998;117:263–271. doi: 10.1104/pp.117.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savouré A, Jaoua S, Hua X-J, Ardiles W, Montagu MV, Verbruggen N. Isolation, characterization, and chromosomal location of a gene encoding the Δ1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995;372:13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- Schindler U, Menkens AE, Beckmann H, Ecker JR, Cashmore AR. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992;11:1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to drought and cold stress. Curr Opin Biotechnol. 1996;7:161–167. doi: 10.1016/s0958-1669(96)80007-3. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene expression and signal transduction in water-stress response. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Cumbes QJ. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry. 1989;28:1057–1060. [Google Scholar]
- Strizhov N, Ábrahám E, Ökrész L, Blickling S, Zilberstein A, Schell J, Koncz C, Szabados L. Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. 1997;12:557–569. doi: 10.1046/j.1365-313x.1997.00557.x. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Lijsebettens MV. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hua X-J, May M, Montagu MV. Environmental and developmental signals modulate proline homeostasis: evidence for a negative transcriptional regulator. Proc Natl Acad Sci USA. 1996;93:8787–8791. doi: 10.1073/pnas.93.16.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton EF, Clark CJ, Boldingh HL. Effects of hydrogen cyanamide on amino acid profiles in kiwifruit buds during budbreak. Plant Physiol. 1991;97:1256–1259. doi: 10.1104/pp.97.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Mundy J, Chua N-H. Four tightly linked rab genes are differentially expressed in rice. Plant Mol Biol. 1989;14:29–39. doi: 10.1007/BF00015652. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. Correlation between the induction of a gene for Δ1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995;7:751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of the level of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 1997;38:1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093. [DOI] [PubMed] [Google Scholar]
- Zhang H-Q, Croes AF. Protection of pollen germination from adverse temperature: a possible role for proline. Plant Cell Environ. 1983;6:471–476. [Google Scholar]