Abstract
In response to oxidative stress, the transcription factor Nrf2 is upregulated and controls activation of many genes that work in concert to defend cells from damages and to maintain cellular redox homeostasis. p53 has been regarded as the guardian of the genome through its pro-oxidant and antioxidant functions. Under low levels of reactive oxygen species (ROS), “normal” amounts of p53 upregulates expression of antioxidant genes, protecting macromolecules from ROS-induced damage. However, at high levels or extended exposure of ROS, p53 expression is enhanced, activating pro-oxidant genes and resulting in p53-dependent apoptosis. We observed a two-phase Nrf2 expression controlled by p53. (i) The induction phase: when p53 expression is relatively low, p53 enhances the protein level of Nrf2 and its target genes to promote cell survival in a p21-dependent manner. (ii) The repression phase: when p53 expression is high, the Nrf2-mediated survival response is inhibited by p53. Our observation leads to the hypothesis that the p53-mediated biphasic regulation of Nrf2 may be key for the tumor-suppressor function of p53 by coordinating cell survival and death pathways. Antioxid. Redox Signal. 17, 1670–1675.
The Role of p53 and Nrf2 in Cell Survival and Death
P53 plays a critical role in the pathogenesis of cancer by maintaining the integrity of the genome. In response to physiological or mild stresses, p53 exerts an antioxidant function to neutralize reactive oxygen species (ROS) and protect the genome from damage by ROS. Low levels of p53 present in normal cells regulate many antioxidant genes to protect cells from ROS-induced damage (8). However, high levels of ROS or prolonged stress upregulates p53 and provokes a pro-oxidant response to further increase ROS, which subsequently elicits the p53-dependent apoptotic processes to eliminate damaged cells (1). Therefore, these multifaceted functions of p53 are crucial in suppressing DNA mutation, maintaining genome integrity, and protecting mammals from tumorigenesis.
Nrf2 mediates a cell survival response by being a master regulator of the cellular antioxidant and defense responses. It upregulates many intracellular antioxidant proteins and detoxifying enzymes through the antioxidant response elements (AREs) located in the promoters of these genes (6). Coordinated upregulation of Nrf2-target genes in response to environmental insults is essential to suppress ROS and thus protect cells from damage, as illustrated by Nrf2-null mice being highly susceptible to chemical carcinogens (7).
Innovation.
Nrf2 controls a cellular defensive response and maintains cellular redox homeostasis. p53 has been regarded as the guardian of the genome through its pro-oxidant and antioxidant functions. Currently, the relationship between Nrf2 and p53 is unknown. To our knowledge, this is the first study to demonstrate that p53 mediates a two-phase Nrf2 response and that Nrf2 is another key player for the tumor-suppressor function of p53. This possibly represents an important new paradigm by which the p53 signaling pathway can induce or inhibit the Nrf2 pathway to determine cell fate. Our current data provide insight to a novel mechanism of how p53 maintains genome integrity by coordinating cell survival and death pathways and suggest that modulating the p53-Nrf2 axis is a potential strategy for cancer intervention.
ROS has been demonstrated as a critical determinant in eliciting a specific function of p53, which dictates cell survival or cell death. Oridonin, a diterpenoid, generates ROS, activates p53, and triggers p53-dependent apoptosis in many types of cancer cells (5). Interestingly, we previously reported that oridonin upregulated Nrf2 at low doses in a dose-dependent manner, while downregulated Nrf2 at high doses (3). These observations implicate that Nrf2 is activated at subtoxic doses by certain compounds to maintain cellular redox homeostasis when cells are under mild reactive stress. On the other hand, cellular apoptotic processes are engaged to remove damaged cells when cells are exposed to highly toxic doses of chemical compounds. At this “point of no return,” the Nrf2-mediated cellular survival mechanism must be disabled through downregulation of Nrf2, allowing initiation of death processes.
Nrf2 Is Regulated by p53 in Two-Phases
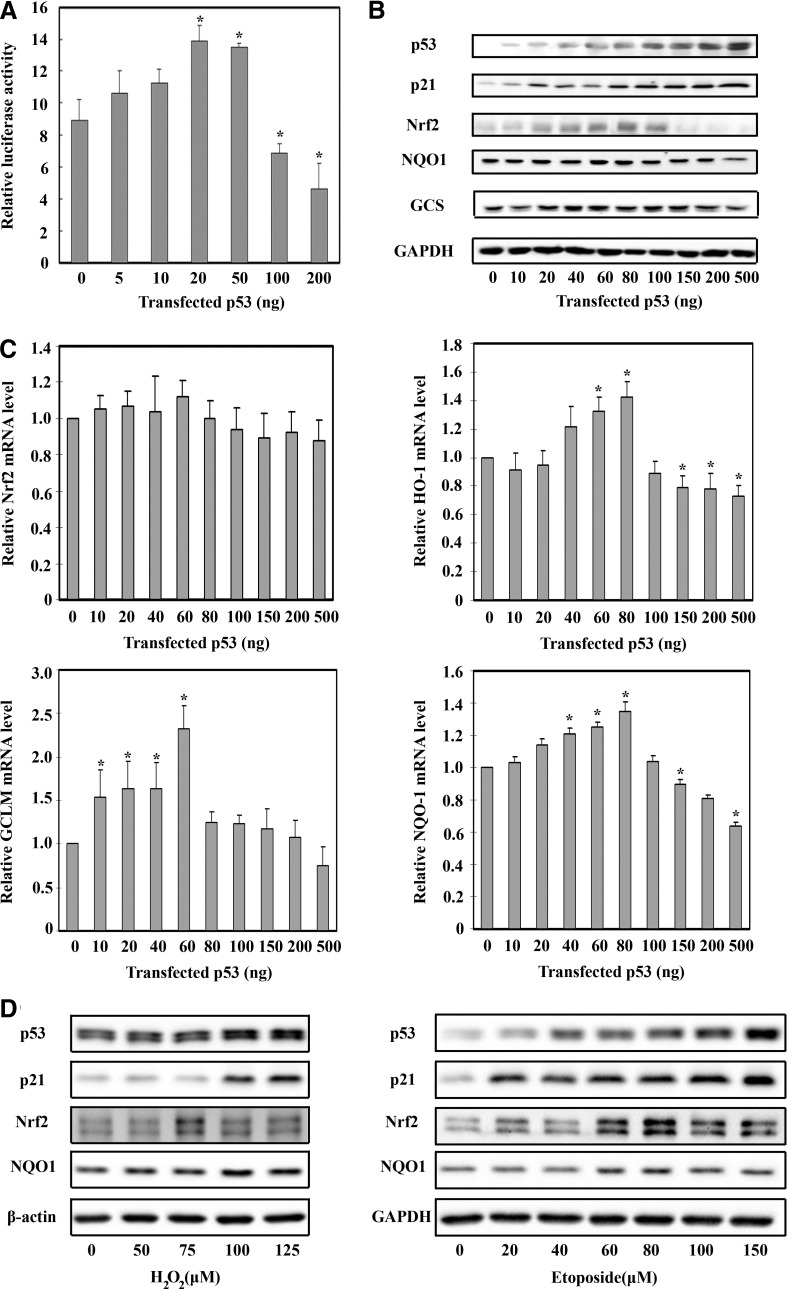
We observed a two-phase dose–response curve of Nrf2 when different amounts of p53 were expressed in a p53-null cell line, Saos-2. p53 enhanced the Nrf2-dependent luciferase activity at first (the induction phase) then decreased the activity (the repression phase) in a dose-dependent manner (Fig. 1A). Further, p53 increased Nrf2 and its target genes (NAD(P)H quinone oxidoreductase [NQO1] and gamma-glutamyl cysteine synthetase [γ-GCS]) when p53 levels were low, followed by a decrease of Nrf2, NQO1, and γ-GCS at high levels (Fig. 1B). As expected, p21 showed a monophasic increase by p53 overexpression (Fig. 1B). Real-time RT-PCR (qRT-PCR) analysis demonstrates that p53 did not modulate Nrf2 at the mRNA level, but mediated a two-phase response in the mRNA expression of heme oxygnase-1 (HO-1), GCLM, and NQO1 (Fig. 1C). A two-phase Nrf2 response was also observed in human renal mesangial cells (HRMC) when endogenous p53 was induced by either H2O2 or etoposide (Fig. 1D).
FIG. 1.
A two-phase Nrf2 response by p53. (A) Saos-2 cells were transfected with the indicated amount of p53, firefly luciferase, and renilla luciferase. Firefly and renilla luciferase activities were measured 36 h after transfection. (B, C) Saos-2 cells were transfected with different amounts of p53 for 36 h and subjected to immunoblot analysis (B) and qRT-PCR (C). (D) Human renal mesangial cells were treated with H2O2 or etoposide for 24 h and subjected to immunoblot analysis. *p<0.05.
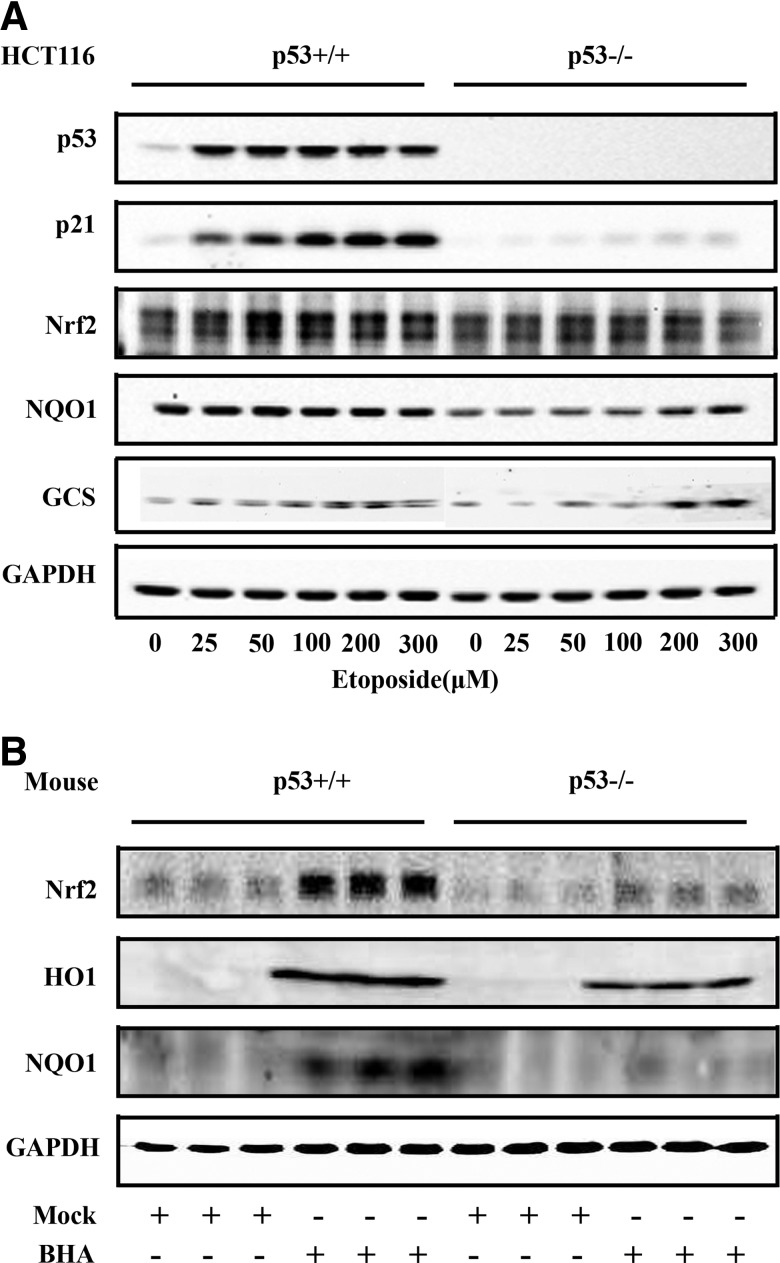
Next, HCT116 p53+/+ and p53−/− cells were used to confirm the two-phase response model. Etoposide treatment gave rise to both the induction and repression phase of Nrf2 expression in p53+/+ cells, whereas the two-phase response was blunted in p53−/− cells (Fig. 2A). A similar two-phase response was also observed with the protein levels of NQO1 and γ-GCS only in p53+/+ cells (Fig. 2A). It is worth mentioning that both basal and induced levels of NQO1 and γ-GCS were lower in p53−/− cells (Fig. 2A). Next, the expression of Nrf2 and its target genes were examined in vivo using p53+/+ and p53−/− mice. Tert-butyl-hydroxyanisole (BHA) was used to induce mild oxidative stress. Both the basal and induced levels of Nrf2, NQO1, and HO-1 in the liver were lower in p53−/− mice than p53+/+ mice (Fig. 2B), indicating that p53 may be important in maintaining both basal and induced levels of Nrf2.
FIG. 2.
p53 is important in maintaining both basal and induced levels of Nrf2. (A) HCT116 p53+/+ and p53−/− cells were treated with etoposide for 24 h and subjected to immunoblot analysis. (B) Wild-type or p53-deficient mice (n=3) were treated with 300 mg/kg BHA for 12 h through intraperitoneal injection. Liver tissues were subjected to immunoblot analysis.
Inhibition of Endogenous p53 Downregulates Nrf2 and Its Downstream Genes
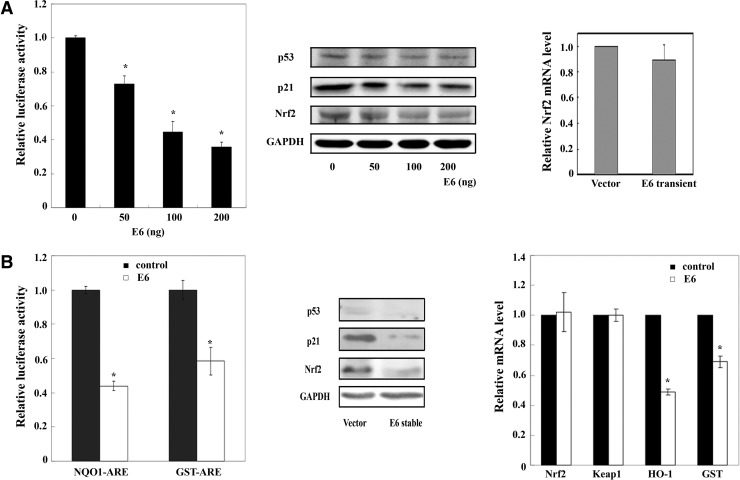
Next, the E6 protein of papillomavirus type 16 was used to transiently decrease endogenous p53. E6 inhibited ARE-luciferase activities (Fig 3A, left panel) and decreased protein levels of p53, p21, and Nrf2 (Fig. 3A, middle panel). E6 had no effect on mRNA expression of Nrf2, indicating that p53-mediated reduction of Nrf2 is at the protein level (Fig. 3A, right panel). As confirmation, an E6 stable cell line was used. Stable overexpression of E6 consistently resulted in a marked reduction in NQO1-ARE or glutathione S-transferase (GST)-ARE luciferase activities (Fig 3B, left panel) and protein levels of p53, p21, and Nrf2 (Fig. 3B, middle panel). Again, reduction of Nrf2 by E6 is post-transcriptional since the mRNA expression of Nrf2 is similar while that of HO-1 and GST-Ya was reduced (Fig. 3B, right panel). In addition, E6 had no effect on the mRNA level of Keap1 (Fig. 3B, right panel).
FIG. 3.
Inhibition of endogenous p53 downregulates Nrf2 and its downstream genes. (A) MEF cells were transiently transfected with E6, firefly luciferase, and renilla luciferase for 48 h. Nrf2 transcriptional activity is presented as firefly luciferase normalized to renilla luciferase (left panel). Expression of p53, p21, Nrf2, and GAPDH was analyzed by immunoblot (middle panel). Nrf2 mRNA was measured by qRT-PCR (right panel). (B) An E6 stable MEF cell line was transfected with either an NQO1-ARE or GST-Ya-ARE firefly luciferase and renilla luciferase. Relative luciferase activity is shown as previously described (left panel). Immunoblot analysis was performed (middle panel). mRNA expression of Nrf2, Keap1, HO-1, and GST were measured using qRT-PCR (right panel). ARE, antioxidant response element; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; HO-1, heme oxygnase-1; MEF, mouse embryonic fibroblast. NQO1, NAD(P)H quinone oxidoreductase. *p<0.05.
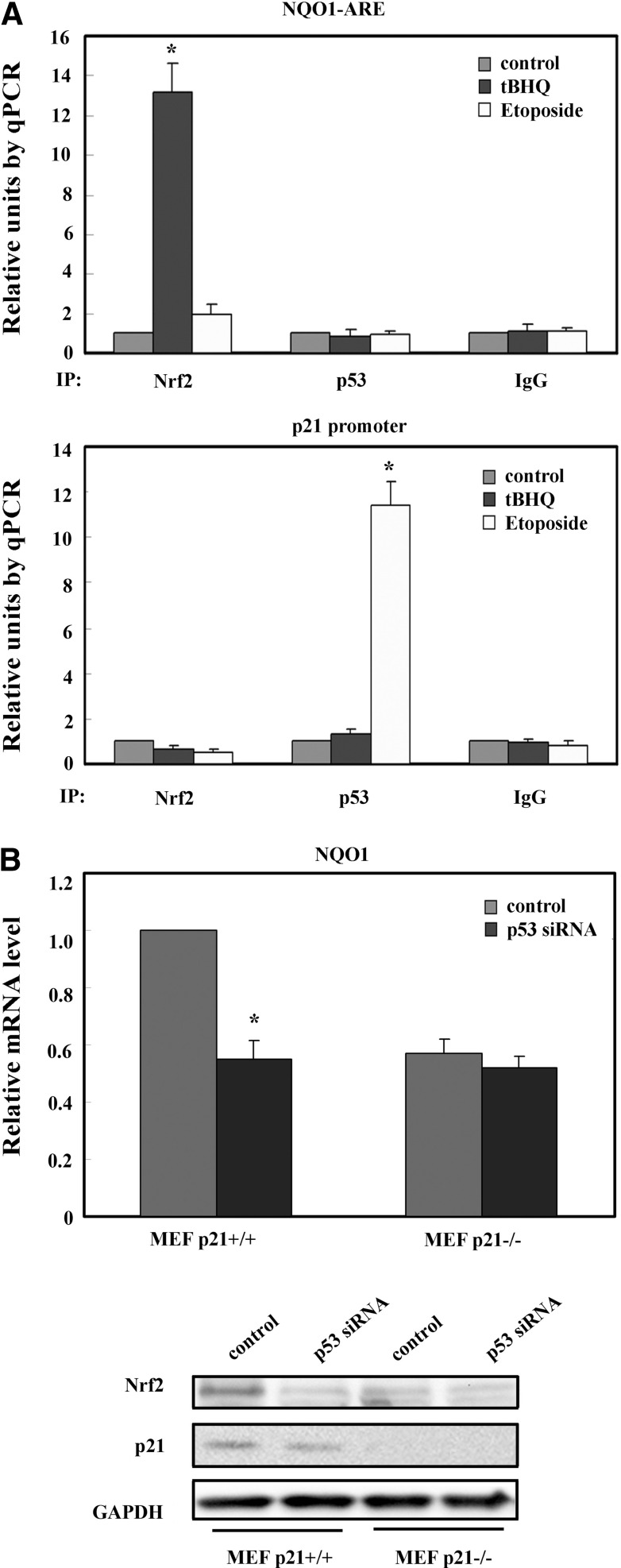
p53-Mediated Downregulation of Nrf2 Requires p21, but Does Not Involve Direct Binding of p53 to the ARE
It has been reported that overexpression of p53 inhibits the transcription of Nrf2 downstream genes, such as x-CT, NQO1 and GST-α1 (4). The authors speculated that p53 binds to a sequence near the ARE, thus repressing Nrf2-dependent transcription by displacing Nrf2 from the ARE (4). As shown in our chromatin immunoprecipitation (ChIP) assay (Fig 4A), tBHQ, an Nrf2 activator, significantly enhanced Nrf2-ARE, but not p53-ARE binding (Fig 4A, top panel). In contrast, etoposide induced p53 binding to the p53 response element, but had no effect on Nrf2 binding to the ARE or p53 response element (Fig. 4A). As expected, etoposide had no effect on p53-ARE binding (Fig. 4A, top panel) and tBHQ had no effect on Nrf2 or p53 binding to the p53 response element (Fig. 4A, bottom panel). These data clearly demonstrate that p53 does not regulate Nrf2 target genes through interaction with the ARE. We then hypothesized that the p53-mediated regulation of Nrf2 expression is through p21, since we previously have shown that p21 competes with Keap1 for Nrf2 binding, thus blocking Keap1-mediated ubiquitination and degradation of Nrf2 (2). Upon transfection of p53-siRNA in mouse embryonic fibroblast (MEF) p21+/+ and p21−/− cells, we observed a reduction in NQO1 mRNA and Nrf2 protein levels only in p21+/+ cells (Fig. 4B). Additionally, there were lower basal levels of NQO1 mRNA and Nrf2 protein in p21−/− cells (Fig. 4B).
FIG. 4.
p53-mediated downregulation of Nrf2 requires p21 but not direct binding to the ARE. (A) Chromatin immunoprecipitation analysis was performed in MEF cells treated with 50 μM tBHQ or 50 μM etoposide for 24 h. (B) MEF p21+/+ and p21−/− cells were transfected with p53-siRNA for 72 h prior to qRT-PCR and immunoblot analysis. *p<0.05.
In summary, we discovered the two-phase regulation of Nrf2 by p53. At low to mild stress conditions, low levels of p53 upregulate the Nrf2-dependent antioxidant response through p21 to reduce ROS and promote cell survival. On the other hand, when cells have been damaged under high stress conditions, elevated p53 levels suppress the Nrf2-mediated cell survival pathway and presumably initiate the apoptotic cell death processes. By doing so, p53 eliminates damaged cells and ensures the integrity of the genome, thus suppressing tumorigenicity. We hypothesize that Nrf2 is another key player for the tumor-suppressor function of p53. The mechanism by which p53 regulates Nrf2 during the repression phase requires further studies.
Notes
Chemicals, antibodies, and transfection reagents
H2O2 (Fisher Scientific), etoposide (Sigma-Aldrich), and BHA (Sigma-Aldrich) were all purchased from commercial sources. Nrf2, p53, p21, NQO1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, and HO-1 and secondary antibodies were all purchased from Santa Cruz Biotechnology. Transfection of cDNA or siRNA was performed using Lipofectamine Plus (Invitrogen) or HiPerFect (Qiagen), respectively, according to the manufacturer's instructions.
Animal treatment, isolation of MEFs and cell culture conditions
Wild-type, p53−/−, and p21−/− mice were purchased from Jackson Laboratory. Male mice at 6 weeks of age were randomly assigned to BHA (300 mg/kg in corn oil, i.p.) or control group (corn oil, i.p.) and euthanized at 12 h following administration. Livers were excised and subjected to immunoblot analysis. MEF cells were isolated from p53 and p21 wild-type and knockout pregnant female mice at embryonic day 13. Saos-2, HCT116, and MEF cells were all maintained in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) and HRMCs (ScienCell Research Laboratories) was maintained in RPMI-1640 supplemented with 2% fetal bovine serum and mesangial cell growth supplement (ScienCell Research Laboratories).
Reporter gene assay, real-time PCR, and ChIP
For the reporter gene assay, cells were transfected with different amounts of p53, along with expression vectors for both ARE-firefly luciferase and thymidine kinase promoter (TK)-renilla luciferase. Luciferase activity was measured using the Promega dual-luciferase reporter gene assay system according to the manufacturer's instructions. Nrf2 transcriptional activity was expressed as firefly luciferase activity normalized to renilla luciferase activity. qRT-PCR was done on total mRNA extracted from cells using TRIzol (Invitrogen). Equal amounts of mRNA were used to generate cDNA using the Transcriptor First Strand cDNA synthesis kit purchased from Roche. qRT-PCR procedures and primer sequences were described previously and the LightCycler 480 system was used (Roche) (9). All reporter gene and qRT-PCR analysis were repeated in three independent experiments and in duplicates. Data are all shown as mean±standard deviation. Statistical analysis was performed using two-tailed Student's t tests to compare means. Significance was set at p<0.05. ChIP assay was performed according to our established protocol (9).
Abbreviations Used
- ARE
antioxidant response element
- BHA
tert-butyl-hydroxyanisole
- ChIP
chromatin immunoprecipitation
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GST
glutathione S-transferase
- HO-1
heme oxygenase-1
- HRMC
human renal mesangial cells
- MEF
mouse embryonic fibroblast
- NQO1
NAD(P)H quinone oxidoreductase
- ROS
reactive oxygen species
- γ-GCS
gamma-glutamyl cysteine synthetase
Acknowledgments
We thank Dr. S.A. Whitman for critical reading and editing of the article. This study was supported by ES015010 and CA154377, awarded to D.D.Z., and ES006694, a center grant.
References
- 1.Bensaad K. Vousden KH. Savior and slayer: the two faces of p53. Nat Med. 2005;11:1278–1279. doi: 10.1038/nm1205-1278. [DOI] [PubMed] [Google Scholar]
- 2.Chen W. Sun Z. Wang XJ. Jiang T. Huang Z. Fang D. Zhang DD. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du Y. Villeneuve NF. Wang XJ. Sun Z. Chen W. Li J. Lou H. Wong PK. Zhang DD. Oridonin confers protection against arsenic-induced toxicity through activation of the Nrf2-mediated defensive response. Environ Health Perspect. 2008;116:1154–1161. doi: 10.1289/ehp.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faraonio R. Vergara P. Di Marzo D. Pierantoni MG. Napolitano M. Russo T. Cimino F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- 5.Gao FH. Liu F. Wei W. Liu LB. Xu MH. Guo ZY. Li W. Jiang B. Wu YL. Oridonin induces apoptosis and senescence by increasing hydrogen peroxide and glutathione depletion in colorectal cancer cells. Int J Mol Med. 2012;29:649–655. doi: 10.3892/ijmm.2012.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes JD. McMahon M. Chowdhry S. Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the keap1-nrf2 pathway. Antioxid Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- 7.Ramos-Gomez M. Kwak MK. Dolan PM. Itoh K. Yamamoto M. Talalay P. Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sablina AA. Budanov AV. Ilyinskaya GV. Agapova LS. Kravchenko JE. Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Z. Zhang S. Chan JY. Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]