Abstract
Background
The incidence of thyroid cancer is increasing worldwide. Up to 30% of thyroid fine-needle aspiration (FNA) biopsies are inconclusive and this is primarily due to several thyroid histologic subtypes with overlapping cytologic features. MicroRNAs (miRNAs) are small noncoding RNAs and have been implicated in carcinogenesis. We hypothesized that there are miRNAs that are differentially expressed between benign and malignant thyroid tumors that are difficult to distinguish by FNA biopsy.
Methods
The expression of 1,263 human miRNAs was profiled in 47 tumor samples representing difficult to diagnose histologic subtypes of thyroid neoplasm (21 benign, 26 malignant). Differentially expressed miRNAs were validated using qRT-PCR. The area under the receiver operating characteristic curve (AUC) was used to determine the diagnostic accuracy of differentially expressed miRNAs.
Results
Supervised hierarchical cluster analysis demonstrated grouping of 2 histologies (papillary and follicular thyroid carcinoma). 34 miRNAs were differentially expressed in malignant compared to benign thyroid neoplasms (p<0.05). 25 of the 34 nonproprietary miRNAs were selected for validation, and 15 of the 25 miRNAs were differentially expressed between benign and malignant samples with p-value <0.05. Seven miRNAs had AUC values > 0.7. miR-7 and miR-126 had the highest diagnostic accuracy with an AUC = 0.81 and 0.77, respectively.
Conclusion
To our knowledge, this is the first study to evaluate the diagnostic accuracy of miRNAs in thyroid histologies that are difficult to distinguish as benign or malignant by FNA biopsy. miR-126 and miR-7 had high diagnostic accuracy and could be helpful adjuncts to thyroid FNA biopsy.
Introduction
Thyroid cancer is the most common endocrine malignancy. It arises from two cell types (follicular and parafollicular cells) with follicular cell-derived thyroid cancer accounting for approximately 95% of all thyroid cancers cases.[1–3] Thyroid cancer incidence has doubled in the last three decades [4–5] and it is estimated that there will be more than 44,000 new cases and 1,690 deaths in 2011 in the United States. [6]
Fine-needle aspiration biopsy (FNAB) and cytologic examination is the gold-standard in the work up of a thyroid nodule to exclude a cancer diagnosis.[7–11] The cytologic finding of a fine-needle aspiration can be grouped into 6 categories (nondiagnostic or unsatisfactory sample, benign, follicular lesion of undetermined significance, follicular neoplasm or suspicious for a follicular neoplasm, suspicious for malignancy, and malignant) based on The Bethesda System for Reporting Thyroid Cytopathology with each category associated with a different malignancy risk (benign: 0–3%; follicular lesion of undetermined significance: 5–15%; follicular neoplasm or suspicious for a follicular neoplasm: 15–30%; suspicious for malignancy: 60–75%; and malignant: 97–99%).[12] Although FNAB has relatively high sensitivity and specificity (65–98% and 72–100%, respectively)[8] for those lesions that are definitively categorized as benign or malignant, up to 10–25% of the lesions are interpreted as indeterminate without a definitive cytological diagnosis.[13–16] Follicular and Hürthle cell neoplasms pose a significant challenge as the cytological features between adenomas and carcinomas are indistinguishable.[17–18] Thyroidectomy is commonly recommended for lesions interpreted as indeterminate or suspicious due to the high risk of malignancy and the extent of surgery may vary depending on the final histology, tumor size of the lesion, and the patients’ risk factors for malignancy and patient preference.[8, 10–12] At a minimum, thyroid lobectomy is recommended for lesions interpreted as follicular neoplasm or suspicious for a follicular neoplasm in order to reach a definitive histological diagnosis. However, given a malignancy risk of 15–30% for these indeterminate lesions, the vast majority of final pathologic diagnoses are benign rendering many of the surgical interventions unnecessary.
Our understanding of the molecular basis of thyroid neoplasm has improved and numerous investigators have evaluated molecular markers to improve the accuracy of thyroid cancer diagnosis given the limitations of thyroid FNA biopsy mentioned above.[3, 13, 19–23] Among these efforts include investigations on clinical and tumor characteristics to create a diagnostic predictor model,[24–25] combined analysis of clinical factors and candidate diagnostic markers[26], and the detection of common somatic mutations and gene rearrangements associated with thyroid cancer.[27–31] Recently there has been increasing interest in examining the expression profile of microRNAs (miRNAs) in thyroid cancers to not only understand tumorigenesis, but also to improve thyroid cancer diagnosis.[32–36] These studies have, however, focused on identifying differentially expressed miRNA in papillary thyroid cancer and not the histologic subtypes that are challenging to diagnose on FNA biopsy.
miRNAs are small non-coding RNAs of approximately 21–23 nucleotides long and they function as regulatory molecules by binding to target messenger RNA (mRNA) in the 3′-untranslated region (3′-UTR) leading to either repression of mRNA translation or promoting degradation. [37–39] miRNAs may function as either tumor suppressors or as oncogenes [40–42] and have been demonstrated to have a tissue specific pattern of expression in several cancer histologies.[39, 43–45] Nikiforova et al demonstrated distinct miRNA expression profile amongst various histopathological types of thyroid tumors and demonstrated a correlation between dysregulated miRNA patterns and the presence of somatic mutation.[33] This study also demonstrated significant correlation between miRNA dysregulation in tissue and FNAB samples.[33] miR-221 and miR-222 have been found to be consistently upregulated in several studies.[32–34, 46–49] Chen et al validated the presence and upregulation of miR-221, miR-222, and miR-146b in papillary thyroid cancer and demonstrated potential diagnostic utility of miR-222 and miR-146b as distinguishing markers for papillary thyroid cancer in FNAB.[32]
Although there have been several investigations which have identified miRNAs that are dysregulated among different histologic types of thyroid neoplasms, there has not been a study that addresses the specific histologies that are difficult to diagnose on FNAB as a whole. Thus, the goals of our study were to perform microRNA profiling on thyroid tissue samples by using microarray analysis to identify miRNAs that are dysregulated between benign and malignant histologies that are difficult to diagnose by FNA biopsy and to validate the result of tissue microarray using qRT-PCR.
Materials and Methods
Tissue samples
Thyroid tissue samples, patient demographics, and histopathological information were obtained on a protocol approved by the Committee on Human Research at the University of California, San Francisco. Samples were obtained at the time of thyroidectomy, snap frozen in liquid nitrogen, and stored at −80°C. Tissue samples were classified as normal, benign [follicular adenoma (FA), multinodular goiter (MNG), Hürthle cell adenoma (HCA)], or malignant [Hürthle cell carcinoma (HCC), papillary thyroid carcinoma (PTC), follicular variant of papillary thyroid carcinoma (FPTC), follicular thyroid carcinoma (FC)] based on secondary review of the procured tissue sample by an endocrine pathologist. Multinodular goiter (MNG) samples were initially interpreted as indeterminate (follicular neoplasm) on preoperative FNA biopsy and permanent histologic examination demonstrated hyperplastic nodule in the setting of multinodular goiter. Normal thyroid tissue was obtained from patient undergoing thyroidectomy for benign or malignant disease from the contralateral thyroid lobe. All the thyroid tissue samples, except normal, included in the analysis were interpreted as either indeterminate (follicular neoplasm/lesion) or suspicious for malignancy on the preoperative thyroid FNAB.
RNA isolation
RNA extraction was performed using TriZol (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA quantity and quality were determined using NanoDrop (Thermo Scientific, Wilmington, DE) and the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA).
miRNA microarray profiling and data analysis
200ng of total RNA from 47 tissue samples (21 benign and 26 malignant) was submitted for microarray profiling. Pooled RNA of all 68 samples that were submitted was used as the reference RNA. Total RNA from each sample and reference RNA was labeled with Hy3 and Hy5 fluorescent labels respectively, using the miRCURY LNA Array power labeling kit (Exiqon) as described by the manufacturer. Fluorescent labeled RNA samples were mixed pair wise and hybridized to the miRCURY LNA array version 11.0 (Exiqon) containing 1,263 probes from miRBase version 14.0. Arrays were scanned using Agilent 62505M Microarray scanner and the spot intensities were measured using ImaGene image analysis software (BioDiscovery, El Segundo, CA). Data was normalized using R scripts (Bioconductor). For each miRNA probe, log2 ratio of Hy3 and Hy5 signals were calculated. A two-tailed t-test was used to compare expression levels between benign and malignant histologies.
Quantitative real-time RT-PCR
Statistically significant differentially expressed miRNAs based on miRNA microarray were validated using quantitative real-time RT-PCR (qRT-PCR) using total RNA from the same 47 tissue samples analyzed on microarray. qRT-PCR was carried out using TaqMan® microRNA assay probes (Applied Biosystems, Foster City, CA). Probes for 25 out of 34 differentially expressed miRNAs were nonproprietary and used for the validation [miR-126 (assay #002228), miR-222 (assay #002276), miR-144 (assay #002676), miR-30e (assay #002223), miR-221 (assay #000524), miR-374a (assay #000563), miR-145 (assay #002278), miR-30c (assay #000419), miR-146b-5p (assay #001097), miR-143 (assay #002249), miR-195 (assay #000494), miR-335 (assay #000546), miR-193a-3p (assay #002250), let-7g (assay #002282), miR-30a (assay #000417), miR-30b (assay #000602), miR-7 (assay #000268), miR-26b (assay #000407), miR-101 (assay #002253), let-7f (assay #000382), miR-451 (assay #001105), miR-32* (assay #002129), miR-22 (assay #000398), miR-30b* (assay #002129), miR-744 (assay #002324)]. cDNA was created using 5ng of total RNA and miRNA specific hairpin primers with TaqMan® microRNA Reverse Transcription Kit (PN 4366597, Applied Biosystems). 5ng of total RNA was used in a 15 μl reaction to create the cDNA template. 2.5 μl of cDNA was used in a 10μl PCR reaction and all qRT-PCR reaction was performed in triplicates using miRNA specific primers (TaqMan® microRNA assay) and TaqMan® Universal PCR Master Mix, No AmpErase UNG (PN #4324018, Applied Biosystems, Foster City, CA) on an ABI 7900HT Fast Real-Time PCR system (Applied Biosystems). Several candidate endogenous controls were tested [U6 (assay#001973), RNU 48 (assay #001006), miR-423-5p (assay #002340), miR-885-5p (assay #002296), miR-519d (assay #002403), miR-625* (assay #002432)] and the control with the lowest variability among all the tissue samples (miR-625*) was selected for the endogenous control. miRNA expression level was calculated using delta cycle threshold (ΔCt), which is the cycle threshold (Ct) of the miRNA of interest subtracted by the Ct value of the endogenous control (miR-625*). Fold difference between benign and malignant samples was determined by using the formula [Fold change = 2−ΔΔCt] and ΔΔCt was calculated by normalizing the ΔCt to the endogenous control sample, in this case was RNA from TPC-1 cell line.
Statistical analysis
The Mann-Whitney U test was used to compare miRNA expression levels by histologic diagnosis. A p-value of less than 0.05 was considered worthy of further evaluation. A Pearson correlation coefficient (r) was calculated on microarray and qRT-PCR data to assess the amount of correlation between the two assays. Area Under the Curve (AUC) was calculated on the 15 most significant miRNAs and receiver operating curve (ROC) was plotted for the top 2 statistically significant miRNAs.
Results
Tissue sample microarray
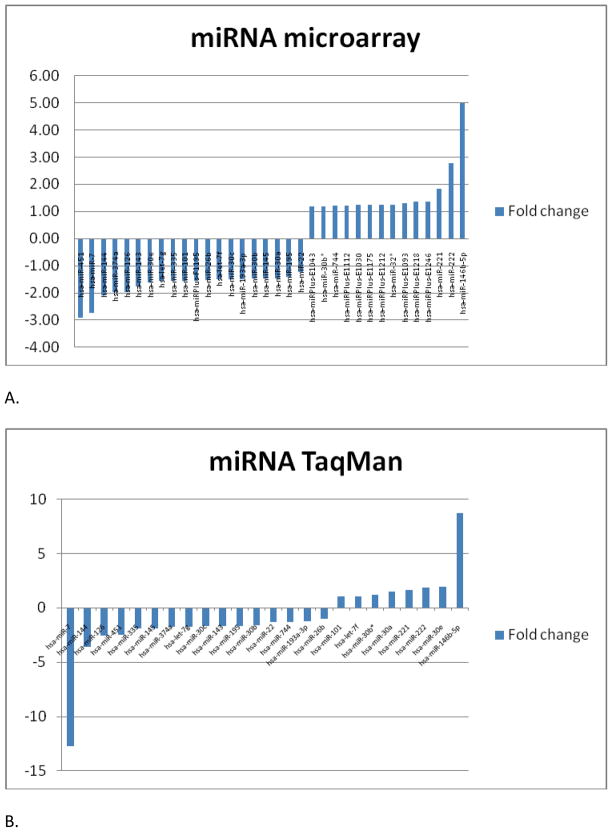
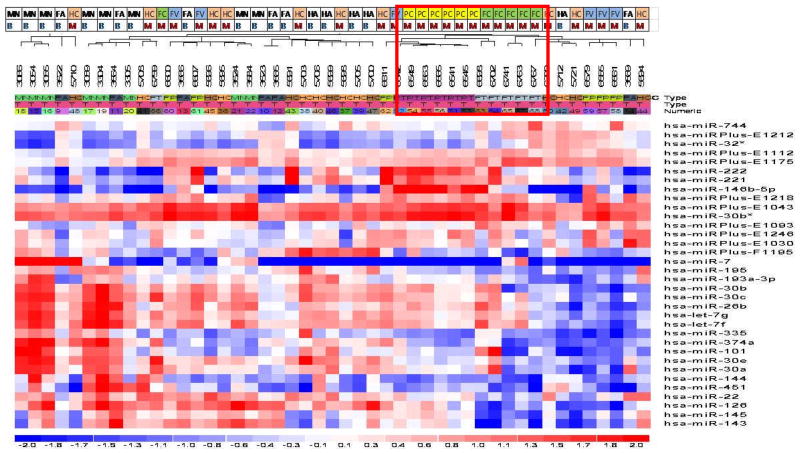
miRNA microarray was performed on a total of 47 tissue samples [21 benign (6 FA; 8 MNG; 7 HCA) and 26 malignant (8 HCC; 6 PTC; 6 FPTC; 6 FC)]. Thirty-four miRNAs were found to be differentially expressed between benign and malignant samples with p value <0.05 (Figure 1A). miR-146b-5p and miR-222 were up-regulated more than 2-fold and miR-451, miR-7, and miR-144 were down-regulated more than 2-fold compared to the benign samples (Table 1). Interestingly, miR-221 and miR-222, which been demonstrated consistently to be upregulated in thyroid malignancy in previous studies, were also found to be upregulated in our study.[32–34, 46–48] Supervised hierarchical clustering of the 34 most differentially expressed miRNAs demonstrated clustering of small groups of papillary thyroid cancer and follicular thyroid cancer but overall there was no clear separation between benign and malignant samples (Figure 2).
Figure 1.
(A) Top 34 miRNAs that were differentially up- or down- regulated between benign and malignant thyroid neoplasm on microarray analysis with p value of <0.05. (B). 24 miRNAs that were validated on TaqMan® qRT-PCR. MiRNA expression level differences between the microarray and qRT-PCR data were similar. miR-32* is not shown as it did not amplify on TaqMan® qRT-PCR.
Table 1.
Differentially expressed miRNAs between benign and malignant on microarray data. 14 miRNAs were up-regulated (A) and 20 were down-regulated (B). The right column represents the fold difference of each miRNAs relative to pooled samples.
| Upregulated miRNA in malignant compared with benign thyroid neoplasm
| ||
|---|---|---|
| miRNA | p-value | Fold change |
| hsa-miR-146b-5p | 3.5E-03 | 5.00 |
| hsa-miR-222 | 6.1E-03 | 2.77 |
| hsa-miR-221 | 2.2E-01 | 1.84 |
| hsa-miRPlus-E1246 | 2.3E-01 | 1.36 |
| hsa-miRPlus-E1218 | 1.6E-02 | 1.35 |
| hsa-miRPlus-E1093 | 3.1E-02 | 1.29 |
| hsa-miR-32* | 1.9E-02 | 1.26 |
| hsa-miRPlus-E1212 | 1.5E-02 | 1.26 |
| hsa-miRPlus-E1175 | 3.0E-02 | 1.25 |
| hsa-miRPlus-E1030 | 3.2E-01 | 1.25 |
| hsa-miRPlus-E1112 | 3.4E-02 | 1.21 |
| hsa-miR-744 | 7.2E-01 | 1.21 |
| hsa-miR-30b* | 2.8E-01 | 1.18 |
| hsa-miRPlus-E1043 | 3.8E-01 | 1.18 |
|
| ||
| A | ||
| Downregulated miRNA in malignant compared with benign thyroid neoplasm
| ||
|---|---|---|
| miRNA | p-value | Fold change |
| hsa-miR-451 | 1.1E-01 | −2.91 |
| hsa-miR-7 | 4.8E-02 | −2.74 |
| hsa-miR-144 | 1.7E-02 | −2.10 |
| hsa-miR-374a | 4.2E-03 | −1.98 |
| hsa-miR-126 | 3.8E-05 | −1.96 |
| hsa-miR-143 | 1.6E-02 | −1.76 |
| hsa-miR-30e | 2.1E-04 | −1.62 |
| hsa-let-7g | 5.0E-03 | −1.57 |
| hsa-miR-335 | 3.5E-03 | −1.56 |
| hsa-miR-101 | 6.2E-02 | −1.55 |
| hsa-miRPlus-F1195 | 1.8E-05 | −1.54 |
| hsa-miR-26b | 3.5E-02 | −1.52 |
| hsa-let-7f | 8.6E-03 | −1.50 |
| hsa-miR-30c | 3.1E-03 | −1.49 |
| hsa-miR-193a-3p | 5.1E-04 | −1.49 |
| hsa-miR-30b | 3.1E-03 | −1.46 |
| hsa-miR-145 | 4.5E-02 | −1.46 |
| hsa-miR-30a | 1.4E-02 | −1.45 |
| hsa-miR-195 | 1.7E-05 | −1.41 |
| hsa-miR-22 | 5.9E-05 | −1.24 |
|
| ||
| B. | ||
Figure 2.
Supervised cluster analysis of the top 34 differentially expressed miRNAs. Each row represents a miRNA and each column represents a sample. The top row indicates the specific tumor types [follicular adenoma (FA), multinodular goiter (MN), Hürthle cell adenoma (HA), Hürthle cell carcinoma (HC), papillary thyroid carcinoma (PC), follicular variant of papillary thyroid carcinoma (FV), follicular thyroid carcinoma (FC)]. There is clustering of PC and FC.
Validation of differentially expressed miRNAs by real-time qRT-PCR
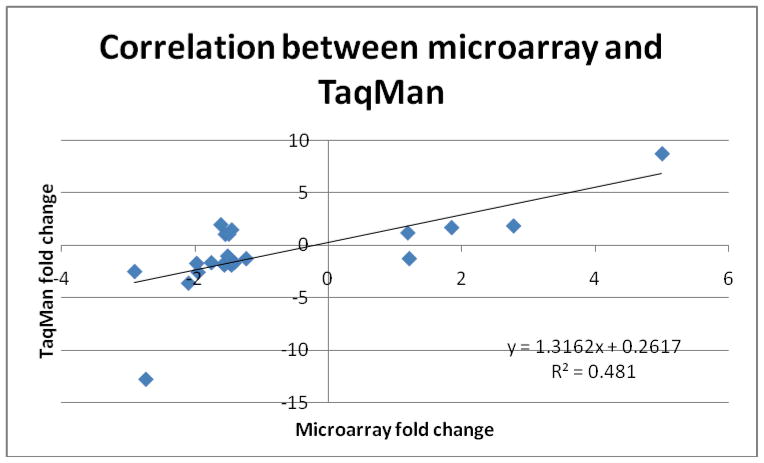
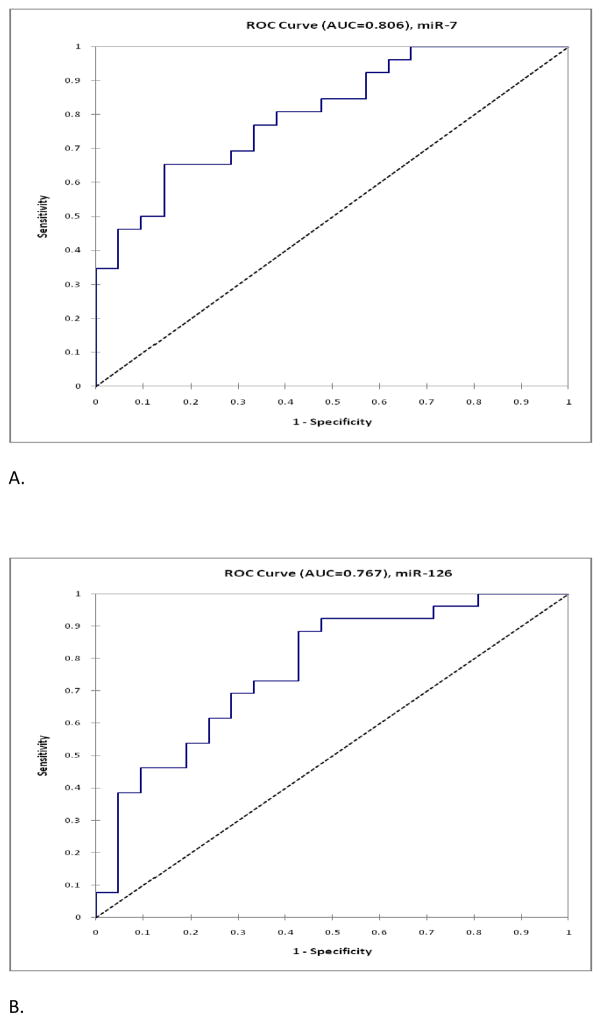
25 miRNAs were nonproprietary and were selected for validation among the 34 differentially expressed miRNAs. There was similar up- and down-regulation profile between microarray and TaqMan® (Figures 1A and 1B) in up to 79% of the miRNAs. The average fold difference values by qRT-PCR TaqMan® for each miRNA were plotted against the fold change values obtained from microarray data and the Pearson correlation coefficient (r) was 0.69 with R2 of 0.48 (Figure 3). Fifteen out of 25 miRNAs were found to be significantly differentially expressed (p<0.05) between benign and malignant samples (Table 2.A). miR-32* did not amplify on TaqMan® PCR. The Area Under the Curve (AUC) was calculated for the 15 most significant miRNAs and nine miRNAs had AUC values greater than 0.70 (Table 2.B). miR-126 and miR-7 had the highest AUC values indicating that these miRNAs had the greatest potential as diagnostic markers (Fig. 4).
Figure 3.
Correlation of fold change values between microarray and qRT-PCR data which demonstrates a good correlation between microarray and qRT-PCR data.
Table 2.
(A) List of miRNAs that were validated on TaqMan® qRT-PCR. The right column shows the p-values for the comparison of between versus malignant and the line is drawn to divide the statistically significant and unsignificant miRNAs. P-value was calculated using Mann-Whitney U test. 15 out of 25 miRNAs were statistically significant. miR-32* value is not shown here as it did not amplify on TaqMan qRT-PCR®. (B) Area under the curve (AUC) values were calculated for the miRNAs that were statistically significant based on the Mann-Whiteney U test. The line is drawn to divide the statistically significant and unsignificant miRNAs.
| miRNA | p-value |
|---|---|
|
| |
| hsa-miR-7 | 0.0004 |
| hsa-miR-126 | 0.0018 |
| hsa-miR-374a | 0.0027 |
| hsa-let-7g | 0.0027 |
| hsa-miR-30c | 0.0058 |
| hsa-miR-195 | 0.0075 |
| hsa-miR-335 | 0.009 |
| hsa-miR-30b | 0.0139 |
| hsa-miR-451 | 0.0165 |
| hsa-miR-145 | 0.0197 |
| hsa-miR-193a-3p | 0.0197 |
| hsa-miR-22 | 0.0247 |
| hsa-miR-144 | 0.0326 |
| hsa-miR-143 | 0.04 |
| hsa-miR-101 | 0.0466 |
|
| |
| hsa-miR-30a | 0.0597 |
| hsa-miR-26b | 0.0657 |
| hsa-miR-744 | 0.0831 |
| hsa-miR-146b-5p | 0.1517 |
| hsa-let-7f | 0.1643 |
| hsa-miR-30e | 0.1709 |
| hsa-miR-222 | 0.2226 |
| hsa-miR-221 | 0.2479 |
| hsa-miR-30b* | 0.5634 |
|
| |
| A. | |
| miRNA | AUC value |
|---|---|
|
| |
| hsa-miR-7 | 0.806 |
| hsa-miR-126 | 0.767 |
| hsa-miR-374a | 0.756 |
| hsa-let-7g | 0.756 |
| hsa-miR-30c | 0.736 |
| hsa-miR-195 | 0.729 |
| hsa-miR-335 | 0.723 |
| hsa-miR-30b | 0.711 |
| hsa-miR-451 | 0.705 |
|
| |
| hsa-miR-145 | 0.7 |
| hsa-miR-193a-3p | 0.7 |
| hsa-miR-22 | 0.692 |
| hsa-miR-144 | 0.688 |
| hsa-miR-143 | 0.676 |
| hsa-miR-101 | 0.67 |
|
| |
| B. | |
Figure 4.
Receiver Operating Curve (ROC) was plotted for (A) miR-7 and (B) miR-126 using the expression profiling represented by ΔCT value on qRT-PCR. Area under the curve (AUC) values are shown on the graph.
Sub-group analysis of the candidate miRNAs
Sub-group analyses of the 2 candidate miRNAs (miR-126 and miR-7) were performed on the miRNA validated by qRT-PCR. Three different comparisons were performed using Mann-Whitney U test: (1) FA versus FC; (2) HCA versus HCC; and (3) FA/HCA versus FC/HCC/FPTC. Both miRNAs were significantly differentially expressed within the first group (p=0.01 and p=0.03, miR-126 and miR-7, respectively) and miR-126 was significantly differentially expressed within the third group (p=0.03). The other comparisons did not reach statistical significance (p=0.64 and p=0.56, miR-126 and miR-7, respectively in Group 2; miR-7 in Group 3 had p-value of 0.13). The same sub-group analyses were performed on the other miRNAs that were validated on the tissue samples and miR-144, -145, and -195 were significantly differentially expressed in Group 1 comparison (p=0.02, p=0.01, p=0.01, respectively) and the remainder of miRNAs did not reach statistical significance.
Discussion
In this study, we identified differentially expressed miRNAs in difficult to diagnose thyroid histologic subtypes in an effort to identify diagnostically useful markers. We found 34 differentially expressed miRNAs by miRNA microarray analysis in a diverse group of benign versus malignant thyroid histologies. 25 of those 34 miRNAs were validated using TaqMan® qRT-PCR and there was good correlation between microarray data and TaqMan® data. 15 out of the 25 miRNAs were confirmed to be differentially expressed by qRT-PCR and nine out of those 15 miRNAs were found to have AUC values >0.7 in the distinction of benign versus malignant lesions. Sub-group analysis indicated that both miR-126 and miR-7 were significantly differentially expressed between follicular adenoma and follicular carcinoma, and miR-126 was significantly differentially expressed in follicular carcinoma, Hürthle cell carcinoma, and follicular variant of papillary thyroid carcinoma as compared to follicular adenoma and Hürthle cell adenoma.
As the incidence of thyroid cancer is increasing, there is a pressing need to improve the accuracy of thyroid cancer diagnosis, in particular for those histologic subtypes with features difficult to diagnose by FNAB. Although the routine use of FNAB significantly reduced the number of unnecessary thyroid surgeries while increasing the yield of malignancy in the thyroidectomy specimen[50–52], it has limitations as up to 30% of FNAB results are inconclusive requiring diagnostic thyroidectomies.[13–16] For these biopsies requiring surgical intervention for definitive histological diagnosis, the vast majority will turn out having benign disease.[51, 53]
Several investigators have identified dysregulated miRNAs in thyroid cancer with implications on diagnostic utility. Pallante et al [34] performed genome-wide miRNA expression profiling comparing normal thyroid tissue and papillary thyroid cancer and found significant upregulation of miR-221, -222, and -181b. They were able to validate this result in FNAB specimens by demonstrating the same trend of upregulation in specimen that were found to be malignant on post-operative histology. Nikiforova et al [33] performed microRNA profiling on a smaller scale using TaqMan microRNA Assays Human Panel designed to detect 158 human microRNAs and analyzed miRNA expression levels in thyroid tumors and normal thyroid tissue. In this study, they were able to demonstrate distinct miRNA expression profiles among different histopathological subtypes, and they concluded that a set of seven miRNAs (miR-187, -221, -222, -146b, -155, -224, and -197) were highly accurate in distinguishing thyroid cancers from hyperplastic nodules. Furthermore, they tested these miRNAs on 62 consecutive FNAB samples and these miRNAs were upregulated in all 8 specimens (7 PTC and 1 FTC oncocytic type) that ultimately turned out to be malignant. A caveat to this study is that only 13 out of 62 patients had surgery (4 FNAB interpreted as malignant and 8 with atypical cytology) with complete histological information. Of the remaining 49 samples, 3 of them had upregulation of 1 to 3 of these miRNAs but because these patients did not undergo surgery it is difficult to interpret the data without complete histologic information. Chen et al[32] studied seven miRNAs (miR-146a, -146b, -155, -187, -221, -222) based on previous literature and were able to confirm upregulation of miR-146b, -221, and -222 in papillary thyroid carcinoma compared to follicular adenoma. They tested additional histologic subtypes and concluded that miR-146b best distinguishes papillary thyroid carcinoma from other non-papillary carcinoma groups. Furthermore, they tested these miRNAs on FNAB specimen and confirmed upregulation of miR-146b in papillary thyroid cancer compared to follicular adenoma and hyperplastic nodules. Interestingly, miR-221 and miR-222 were only significantly upregulated in the comparison of papillary thyroid carcinoma versus follicular adenoma but not between papillary thyroid carcinoma versus hyperplastic nodule. There was no significant difference found in these miRNAs among the classical papillary thyroid carcinoma versus the follicular variants. While all of these studies demonstrate differential expression of miRNAs among diverse groups of thyroid histologies, none have specifically evaluated if a set of miRNAs in the difficult to diagnose histologic subtypes of thyroid neoplasm have diagnostic utility as an adjunct to thyroid FNAB. This is important because thyroid FNAB result that are indeterminate/suspicious on cytologic examination are mostly made up histologic subtypes of thyroid tissue samples we studied (hyperplastic nodule, follicular adenoma, Hürthle cell adenoma, Hürthle cell carcinoma, follicular thyroid cancer, follicular variant of papillary thyroid carcinoma, papillary thyroid cancer).
MiRNA profiling performed in our study confirmed the upregulation of 3 miRNAs (miR-146b, -221, and miR-222) observed in other studies to be upregulated in papillary thyroid carcinoma, however, their expression was not statistically significant on TaqMan® qRT-PCR validation in the difficult to diagnose histologic groups. Chou et al[54] demonstrated that up-regulation of these 3 miRNAs has significant association with extrathyroidal invasion and demonstrated miR-146b level to be much higher in papillary thyroid carcinoma with BRAF mutation than tumors without the mutation. They also demonstrated upregulation of miR-146b and miR-221 in high risk papillary thyroid carcinoma samples. Interestingly, miR-221 has also been shown to be upregulated in normal thyroid tissue from patients with history of papillary thyroid cancer [46] indicating that it might play a role in early stage of thyroid tumorigenesis. More recently, Visone et al [36] have identified CDKN1B (p27kip1) as a putative target of miR-221 and miR-222, indicating potential therapeutic usage of these miRNAs.
The miRNAs that were significantly differentially expressed in our study were miR-7 and miR-126 and were both found to be downregulated in malignant thyroid tumors compared to benign tumors. These miRNAs have not previously been described in thyroid malignancy, however, there are some studies describing their potential role as tumor suppressors in other cancers. Downregulation of miR-126 has been observed in colorectal cancer,[55] gastric cancer,[45] and metastatic breast cancer.[56] MiR-126 over-expression has been shown to result in decreased in vitro cell proliferation in non-small cell lung cancer[57] and breast cancer cells.[58] miR-126 is located at intron 6 of chromosome 9 and is speculated to target epidermal growth factor-like domain 7 (EGFL7) which is thought to play a role in angiogenesis.[59–62] Downregulation of miR-7 has been demonstrated in glioblastoma,[63] neuroblastoma,[64] and squamous cell cancer of the tongue.[65] miR-7 is also located on chromosome 9 and is speculated to control epidermal growth factor receptor (EGFR) signaling in human cancers.[65–67]
To our knowledge, this is the first study to demonstrate downregulation of miR-7 and miR-126 in thyroid cancer. In addition, our study is the first to focus on difficult to diagnose histologic subtypes by systematically identifying differentially expressed miRNAs and validating the result by qRT-PCR. Several studies have attempted to identify differentially expressed miRNA between the broad grouping of benign versus malignant, however, the malignant group was not inclusive of all the difficult to diagnose histological subtypes (i.e. Hürthle cell carcinoma, follicular variant of papillary thyroid carcinoma, and follicular thyroid carcinoma). Papillary thyroid cancer samples were included in the malignant category along with the more difficult to diagnose subtypes (i.e. Hürthle cell carcinoma, follicular variant of papillary thyroid carcinoma, and follicular thyroid carcinoma) as it is the most common type of thyroid cancer. More importantly, papillary thyroid carcinoma samples were included in the analysis because, although the overall malignancy rate among indeterminate lesions are relatively low (5–16%)[12], it is the most common malignant histologic diagnosis made among the indeterminate FNA cytologic results.[53, 68–69] Thus, we believe including this combination of histologies, as in our study, is more representative of what is seen in the clinical setting of FNA biopsy result with indeterminate or suspicious features.
In summary, there are a number of miRNAs that are differentially expressed between benign and malignant thyroid tumors. This study demonstrated two (miR-7 and miR-126) significantly differentially expressed miRNAs that were found on microarray analysis and validated on TaqMan® qRT-PCR. The sub-group analysis confirmed that these 2 microRNAs are significantly differentially expressed between follicular adenoma and follicular carcinoma samples. However, as the number of samples was compromised by performing the sub-group analysis, this information needs to be further validated using FNAB samples at a larger scale. The expression of these miRNAs may serve as an adjunct to other diagnostic markers, such as clinical factors, gene rearrangement status and somatic mutation testing, in order to definitively classify difficult to diagnose histologic subtypes pre-operatively to obviate diagnostic thyroid surgeries. Performing functional studies using these miRNAs may also provide additional information on thyroid tumorigenesis and potentially have therapeutic implications. This study is an ongoing effort in identifying differentially expressed miRNAs in difficult to diagnose thyroid histologic subtypes and the downregulation of these miRNAs needs to be validated in additional tumor samples, particularly in FNAB samples that were pre-operatively classified as inconclusive, in order to test their utility in clinical setting.
Synopsis.
Expression profiling of difficult to diagnose thyroid histologic subtypes was performed using microRNA microarray. The result was validated using qRT-PCR and identified 2 candidate microRNAs that may be helpful adjunct markers to thyroid fine-needle aspiration biopsy.
Footnotes
Disclosure: Nothing to disclose
Reference List
- 1.Pallante P, Visone R, Croce CM, Fusco A. Deregulation of microRNA expression in follicular cell-derived human thyroidcarcinomas. Endocr Relat Cancer. 2010;17(1):F91–104. doi: 10.1677/ERC-09-0217. [DOI] [PubMed] [Google Scholar]
- 2.Shibru D, Chung KW, Kebebew E. Recent developments in the clinical application of thyroid cancer biomarkers. Curr Opin Oncol. 2008;20(1):13–8. doi: 10.1097/CCO.0b013e3282f27e49. [DOI] [PubMed] [Google Scholar]
- 3.Vriens MR, Schreinemakers JM, Suh I, Guerrero MA, Clark OH. Diagnostic markers and prognostic factors in thyroid cancer. Future Oncol. 2009;5(8):1283–93. doi: 10.2217/fon.09.85. [DOI] [PubMed] [Google Scholar]
- 4.Altekruse SF, KC, Krapcho M, Neyman N, Aminou R, Waldron W, Ruhl J, Howlader N, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Cronin K, Chen HS, Feuer EJ, Stinchcomb DG, Edwards BK. SEER Cancer Statistics Review, 1975–2007. National Cancer Institute; 2010. [cited 2010 December 13]; Available from: http://seer.cancer.gov/statfacts/html/thyro.html. [Google Scholar]
- 5.Sipos JA, Mazzaferri EL. Thyroid cancer epidemiology and prognostic variables. Clin Oncol (R Coll Radiol) 2010;22(6):395–404. doi: 10.1016/j.clon.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Institute, N.C. Thyroid Cancer. [cited 2010 December 13]; Available from: http://www.cancer.gov/cancertopics/types/thyroid.
- 7.Cibas ES, Alexander EK, Benson CB, de Agustin PP, Doherty GM, Faquin WC, et al. Indications for thyroid FNA and pre-FNA requirements: a synopsis of the National Cancer Institute Thyroid Fine-Needle Aspiration State of the Science Conference. Diagn Cytopathol. 2008;36(6):390–9. doi: 10.1002/dc.20827. [DOI] [PubMed] [Google Scholar]
- 8.Gharib H, Papini E, Paschke R, Duick DS, Valcavi R, Hegedus L, et al. American Association of Clinical Endocrinologists, Associazione Medici Endocrinologi, and European Thyroid Association medical guidelines for clinical practice for the diagnosis and management of thyroid nodules: executive summary of recommendations. J Endocrinol Invest. 2010;33(5 Suppl):51–6. [PubMed] [Google Scholar]
- 9.Cibas ES. Fine-needle aspiration in the work-up of thyroid nodules. Otolaryngol Clin North Am. 2010;43(2):257–71. vii–viii. doi: 10.1016/j.otc.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Miller MC. The patient with a thyroid nodule. Med Clin North Am. 2010;94(5):1003–15. doi: 10.1016/j.mcna.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Cooper DS, Doherty GM, Haugen BR, Kloos RT, Lee SL, Mandel SJ, et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19(11):1167–214. doi: 10.1089/thy.2009.0110. [DOI] [PubMed] [Google Scholar]
- 12.Layfield LJ, Cibas ES, Baloch Z. Thyroid fine needle aspiration cytology: a review of the National Cancer Institute state of the science symposium. Cytopathology. 2010;21(2):75–85. doi: 10.1111/j.1365-2303.2010.00750.x. [DOI] [PubMed] [Google Scholar]
- 13.Kato MA, Fahey TJ., 3rd Molecular markers in thyroid cancer diagnostics. Surg Clin North Am. 2009;89(5):1139–55. doi: 10.1016/j.suc.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Wang CC, Friedman L, Kennedy GC, Wang H, Kebebew E, Steward DL, et al. A Large Multicenter Correlation Study of Thyroid Nodule Cytopathology and Histopathology. Thyroid. 2010 doi: 10.1089/thy.2010.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis CM, Chang KP, Pitman M, Faquin WC, Randolph GW. Thyroid fine-needle aspiration biopsy: variability in reporting. Thyroid. 2009;19(7):717–23. doi: 10.1089/thy.2008.0425. [DOI] [PubMed] [Google Scholar]
- 16.Alexander EK. Approach to the patient with a cytologically indeterminate thyroid nodule. J Clin Endocrinol Metab. 2008;93(11):4175–82. doi: 10.1210/jc.2008-1328. [DOI] [PubMed] [Google Scholar]
- 17.Suh I, Vriens MR, Guerrero MA, Griffin A, Shen WT, Duh QY, et al. Serum thyroglobulin is a poor diagnostic biomarker of malignancy in follicular and Hurthle-cell neoplasms of the thyroid. Am J Surg. 2010;200(1):41–6. doi: 10.1016/j.amjsurg.2009.08.030. [DOI] [PubMed] [Google Scholar]
- 18.Davidov T, Trooskin SZ, Shanker BA, Yip D, Eng O, Crystal J, et al. Routine second-opinion cytopathology review of thyroid fine needle aspiration biopsies reduces diagnostic thyroidectomy. Surgery. 2010;148(6):1294–9. doi: 10.1016/j.surg.2010.09.029. discussion 1299–301. [DOI] [PubMed] [Google Scholar]
- 19.Kouniavsky G, Zeiger MA. Thyroid tumorigenesis and molecular markers in thyroid cancer. Curr Opin Oncol. 2010;22(1):23–9. doi: 10.1097/CCO.0b013e328333846f. [DOI] [PubMed] [Google Scholar]
- 20.Gomez Saez JM. Diagnostic usefulness of tumor markers in the thyroid cytological samples extracted by fine-needle aspiration biopsy. Endocr Metab Immune Disord Drug Targets. 2010;10(1):47–56. doi: 10.2174/187153010790828000. [DOI] [PubMed] [Google Scholar]
- 21.Chudova D, Wilde JI, Wang ET, Wang H, Rabbee N, Egidio CM, et al. Molecular classification of thyroid nodules using high-dimensionality genomic data. J Clin Endocrinol Metab. 2010;95(12):5296–304. doi: 10.1210/jc.2010-1087. [DOI] [PubMed] [Google Scholar]
- 22.Stang MT, Carty SE. Recent developments in predicting thyroid malignancy. Curr Opin Oncol. 2009;21(1):11–7. doi: 10.1097/CCO.0b013e32831db2af. [DOI] [PubMed] [Google Scholar]
- 23.Yip L, Kebebew E, Milas M, Carty SE, Fahey TJ, 3rd, Parangi S, et al. Summary statement: utility of molecular marker testing in thyroid cancer. Surgery. 2010;148(6):1313–5. doi: 10.1016/j.surg.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raparia K, Min SK, Mody DR, Anton R, Amrikachi M. Clinical outcomes for “suspicious” category in thyroid fine-needle aspiration biopsy: Patient’s sex and nodule size are possible predictors of malignancy. Arch Pathol Lab Med. 2009;133(5):787–90. doi: 10.5858/133.5.787. [DOI] [PubMed] [Google Scholar]
- 25.Banks ND, Kowalski J, Tsai HL, Somervell H, Tufano R, Dackiw AP, et al. A diagnostic predictor model for indeterminate or suspicious thyroid FNA samples. Thyroid. 2008;18(9):933–41. doi: 10.1089/thy.2008.0108. [DOI] [PubMed] [Google Scholar]
- 26.Mathur A, Weng J, Moses W, Steinberg SM, Rahabari R, Kitano M, et al. A prospective study evaluating the accuracy of using combined clinical factors and candidate diagnostic markers to refine the accuracy of thyroid fine needle aspiration biopsy. Surgery. 2010;148(6):1170–7. doi: 10.1016/j.surg.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moses W, Weng J, Sansano I, Peng M, Khanafshar E, Ljung BM, et al. Molecular testing for somatic mutations improves the accuracy of thyroid fine-needle aspiration biopsy. World J Surg. 2010;34(11):2589–94. doi: 10.1007/s00268-010-0720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapio MR, Posca D, Raggioli A, Guerra A, Marotta V, Deandrea M, et al. Detection of RET/PTC, TRK and BRAF mutations in preoperative diagnosis of thyroid nodules with indeterminate cytological findings. Clin Endocrinol (Oxf) 2007;66(5):678–83. doi: 10.1111/j.1365-2265.2007.02800.x. [DOI] [PubMed] [Google Scholar]
- 29.Pelizzo MR, Boschin IM, Barollo S, Pennelli G, Toniato A, Zambonin L, et al. BRAF analysis by fine needle aspiration biopsy of thyroid nodules improves preoperative identification of papillary thyroid carcinoma and represents a prognostic factor. A mono-institutional experience. Clin Chem Lab Med. 2010 doi: 10.1515/CCLM.2011.031. [DOI] [PubMed] [Google Scholar]
- 30.Nikiforova MN, Nikiforov YE. Molecular diagnostics and predictors in thyroid cancer. Thyroid. 2009;19(12):1351–61. doi: 10.1089/thy.2009.0240. [DOI] [PubMed] [Google Scholar]
- 31.Nikiforov YE, Steward DL, Robinson-Smith TM, Haugen BR, Klopper JP, Zhu Z, et al. Molecular testing for mutations in improving the fine-needle aspiration diagnosis of thyroid nodules. J Clin Endocrinol Metab. 2009;94(6):2092–8. doi: 10.1210/jc.2009-0247. [DOI] [PubMed] [Google Scholar]
- 32.Chen YT, Kitabayashi N, Zhou XK, Fahey TJ, 3rd, Scognamiglio T. MicroRNA analysis as a potential diagnostic tool for papillary thyroid carcinoma. Mod Pathol. 2008;21(9):1139–46. doi: 10.1038/modpathol.2008.105. [DOI] [PubMed] [Google Scholar]
- 33.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93(5):1600–8. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 35.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26(54):7590–5. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 36.Visone R, Russo L, Pallante P, De Martino I, Ferraro A, Leone V, et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr Relat Cancer. 2007;14(3):791–8. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 37.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptionalgene regulation. Oncogene. 2006;25(46):6163–9. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 38.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmerman AL, Wu S. MicroRNAs, cancer and cancer stem cells. Cancer Lett. 2011;300(1):10–9. doi: 10.1016/j.canlet.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3–4):369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 41.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16(1):4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Hammond SM. MicroRNAs as tumor suppressors. Nat Genet. 2007;39(5):582–3. doi: 10.1038/ng0507-582. [DOI] [PubMed] [Google Scholar]
- 43.Osada H, Takahashi T. let-7 and miR-17–92: small-sized major players in lung cancer development. Cancer Sci. 2011;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 44.Meyer-Rochow GY, Jackson NE, Conaglen JV, Whittle DE, Kunnimalaiyaan M, Chen H, et al. MicroRNA profiling of benign and malignant pheochromocytomas identifies novel diagnostic and therapeutic targets. Endocr Relat Cancer. 2010;17(3):835–46. doi: 10.1677/ERC-10-0142. [DOI] [PubMed] [Google Scholar]
- 45.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 46.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, et al. The role of microRNA genesin papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheu SY, Grabellus F, Schwertheim S, Worm K, Broecker-Preuss M, Schmid KW. Differential miRNA expression profiles in variants of papillary thyroid carcinoma and encapsulated follicular thyroid tumours. Br J Cancer. 2010;102(2):376–82. doi: 10.1038/sj.bjc.6605493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18(3):163–73. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- 49.Mazeh H, Mizrahi I, Halle D, Ilyayev N, Stojadinovic A, Trink B, et al. Development of a MicroRNA-Based Molecular Assay for the Detection of Papillary Thyroid Carcinoma in Aspiration Biopsy Samples. Thyroid. 2011;21(2):111–8. doi: 10.1089/thy.2010.0356. [DOI] [PubMed] [Google Scholar]
- 50.Hamberger B, Gharib H, Melton LJ, 3rd, Goellner JR, Zinsmeister AR. Fine-needle aspiration biopsy of thyroid nodules. Impact on thyroid practice and cost of care. Am J Med. 1982;73(3):381–4. [PubMed] [Google Scholar]
- 51.Castro MR, Gharib H. Thyroid fine-needle aspiration biopsy: progress, practice, and pitfalls. Endocr Pract. 2003;9(2):128–36. doi: 10.4158/EP.9.2.128. [DOI] [PubMed] [Google Scholar]
- 52.Hadi M, Gharib H, Goellner JR, Heerden JA. Has fine-needle aspiration biopsy changed thyroid practice? Endocr Pract. 1997;3(1):9–13. doi: 10.4158/EP.3.1.9. [DOI] [PubMed] [Google Scholar]
- 53.Nayar R, Ivanovic M. The indeterminate thyroid fine-needle aspiration: experience from an academic center using terminology similar to that proposed in the 2007 National Cancer Institute Thyroid Fine Needle Aspiration State of the Science Conference. Cancer Cytopathol. 2009;117(3):195–202. doi: 10.1002/cncy.20029. [DOI] [PubMed] [Google Scholar]
- 54.Chou CK, Chen RF, Chou FF, Chang HW, Chen YJ, Lee YF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489–94. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 55.Li XM, Wang AM, Zhang J, Yi H. Down-regulation of miR-126 expression in colorectal cancer and its clinical significance. Med Oncol. 2010 doi: 10.1007/s12032-010-9637-6. [DOI] [PubMed] [Google Scholar]
- 56.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun. 2010;391(3):1483–9. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 58.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, et al. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377(1):136–40. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 59.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–75. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 60.Nikolic I, Plate KH, Schmidt MH. EGFL7 meets miRNA-126: an angiogenesis alliance. J Angiogenes Res. 2010;2(1):9. doi: 10.1186/2040-2384-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379(3):726–31. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- 62.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2(52):pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008;68(10):3566–72. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- 64.Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394(4):921–7. doi: 10.1016/j.bbrc.2010.03.076. [DOI] [PubMed] [Google Scholar]
- 65.Jiang L, Liu X, Chen Z, Jin Y, Heidbreder CE, Kolokythas A, et al. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinomacells. Biochem J. 2010;432(1):199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68(20):8195–200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70(21):8822–31. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- 68.Theoharis CG, Schofield KM, Hammers L, Udelsman R, Chhieng DC. The Bethesda thyroid fine-needle aspiration classification system: year 1 at an academic institution. Thyroid. 2009;19(11):1215–23. doi: 10.1089/thy.2009.0155. [DOI] [PubMed] [Google Scholar]
- 69.Yang J, Schnadig V, Logrono R, Wasserman PG. Fine-needle aspiration of thyroid nodules: a study of 4703 patients with histologic and clinical correlations. Cancer. 2007;111(5):306–15. doi: 10.1002/cncr.22955. [DOI] [PubMed] [Google Scholar]