Abstract
MeCP2 (methyl CpG binding protein 2) is a key player in recognizing methylated DNA and interpreting the epigenetic information encoded in different DNA methylation patterns. The functional significance of MeCP2 to the mammalian nervous system is highlighted by the discovery that mutations in the MECP2 gene cause Rett syndrome (RTT), a devastating neurological disease that shares many features with autism. Synaptic scaling is a form of non-Hebbian homeostatic plasticity that allows neurons to regulate overall excitability in response to changes in network neuronal activity levels. While it is known that neuronal activity can induce phosphorylation of MeCP2 and that MeCP2 can regulate synaptic scaling, the molecular link between MeCP2 phosphorylation and synaptic scaling remains undefined. We show here that MeCP2 phosphorylation is specifically required for bicuculline-induced synaptic scaling down in mouse hippocampal neurons and this phenotype is mediated by mGluR5 (metabotropic glutamate receptor 5). Our results reveal an important function of MeCP2 in regulating neuronal homeostasis and may eventually help us understand how MECP2 mutations cause RTT.
Introduction
DNA methylation and histone tail modifications have been studied extensively as the two major components of epigenetic codes. Methyl-DNA binding proteins have been shown to function as the molecular linker between these two codes. The functional significance of such a linker is highlighted by the discovery that mutations in the methyl-CpG binding protein 2 gene (MECP2)—the founding member of the methyl-CpG binding domain-containing protein (MBD) gene family—cause Rett syndrome (Amir et al., 1999), an autism spectrum developmental disorder (Hagberg, 1985). MeCP2 was initially identified as a protein that specifically binds to methylated DNA (Lewis et al., 1992; Nan et al., 1997). Although originally described as a transcription repressor through its interaction with a core repressor complex containing mSin3A and histone deacetylases (Jones et al., 1998; Nan et al., 1998), recent evidence suggests MeCP2 can also activate gene transcription through its interaction with CREB (cAMP response element-binding protein) and coactivators (Chahrour et al., 2008). Several posttranslational modifications have been identified on MeCP2 (Zhou et al., 2006; Tao et al., 2009; Gonzales et al., 2012). Among these, phosphorylation at serine 421 (S421) has been most extensively studied. Many studies have clearly demonstrated that S421 phosphorylation can be induced in neurons by neuronal activity both in vitro and in vivo (Chen et al., 2003; Zhou et al., 2006; Géranton et al., 2007, 2008; Murgatroyd et al., 2009; Deng et al., 2010; Li et al., 2011; Mao et al., 2011), and the loss of S421 phosphorylation significantly changes various aspects of brain functions (Cohen et al., 2011; Li et al., 2011). Most recently, MeCP2 has been implicated in synaptic scaling (Qiu et al., 2012), a non-Hebbian form of synaptic plasticity (Turrigiano et al., 1998). However, it is not known what downstream effectors mediate the MeCP2 effect on synaptic scaling. In addition, although S421 phosphorylation is observed during the process of synaptic scaling (Qiu et al., 2012), it is not clear whether such phosphorylation is required for synaptic scaling. In this study, we show that S421 and/or S424 phosphorylation are required specifically for bicuculline-induced synaptic scaling down, but not tetrodotoxin (TTX)-induced synaptic scaling up. Moreover, such effect could be mediated by metabotropic glutamate receptor 5 (mGluR5). By linking posttranslational modification of MeCP2 with homeostatic synaptic plasticity and beginning to reveal its molecular mechanism, our work helps to understand what MeCP2 does to maintain normal neuronal function.
Materials and Methods
DNA construct.
Grm5 isoform b was subcloned into the Lox-Syn Lentivirus vector (Gascón et al., 2008) to overexpress mGluR5 in cultured hippocampal neurons. This vector contains two neuronal specific synapsin promoters. One promoter controls the expression of mGluR5 and the other controls the expression of DsRed.
Antibodies and drugs.
The following antibodies and drugs were used: anti-mGluR5 (Millipore, AB5675), anti-mGluR1 (BD Biosciences, 556331), anti-β-Actin (Sigma, A5441), anti-GluA1 (Millipore, clone RH95, MAB2263), anti-GluA2 (Millipore, MAB397), anti-MAP2 (anti-microtubule-associated protein 2; Abcam, ab32454), Alexa Fluor-conjugated secondary antibodies (Invitrogen), DyLight 680/800-conjugated secondary antibody (Thermo Fisher Scientific), tetrodotoxin citrate (Ascent, Asc-055), d-AP5 (Ascent, Asc-003), (−)-bicuculline methobromide (Enzo, BML-EA109, and Sigma, B7561), and (R,S)-3,5-DHPG (3,4-dihydroxyphenylglycol; Tocris Bioscience).
Primary culture of hippocampal neuron.
Mecp2S421A;S424A/+ female and wild-type male were mated to generate Mecp2S421A;S424A/y and Mecp2+/y pups. Genotyping assay was performed as described previously (Li et al., 2011). Dissociated hippocampal neurons were isolated from postnatal day 0–1 pups of both genotypes. The hippocampal neurons used in Figure 1 were isolated with Trypsin. The hippocampal neurons used in the rest of the study were isolated with the Papain Dissociation System (Worthington Biochemical Corporation LK003153). For the electrophysiology and AMPA receptor surface staining experiment, hippocampal neurons were plated at a density of 2.5 × 104 cm−2 on 12 mm coverslips coated with 100 μg/ml poly-l-lysine (Sigma P2636), and cultured in Neurobasal A (Invitrogen 10888) containing 2% B27 (v/v, Invitrogen 12587-010), 1% FBS (v/v, Invitrogen 26140), 25 μm glutamate, and 0.5 mm GlutaMax (Invitrogen 35050-061). Half of the media was changed every 4 d with Neurobasal A containing 2% B27 and 0.5 mm GlutaMax. Ara C (10 μm; Sigma C1768) was added to the culture media to inhibit glial cell growth on 8 days in vitro (DIV). In experiments involving drug treatments, the treatments started at 16 DIV. For the Western blot analysis and the total RNA purification, 6–8 × 104 cm−2 hippocampal neurons were cultured in 6 well plates with Neurobasal A media containing 2% B27, 25 μm glutamate, and 0.5 mm GlutaMax, with media changed as described above. Ara C (10 μm) was added starting from 8 DIV. The neurons were harvested on 18 DIV.
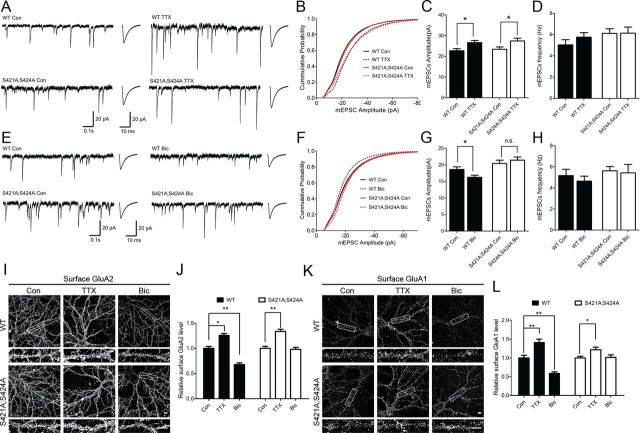
Figure 1.
Mecp2S421A;S424A/y neurons exhibit intact TTX-induced synaptic scaling up, but impaired bicuculline-induced synaptic scaling down. A, Representative whole-cell recording sample traces, and average mEPSC waveforms from cultured Mecp2S421A;S424A/y and wild-type (WT) hippocampal neurons (18 DIV) treated with either control solution or TTX (1 μm) for 48–72 h. B, Cumulative probability distribution of mEPSC amplitude from cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18 DIV) treated with either control solution or TTX (1 μm) for 48–72 h. C, Quantification of mean mEPSC amplitude for each population in A (n = 49–54 cells each group). D, Quantification of mean mEPSC frequencies for each population in A. E, Representative whole-cell recording sample traces, and average mEPSC waveforms from cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18 DIV) treated with control solution or bicuculline (40 μm) for 48–72 h. F, Cumulative probability distribution of mEPSC amplitude from cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18 DIV) treated with control solution or bicuculline (40 μm) for 48–72 h. G, Quantification of mean mEPSC amplitude for each population in E (n = 47–61 cells each group). H, Quantification of mean mEPSC frequencies for each population in E. I–L, Representative images and quantification of surface GluA2 and GluA1 immunoreactivity in Mecp2S421A;S424A/y and wild-type cultured hippocampal neurons (18 DIV) treated with control solution, TTX (1 μm), or bicuculline (40 μm) for 48 h. Scale bar, 10 μm (n = 15–20 cells each group). The bar graph shows the mean ± SEM. *p < 0.05, **p < 0.01.
Drug treatment, lentivirus preparation, and infection.
For the electrophysiology experiment, 40 μm bicuculline or 1 μm TTX was added to the culture media for hippocampal neurons on 16–18 DIV. Miniature EPSCs (mEPSCs) were recorded 48–72 h after drug treatment. For surface staining, the same concentration of bicuculline or TTX was used on 16 DIV, and immunocytochemistry was performed on 18 DIV, 48 h after the drug treatment. For the DHPG pharmacological rescue experiment, hippocampal neurons were preincubated with 100 μm (R,S)-3,5-DHPG for 30 min. After the DHPG was washed out with the neuronal culture media, the neurons were incubated with 40 μm bicuculline for 48 h.
Lentiviral particles were produced by HEK293FT cells (Invitrogen) cotransfected with the virus packaging vectors as described previously (Gascón et al., 2008). Briefly, HEK293FT cells were triple transfected using the calcium phosphate method with the lentiviral expression vector and two packaging vectors: pCAG-VSV-G, a plasmid expressing the VSV-G envelope gene, and pCMVΔR8.91, a plasmid expressing the gag/pol genes. The supernatants containing the viral particles were collected 48–60 h after transfection and concentrated by ultracentrifuge (SW32 rotor, centrifuge at 25,000 rpm, 4°C for 1.5 h). Lentivirus was added to the media of hippocampal neurons at 4 DIV.
Real-time quantitative PCR and primers.
Total RNA was extracted from 18 DIV cultured hippocampal neurons using TRIzol Reagent (Invitrogen). The qScript cDNA SuperMix kit (Quanta BioSciences) was used for cDNA synthesis. Real-time quantitative PCR was performed on a StepOne Plus Real-Time PCR System (Applied Biosystems) using RT2 SYBR Green ROX qPCR Mastermix (Qiagen). Fold change was calculated with the 2−ΔCt method after normalization to Gapdh. Mouse Grm5: 5′-aggacagataaaggtgatcc-3′ (forward), 5′-tcacaacctgtcaagtcgtc-3′ (reverse); Mouse Grm1: 5′-acagaagggaattacggcgaga-3′ (forward), 5′-atcaaagctcttctcgccagca-3′ (reverse); Mouse Gria1: 5′-agcgagtcgaagcggatgaa-3′, (forward), 5′-ctctgccattctctccactg-3′ (reverse); Mouse Gria2: 5′-atgggaaagctgatattgcca-3′ (forward), 5′-catgatagagattccaaggc-3′ (reverse); Mouse Mecp2: 5′-gagagactggaggaaaagtc-3′ (forward), 5′-agtggctcatgcttgccttc-3′ (reverse); Mouse Gapdh: 5′-aatgggaagcttgtcatcaacg-3′ (forward), 5′-gaagacaccagtagactccacgacata-3′ (reverse). Paired t tests were performed for statistical analysis.
Western blot analysis.
Western blot analysis was performed as described previously (Ayala et al., 2012). The 18 DIV hippocampal culture neurons were washed with PBS once and then lysed in Laemmli Sample Buffer (Bio-Rad) with 50 mm dithiothreitol. Sonication was used to lyse the cells and shear the genomic DNA. To minimize the aggregation of mGluR proteins, lysates were never boiled, and were subjected to SDS-PAGE the same day they were prepared. The Odyssey Western Detection method (LI-COR Biosciences) was used for signal detection. The integrated pixel intensity values for the target protein bands were analyzed with NIH ImageJ software, and then normalized to the intensity of the control β-actin from the same samples. Paired t test was performed for statistical analysis.
Immunocytochemistry, imaging, and data analysis.
Antibodies were applied in NDS buffer (10% normal donkey serum in PBS buffer, pH 7.4). All the experimental processes were performed at room temperature unless stated otherwise. PBS washing was performed 3 times after the paraformaldehyde fixation and the antibody incubation. Surface staining was performed as described previously (Shepherd et al., 2006). Briefly, 2.5 μg of NT-GluA1 mouse monoclonal antibody or 1 μg of GluA2 extracellular region mouse monoclonal antibody was added to the neuronal culture media with 10 mm HEPES, pH 7.4. To label GluA1-containing AMPA receptors, the neurons were incubated at 10°C for 20 min, with gentle rocking. To label GluA2-containing AMPA receptors, the neurons were put in a CO2 incubator for 15 min, and gently mixed well every 5 min. Once the incubation was done, the neurons were quickly washed with fresh prewarmed Neurobasal A three times and then fixed with 4% paraformaldehyde, 4% sucrose in PBS solution for 15 min. After incubation with 10% normal donkey serum in PBS for 1 h, the neurons were exposed to 1:600 diluted donkey anti-mouse Alexa Fluor 488 or 647 secondary antibody for 1 h. The neurons were permeabilized with 0.3% Triton X-100 in PBS for 30 min. Following blockage with 10% normal donkey serum in PBS for 1 h, the neurons were incubated with 1:1000 diluted MAP2 antibodies for 1 h. The neurons were then exposed to 1:600 diluted Alexa Fluor 568 or 488 secondary antibody for 1 h. The coverslips were mounted on precleaned slides with Fluoromount-G (Southern Biotech).
Surface GluA2 and GluA1 images were captured on a Nikon A1RSI confocal microscope. For each experimental series, all the images were acquired with identical settings for laser power, photomultiplier gain, and offset. Z-stacks were collapsed in a maximum projection and analyzed with NIH ImageJ software. Excitatory pyramidal neurons were identified by morphology using MAP2 immunoactivity or DsRed fluorescence. Proximal dendritic segments (50–80 μm, 30 μm from soma) were selected for the analysis. All the images were thresholded using a single threshold which is determined by the background level of the control condition in each experiment. The integrated intensity of GluA2 or GluA1 immunoreactivity was obtained for each dendritic segment from ImageJ software. The average pixel intensity from each region was calculated using the integrated intensity divided by the dendritic length. The average surface GluA2 or GluA1 level in each group was then normalized to the level in their control group. Student's t test was used for the statistical analysis.
Electrophysiology.
Whole-cell patch-clamp was performed at room temperature. The recording chamber was continuously perfused with external recording solution contained (in mm): 120 NaCl, 3 KCl, 15 HEPES, 1 MgCl2, 2 CaCl2, 20 glucose, pH7.4 (300 ± 5 mmol/kg). To record the AMPA receptor media mEPSCs only, the bath solution also contained 1 μm TTX, 20 μm bicuculline, and 50 μm d-AP5. The patch pipette (3–5 MΩ) solution contained (in mm): 140 K-gluconate, 7.5 KCl, 10 HEPES-K, 0.5 EGTA-K, 4 Mg-ATP, and 0.3 Li-GTP, pH 7.4 (290 ± 5 mmol/kg). Hippocampal culture neurons were visualized using an Olympus Optical BX51WI microscope. Pyramidal neurons were chosen according to the cell morphology (Turrigiano et al., 1998). Lentivirus-infected neurons were visualized using Olympus reflected fluorescence system with Cy3 filter cube. Infected neurons with too strong or too weak DsRed fluorescence were excluded. Only the pyramidal neurons with similar DsRed fluorescent intensity were recorded. The mEPSCs were recorded 3 min after the whole-cell voltage-clamp configuration was established and lasted for 5 min on a gap-free mode. The neurons were held at −70 mV. Whole-cell capacitance compensation was applied. Recordings with resting membrane potential (Vr) higher than −50 mV and series resistance (Rs) higher than 15 MΩ were excluded. Raw data were amplified with a Multiclamp 700B amplifier, acquired with pClamp10.2 software (Molecular Devices). Signals were filtered at 2 Hz and sampled at 10 kHz by Digidata 1440A (Molecular Devices). mEPSCs were analyzed using the Template Search tool of the Clampfit10.2 software (Molecular Devices). To create the template, several well-shaped mEPSCs traces were picked from the wild-type hippocampal pyramidal neurons recording and averaged to the template window. Only the first 200 events were used to do the data analysis. The mEPSCs events were accepted manually. All group data are shown as mean ± SEM. For the mEPSCs average event and the cumulative curve preparation, all the events from all of the recorded neurons were included. The average mEPSCs event was gained by using the concatenate files command and average traces command of Clampfit10.2. The sample traces and the average event traces were transferred out, and the traces were made by using SigmaPlot11.0 software (Scientific Computing). All results were analyzed via t test with repeated measures. Significance for all tests was set at p < 0.05.
Results
Loss of MeCP2 phosphorylation at S421 and S424 specifically affects bicuculline-induced synaptic scaling down, but not TTX-induced synaptic scaling up
Synaptic scaling is a non-Hebbian form of homeostatic synaptic plasticity that plays an essential role in adjusting neuronal excitability in response to chronic network activity changes. Since there is strong evidence that neuronal activity induces phosphorylation of MeCP2, we explored the possibility that such phosphorylation may also affect synaptic scaling. Hippocampal neurons were isolated from postnatal day 0–1 (P0–P1) Mecp2S421A;S424A/y pups and their wild-type littermates and cultured for 16 DIV, at which time cultures from both genotypes were treated for 48–72 h with either tetrodotoxin (TTX), or bicuculline, or left untreated. Consistent with previous studies, the amplitudes of mEPSCs in wild-type neurons increased significantly in response to the TTX treatment (Fig. 1A–C) and decreased significantly in response to the bicuculline treatment (Fig. 1E–G). Similar to the observation in wild-type neurons, the amplitudes of mEPSCs in the Mecp2S421A;S424A/y neurons increased significantly after the TTX treatment (Fig. 1A–C). Different from the observation in wild-type neurons, the amplitudes of mEPSCs in the Mecp2S421A;S424A/y neurons remained unchanged after the bicuculline treatment (Fig. 1E–G). To independently confirm our electrophysiological recording results, we examined the surface level AMPA receptors in live Mecp2S421A;S424A/y and wild-type hippocampal neurons. Consistent with the recordings, the TTX treatment led to significantly increased level of GluA2 (Fig. 1I,J) and GluA1 (Fig. 1K,L) on the cell surface (sGluA2 and sGluA1) of both the Mecp2S421A;S424A/y and wild-type hippocampal neurons; the bicuculline treatment led to significantly decreased level of sGluA2 (Fig. 1I,J) and sGluA1 (Fig. 1K,L) in the wild-type, but not the Mecp2S421A;S424A/y hippocampal neurons. No statistically significant changes were detected in mEPSC frequencies across all conditions (Fig. 1D,H). Together, these results demonstrate that the loss of phosphorylation at S421 and S424 specifically impairs the bicuculline-induced synaptic scaling down.
mGluR5 expression is reduced in Mecp2S421A;S424A/y hippocampal neurons
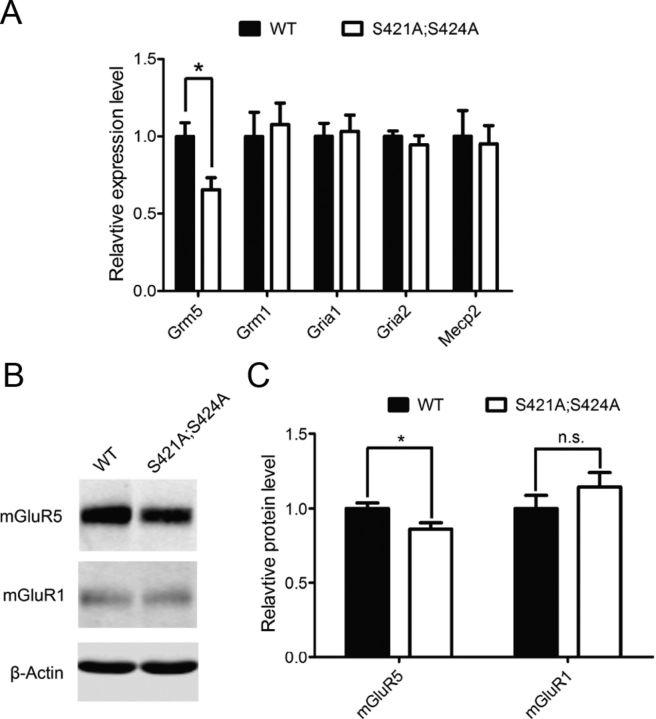
To reveal which downstream effector proteins mediate the effect of loss of MeCP2 phosphorylation on synaptics scaling, we examined the RNA level of several glutamate receptors known to be involved in synaptic plasticity by quantitative PCR. While the RNA levels of Grm1 (metabotropic glutamate receptor 1), Gria1 (ionotropic glutamate receptor 1) and Gria2 (ionotropic glutamate receptor 2) are indistinguishable between the Mecp2S421A;S424A/y and wild-type hippocampal neurons, the RNA level of Grm5 (metabotropic glutamate receptor 5) was significantly lower in Mecp2S421A;S424A/y hippocampal neurons than in wild-type at 18 DIV (Fig. 2A). Mecp2 was included as a control for unchanged RNA level between the two genotypes. In addition, Western blot analysis was performed to look at the level of mGluR5 protein. Consistent with the observation at the RNA level, mGluR5 protein was significantly lower in the in Mecp2S421A;S424A/y hippocampal neurons than in wild-type at 18 DIV (Fig. 2B,C), while the mGluR1 protein level was similar between the two genotypes.
Figure 2.
Reduced mGluR5 expression in Mecp2S421A;S424A/y hippocampal neurons. A, Relative mRNA level of Grm5, Grm1, Gria1, Gria2, and Mecp2 in cultured hippocampal neurons (18 DIV) derived from Mecp2S421A;S424A/y mice and their wild-type (WT) littermates (n = 8 in each genotype). B, C, Western blot and quantification showing relative protein level of mGluR5 and mGluR1 in cultured hippocampal neurons (18 DIV) derived from Mecp2S421A;S424A/y mice and their wild-type littermates (n = 15 in each genotype). The bar graph shows the mean ± SEM. *p < 0.05.
Pharmacological activation of group I mGluRs rescues the loss of bicuculline-induced synaptic scaling down in Mecp2S421A;S424A/y hippocampal neurons
Since the activation of group I metabotropic glutamate receptors has been shown to lead to downregulation of sGluA2 and sGluA1 in cultured embryonic cortical neurons after bicuculline treatment (Hu et al., 2010), it is possible that the reduced Grm5/mGluR5 expression may help explain the lack of bicuculline-induced synaptic scaling down in the Mecp2S421A;S424A/y hippocampal neurons. To test this hypothesis, we first attempted to rescue the synaptic scaling down deficit with pharmacological activation of the group I metabotropic glutamate receptors. In this series of experiments, the Mecp2S421A;S424A/y and wild-type hippocampal neurons at 16–18 DIV were divided into four groups: the first group received no treatment; the second group received bicuculline treatment for 48 h; the third group was treated with DHPG, a selective agonist of group I metabotropic glutamate receptors, for 30 min; the fourth group was pretreated with DHPG for 30 min before they were treated with bicuculline for 48 h. Whole-cell patch-clamp recordings and live immunostainings from the first and second groups showed that, similar to results in Figure 1, chronic bicuculline treatment induced a significant decrease in the mEPSC amplitudes and sGluA2 level in the wild-type, but not the Mecp2S421A;S424A/y hippocampal neurons (Fig. 3A–C,I,J). As expected, results from the third and fourth groups showed that transient activation of the group I metabotropic glutamate receptors before chronic bicuculline treatment restored the bicuculline-induced synaptic scaling down in the Mecp2S421A;S424A/y hippocampal neurons as measured by both the mEPSC amplitudes and the sGluA2 levels (Fig. 3E–G). Moreover, such a transient activation had no effect on the bicuculline-induced synaptic scaling down in the wild-type neurons, suggesting the effect was specific for the mutant neurons (Fig. 3E–G). Again, no statistically significant changes were detected in mEPSC frequencies across all conditions (Fig. 3D,H).
Figure 3.
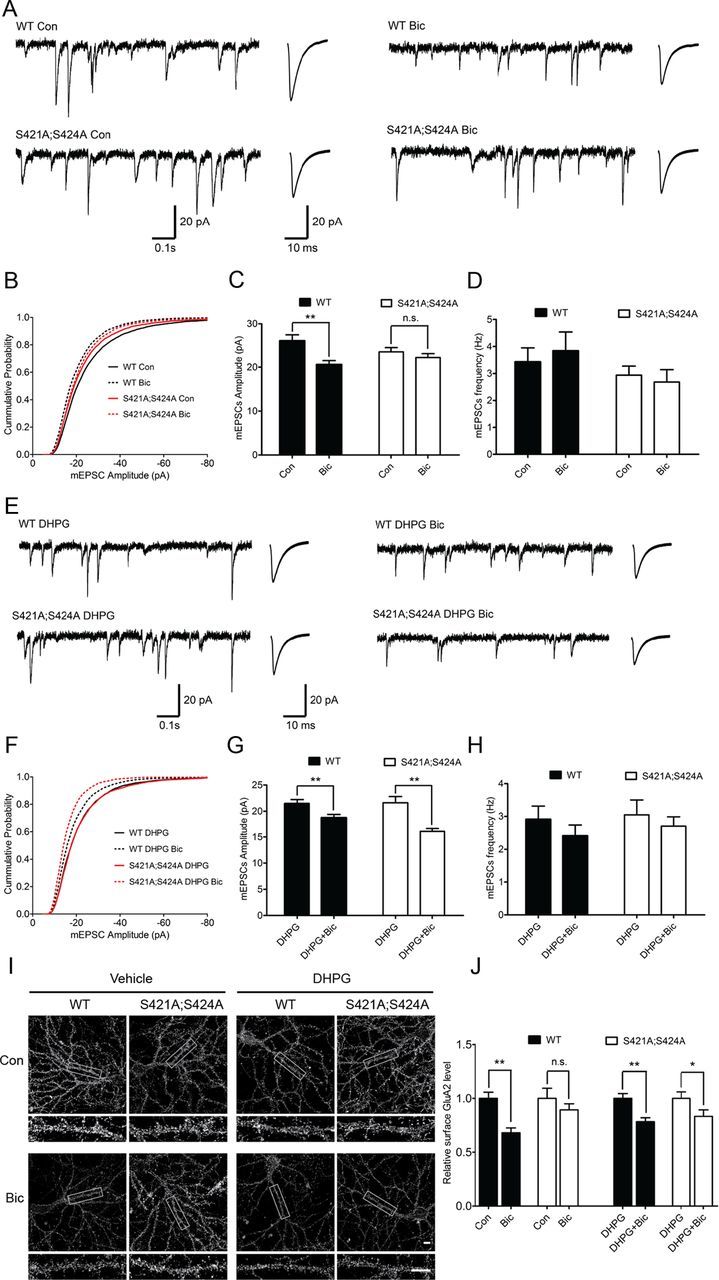
Pretreatment with group I mGluR agonist restores bicuculline-induced synaptic scaling down in Mecp2S421A;S424A/y hippocampal neurons. A–D, Representative whole-cell recording sample traces, average mEPSC waveforms, cumulative probability distribution, and quantification of mEPSC amplitudes and frequencies from cultured from cultured Mecp2S421A;S424A/y and wild-type (WT) hippocampal neurons (18–20 DIV) treated for 48 h with either control solution or bicuculline (40 μm) (n = 22–32 cells in each group). These data were not included in Figure 1. E–H, Representative whole-cell recording sample traces, average mEPSC waveforms, cumulative probability distribution, and quantification of mEPSC amplitudes and frequencies from cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18–20 DIV) pretreated with 30 min DHPG (100 μm), followed by 48 h treatment of either control solution or bicuculline (40 μm) (n = 22–32 cells in each group). I, J, Representative images and quantification of surface GluA2 immunoreactivity in cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18 DIV) with or without 30 min DHPG (100 μm) pretreatment, followed by 48 h treatment of either control solution or bicuculline (40 μm). Scale bar, 10 μm (n = 20 cells each group). The bar graph shows the mean ± SEM. *p < 0.05, **p < 0.01.
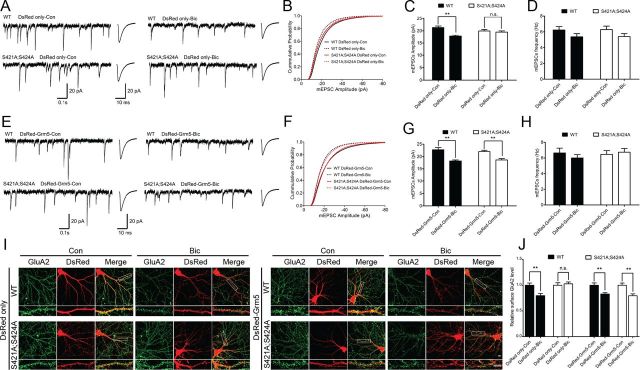
Overexpression of mGluR5 rescues the loss of bicuculline-induced synaptic scaling down in Mecp2S421A;S424A/y hippocampal neurons
Since DHPG activates both mGluR1 and mGluR5, and only mGluR5 level was reduced in the cultured Mecp2S421A;S424A/y hippocampal neurons, the restoration of bicuculline-induced synaptic scaling down by DHPG pretreatment in these neurons could not be specifically attributed to increased mGluR5 activation. To further distinguish between mGluR5 and mGluR1, we used lentivirus-mediated expression to overexpress mGluR5 in both the Mecp2S421A;S424A/y and wild-type hippocampal neurons. Two types of lentiviruses were produced for this series of experiments: the first one expressed only DsRed, a fluorescence marker; the second one expressed both mGluR5 and DsRed. In hippocampal neurons infected with lentivirus expressing only DsRed, chronic bicuculline treatment induced a significant decrease in the mEPSC amplitudes and sGluA2 level in the wild-type, but not the Mecp2S421A;S424A/y neurons (Fig. 4A–C,I,J), suggesting neither lentivirus infection nor expression of DsRed interfered with the normal synaptic scaling process in the wild-type neurons or the abnormal synaptic scaling in the mutant neurons. In contrast, in hippocampal neurons infected with lentivirus expressing both mGluR5 and DsRed, chronic bicuculline treatment induced a significant decrease in the mEPSC amplitudes and sGluA2 level in both the wild-type and the Mecp2S421A;S424A/y neurons (Fig. 4E–G,I,J), suggesting the overexpression of mGluR5 specifically rescued the deficit in the Mecp2S421A;S424A/y neurons. Finally, no statistically significant changes were detected in mEPSC frequencies across all conditions (Fig. 4D,H).
Figure 4.
Overexpression of mGluR5 restores bicuculline-induced synaptic scaling down in Mecp2S421A;S424A/y hippocampal neurons. A–D, Representative whole-cell recording sample traces, average mEPSC waveforms, cumulative probability distribution, and quantification of mEPSC amplitudes and frequencies from cultured Mecp2S421A;S424A/y and wild-type (WT) hippocampal neurons (18–20 DIV) that were transfected with lentivirus expressing only DsRed and treated with either control solution or bicuculline (40 μm) for 48 h (n = 48–52 cells in each group). E–H, Representative whole-cell recording sample traces, average mEPSC waveforms, cumulative probability distribution, and quantification of mEPSC amplitudes and frequencies from cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18–20 DIV) that were transfected with lentivirus expressing both mGluR5 and DsRed and treated with either control solution or bicuculline (40 μm) for 48 h (n = 48–52 cells in each group). I, J, Representative images and quantification of surface GluA2 immunoreactivity in cultured Mecp2S421A;S424A/y and wild-type hippocampal neurons (18 DIV) that were transfected with DsRed or DsRed-Grm5 containing lentivirus and treated with either control solution or bicuculline (40 μm) for 48 h. Scale bar,10 μm (n = 20 cells in each group). The bar graph shows the mean ± SEM. *p < 0.05, **p < 0.01.
Discussion
MeCP2 phosphorylation has been shown to regulate its association with gene promoters, gene transcription, neuronal morphology, synapse formation, synaptic plasticity and animal behavior (Chen et al., 2003; Zhou et al., 2006; Cohen et al., 2011; Li et al., 2011). Here we provide the first evidence that MeCP2 phosphorylation is also required for bicuculline-induced synaptic scaling down, further expanding the long list of important neuronal processes it is involved in. Moreover, our results show that Grm5/mGluR5 expression is lower in cultured neonatal Mecp2S421A;S424A/y hippocampal neurons, and that both transient pharmacological activation of group I mGluRs and specific overexpression of mGluR5 are sufficient to rescue the deficit in bicuculline-induced synaptic scaling down in these neurons. These findings reveal the first downstream effector protein linking the nuclear function of MeCP2 phosphorylation with neurotransmitter receptors at the synapse, advancing our understanding of the role of MeCP2 in synaptic scaling at the molecular level.
The group I mGluRs are essential components of the synapse. Although they have been implicated in synaptic scaling (Hu et al., 2010), they are better known for regulating long-term depression (LTD) in the adult hippocampus. Our current study focused on their expression in cultured neonatal hippocampal neurons and their function in synaptic scaling when MeCP2 cannot be phosphorylated at S421 and S424. It will be interesting to examine their expression levels in the intact hippocampus and study LTD in adult Mecp2S421A;S424A/y mice in the future. In addition, it is worth noting that mGluR5 expression level is much higher than mGluR1 in the cultured neonatal hippocampal neurons (Fig. 2B), which may help explain why mGluR5 appears to be a more significant player than mGluR1 in this experimental system.
We previously showed that the number of excitatory synapses is significantly higher in cultured neonatal Mecp2S421A;S424A/y hippocampal neurons than in the wild-type, which in theory may lead to an elevated baseline network activity level and a different set point for synaptic homeostasis in the phospho-mutant neurons that is refractory to chronic bicuculline treatment. We do not favor this explanation, because both the frequency and amplitude of mEPSCs at the baseline level are indistinguishable between the phospho-mutant and the wild-type hippocampal neurons (Fig. 1E–H). To reconcile the observations of increased number of structural synapse and the unchanged number of functional synapse in the Mecp2S421A;S424A/y hippocampal neurons, one interesting explanation is that many of these may be silent synapses that contain only NMDA receptors. This hypothesis can be experimentally tested in the future by recording NMDA-mEPSCs.
Since MeCP2 is associated with chromatin and widely distributed across the genome (Skene et al., 2010; Cohen et al., 2011), it is unlikely that Grm5/GluR5 is the only gene/protein whose expression is altered in the absence of MeCP2 phosphorylation. To completely understand how the loss of MeCP2 phosphorylation causes the impairment of bicuculline-induced synaptic scaling down, it is important to identify more gene/protein expression changes in the Mecp2 phospho-mutant hippocampal neurons and characterize their role in regulating synaptic scaling downstream of MeCP2.
Footnotes
This work was supported by NIH–NICHD Grants R01 HD064743 to Q.C. and P30 HD03352 to the Waisman Center. H.L. was supported by a pre-doctoral fellowship from the Stem Cell and Regenerative Medicine Center at the University of Wisconsin-Madison. We thank Fei Yu for technical support.
The authors declare no competing financial interests.
References
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ayala R, Kett LR, Leach TL, Young AB, Dunah AW, Orlando LR. Metabotropic glutamate receptor 1 (mGluR1): antibody specificity and receptor expression in cultured primary neurons. J Neurosci Methods. 2012;204:221–226. doi: 10.1016/j.jneumeth.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascón S, Paez-Gomez JA, Diáz-Guerra M, Scheiffele P, Scholl FG. Dual-promoter lentiviral vectors for constitutive and regulated gene expression in neurons. J Neurosci Methods. 2008;168:104–112. doi: 10.1016/j.jneumeth.2007.09.023. [DOI] [PubMed] [Google Scholar]
- Géranton SM, Morenilla-Palao C, Hunt SP. A role for transcriptional repressor methyl-CpG-binding protein 2 and plasticity-related gene serum- and glucocorticoid-inducible kinase 1 in the induction of inflammatory pain states. J Neurosci. 2007;27:6163–6173. doi: 10.1523/JNEUROSCI.1306-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géranton SM, Fratto V, Tochiki KK, Hunt SP. Descending serotonergic controls regulate inflammation-induced mechanical sensitivity and methyl-CpG-binding protein 2 phosphorylation in the rat superficial dorsal horn. Mol Pain. 2008;4:35. doi: 10.1186/1744-8069-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales ML, Adams S, Dunaway KW, Lasalle JM. Phosphorylation of distinct sites in MeCP2 modifies cofactor associations and the dynamics of transcriptional regulation. Mol Cell Biol. 2012;32:2894–2903. doi: 10.1128/MCB.06728-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg B. Rett's syndrome: prevalence and impact on progressive severe mental retardation in girls. Acta Paediatr Scand. 1985;74:405–408. doi: 10.1111/j.1651-2227.1985.tb10993.x. [DOI] [PubMed] [Google Scholar]
- Hu JH, Park JM, Park S, Xiao B, Dehoff MH, Kim S, Hayashi T, Schwarz MK, Huganir RL, Seeburg PH, Linden DJ, Worley PF. Homeostatic scaling requires group I mGluR activation mediated by Homer1a. Neuron. 2010;68:1128–1142. doi: 10.1016/j.neuron.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li H, Zhong X, Chau KF, Williams EC, Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat Neurosci. 2011;14:1001–1008. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao LM, Horton E, Guo ML, Xue B, Jin DZ, Fibuch EE, Wang JQ. Cocaine increases phosphorylation of MeCP2 in the rat striatum in vivo: a differential role of NMDA receptors. Neurochem Int. 2011;59:610–617. doi: 10.1016/j.neuint.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Sylwestrak EL, Lieberman DN, Zhang Y, Liu XY, Ghosh A. The Rett syndrome protein MeCP2 regulates synaptic scaling. J Neurosci. 2012;32:989–994. doi: 10.1523/JNEUROSCI.0175-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent BDNF transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]