Abstract
Stuttering is a common neurologic disorder in children that can persist into adulthood. Although stuttering displays high heritability, Mendelian segregation typically does not occur, and linkage studies have produced limited success. A genome-wide single nucleotide polymorphism (SNP) linkage scan in a consanguineous Pakistani family followed by targeting genotyping using microsatellite markers revealed linkage on chromosome 16q. The highest linkage scores were obtained under a modified recessive model of inheritance, with a maximum multipoint LOD score of 4.42 at marker D16S3043.
Keywords: Stuttering, linkage, complex trait, consanguineous families
Stuttering is a common neurologic disorder that displays many characteristics of a complex genetic trait, and linkage studies have failed to generate strong linkage, with little replication of findings across studies (Kang and Drayna 2011). We sought to gain increased power to identify causative genes in this disorder by performing linkage in highly consanguineous families with many cases of persistent stuttering.
Research subjects were enrolled in Lahore and surrounding areas of Punjab, Pakistan, with written informed consent approved by Institutional Review Boards at the National Institutes of Health (Protocol #97-DC-0057) and the Centre of Excellence in Molecular Biology (CEMB), at the University of Punjab. Subjects provided blood for DNA extraction and recorded speech samples. Speech samples were diagnosed using the Stuttering Severity Index, 3rd Edition (SSI-3), as previously described (Raza et al. 2010), with individuals displaying ≥4% stuttering dysfluencies, measured by either words or syllables, defined as affected. All individuals scored as affected were persistent stutterers. A genome-wide linkage scan was performed on the Illumina Human Likage-24 Chip (containing 5913 SNPS, ftp://ftp.illumina.com). SNP genotypes were qualified using seven genotyping data analysis parameters (cluster separation, call frequency, AB R mean, AB T mean, Mendelian inheritance, heterozygote excess, and minor allele frequencies, http://www.illumina.com/Documents/products/technotes/technote_infinium_genotyping_data_analysis.pdf), after which 5448 SNPs met criteria for linkage analysis.
Linkage analysis was performed using SuperLink v1.6 from the EasyLinkage package (http://compbio.charite.de/genetik/hoffmann/easyLINKAGE/) (Hoffmann and Lindner 2005) for the initial pairwise scan, followed by MLINK for fine mapping with microsatellite markers (Cottingham et al. 1993). Superlink online v1.5 was used to compute multipoint LOD scores (Silberstein et al. 2006), and SLINK was used to calculate simulated LOD scores using a single locus power calculation (1000 replicates with 23870 random seeds, Ott 1989).
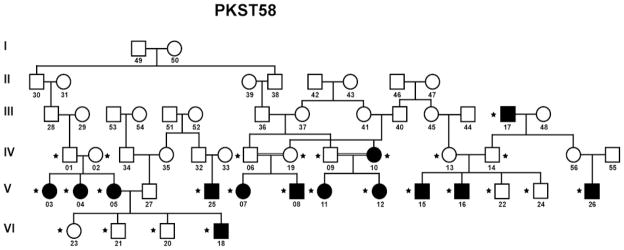
The pedigree of family PKST58 is shown in Figure 1. DNA samples were obtained from 26 family members (starred) who were genotyped, 14 of whom were classified as affected. Parametric analyses were performed with the disease allele frequency set at 0.01 and Caucasian allele frequencies provided by Illumina (ftp://ftp.illumina.com). The results from these analyses gave suggestive linkage scores (LOD scores 2.1 – 2.76) in several regions, including chromosomes 16, 6, 11, 9, and 21.
Figure 1.
Pedigree of Pakistani stuttering family PKST58. Filled symbols represent affected individuals. Double horizontal lines represent consanguineous marriage. Stars indicate individuals for whom DNAs are available.
We selected microsatellite markers spanning these suggestive linkage loci from UCSC genome browser (hg18 assembly) for additional genotyping and fine mapping. The results of these analyses are shown in Table 1. Parametric analysis under a modified recessive model (liability classes defined as 0.0 0.03 0.99) gave strong evidence for linkage for several markers on chromosome 16q (Table 1a), while the LOD scores on other suggestive SNP loci were greatly reduced (chromosomes 6, 9, 11, and 21) (data not shown). Analysis under other models generated reduced evidence for linkage (Table 1b). Multipoint analysis using Superlink online v1.5 under a modified recessive model generated a maximum score of 4.42 at D16S3043. Although these results demonstrate strong evidence for linkage at chromosome 16q12.1–16q23.1, the maximum simulated LOD score obtainable from the available family members under a simple Mendelian recessive single locus model is 6.36. The affected individuals from the non-consanguineous portions of the pedigree did not carry the stuttering-associated haplotype on chromosome 16q. This suggested that these parts of the pedigree might support significant linkage at another locus, perhaps under another mode of inheritance. We therefore performed linkage analysis under different modes of inheritance and varied penetrance for chromosome 6, 9, 11 and 21 excluding the individuals carrying the disease-associated haplotype. No significant evidence of linkage was observed in these analyses, in which the highest LOD score was 2.51, obtained at marker D9S2026 under a fully recessive model of inheritance.
Table 1a.
Single point LOD scores obtained in family PKST58 under a recessive model with modified penetrance *
| Marker ID | hg 18 Physical position | Single point LOD score at recombination fraction theta (θ) | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Mb | 0 | 0.01 | 0.05 | 0.1 | 0.15 | 0.2 | 0.25 | |
| D16S2623 | 50.66 | −0.12 | 0.06 | 0.55 | 0.88 | 1 | 0.98 | 0.88 |
| D16S3132 | 58.75 | 0.59 | 0.67 | 0.83 | 0.85 | 0.81 | 0.72 | 0.62 |
| D16S3089 | 59.06 | 3.64 | 3.64 | 3.45 | 3.05 | 2.58 | 2.08 | 1.58 |
| D16S3143 | 61.13 | 2.53 | 2.52 | 2.36 | 2.02 | 1.64 | 1.23 | 0.85 |
| GATA140E03 | 62.35 | 3.23 | 3.16 | 2.87 | 2.47 | 2.06 | 1.64 | 1.25 |
| D16S3043 | 64.04 | 3.25 | 3.26 | 3.1 | 2.74 | 2.31 | 1.86 | 1.42 |
| D16S3031 | 64.33 | 1.89 | 1.95 | 1.99 | 1.83 | 1.59 | 1.3 | 1 |
| D16S3107 | 66.21 | 1.93 | 2.02 | 2.13 | 2 | 1.74 | 1.44 | 1.11 |
| D16S3025 | 67.12 | 4.35 | 4.25 | 3.82 | 3.28 | 2.73 | 2.18 | 1.65 |
| D16S3067 | 67.66 | 2.27 | 2.37 | 2.43 | 2.24 | 1.93 | 1.57 | 1.2 |
| D16S3095 | 68.5 | 2.81 | 2.74 | 2.44 | 2.06 | 1.68 | 1.3 | 0.94 |
| D16S752 | 69.89 | 2.13 | 2.17 | 2.14 | 1.91 | 1.59 | 1.24 | 0.9 |
| D16S3106 | 70.74 | 3.37 | 3.37 | 3.2 | 2.84 | 2.4 | 1.93 | 1.47 |
| D16S3018 | 72.73 | 1.86 | 1.91 | 1.88 | 1.69 | 1.43 | 1.16 | 0.9 |
| D16S515 | 75.07 | −1.33 | −1.03 | −0.39 | −0.04 | 0.12 | 0.18 | 0.18 |
Analysis parameters: wt/wt=0, wt/mt=0.03, mt/mt=0.99.
Table 1b.
Comparison of single point LOD scores in PKST58 under different inheritance models
| Marker ID | hg18 | Maximum single point LOD score under different inheritance models | |||
|---|---|---|---|---|---|
|
| |||||
| Physical position | Recessive with modified penetrance | Strict recessive model | Additive model | Dominant model | |
|
| |||||
| Mb | 0.00 0.03 0.99 | 0.00 0.00 0.99 | 0.00 0.40 0.80 | 0.00 0.99 0.99 | |
| D16S2623 | 50.66 | −0.12 | −5.84 | 0.44 | −2.43 |
| D16S3132 | 58.75 | 0.59 | −1.33 | 0.97 | −0.11 |
| D16S3089 | 59.06 | 3.64 | 3.47 | 2.45 | −0.4 |
| D16S3143 | 61.13 | 2.53 | 2.61 | 1.69 | −0.01 |
| GATA140E03 | 62.35 | 3.23 | 2.78 | 1.68 | −1.44 |
| D16S3043 | 64.04 | 3.25 | 2.12 | 2.29 | −0.57 |
| D16S3031 | 64.33 | 1.89 | −1.93 | 2.33 | 2.35 |
| D16S3107 | 66.21 | 1.93 | −4.09 | 2.14 | −2.49 |
| D16S3025 | 67.12 | 4.35 | 3.50 | 2 | −2.66 |
| D16S3067 | 67.66 | 2.27 | −5.24 | 1.97 | −2.41 |
| D16S3095 | 68.5 | 2.81 | 0.45 | 1.62 | −0.06 |
| D16S752 | 69.89 | 2.13 | −1.53 | 1.86 | 0 |
| D16S3106 | 70.74 | 3.37 | 2.23 | 2.18 | −2.26 |
| D16S3018 | 72.73 | 1.86 | −3.65 | 2.25 | −0.65 |
| D16S515 | 75.07 | −1.33 | −7.93 | 0.31 | −0.92 |
Overall, these results provide evidence for a new locus for stuttering and indicate that consanguineous families can generate clear positional information for genes underlying complex traits.
Acknowledgments
We thank Ms. Bushra Raza for assistance with stuttering diagnosis, and A. Schaffer, T. Friedman, and R. Morell for helpful comments on the manuscript. We particularly thank the family members who participated in this research study. This work was supported by the Higher Education Commission and the Ministry of Science and Technology, Islamabad, Pakistan and by the National Institute on Deafness and Other Communication Disorders/National Institutes of Health Intramural grant # Z01-000046-11.
Footnotes
Ethical Standards
The experiments reported here comply with the current laws of Pakistan and the United States.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53 (1):252–263. [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH. easyLINKAGE-Plus--automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21(17):3565–3567. doi: 10.1093/bioinformatics/bti571. bti571 [pii] [DOI] [PubMed] [Google Scholar]
- Kang C, Drayna D. Genetics of speech and language disorders. Annu Rev Genomics Hum Genet. 2011;12:145–164. doi: 10.1146/annurev-genom-090810-183119. [DOI] [PubMed] [Google Scholar]
- Ott J. Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci U S A. 1989;86 (11):4175–4178. doi: 10.1073/pnas.86.11.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza MH, Riazuddin S, Drayna D. Identification of an autosomal recessive stuttering locus on chromosome 3q13.2-3q13.33. Hum Genet. 2010;128(4):461–463. doi: 10.1007/s00439-010-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D. Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. Am J Hum Genet. 2006;78(6):922–935. doi: 10.1086/504158. S0002-9297(07)63915-9 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]