Abstract
RNA interference (RNAi) holds great promise as a strategy to further our understanding of gene function in the central nervous system (CNS) and as a therapeutic approach for neurological and neurodegenerative diseases. However, the potential for its use is hampered by the lack of siRNA delivery vectors which are both safe and highly efficient. Cyclodextrins have been shown to be efficient and low toxicity gene delivery vectors in various cell types in vitro. However, to date, they have not been exploited for delivery of oligonucleotides to neurons. To this end, a modified β-cyclodextrin (CD) vector was synthesized, which complexed siRNA to form cationic nanoparticles of less than 200 nm in size. Furthermore, it conferred stability in serum to the siRNA cargo. The in vitro performance of the CD in both immortalized hypothalamic neurons and primary hippocampal neurons was evaluated. The CD facilitated high levels of intracellular delivery of labeled siRNA, while maintaining at least 80% cell viability. Significant gene knockdown was achieved, with a reduction in luciferase expression of up to 68% and a reduction in endogenous glyceraldehyde phosphate dehydrogenase (GAPDH) expression of up to 40%. To our knowledge, this is the first time that a modified CD has been used as a safe and efficacious vector for siRNA delivery into neuronal cells.
Keywords: Nanotechnology, cyclodextrins, click chemistry, siRNA delivery, neurons, gene knockdown
RNA interference (RNAi) has advanced rapidly since it was first discovered in mammalian cells1 to clinical application and most recently, the first evidence of systemic administration of a targeted nonviral siRNA delivery system to humans was reported.2 RNAi is a naturally occurring biological process which results in specific silencing of a target gene by short sequences of RNA known as small interfering RNAs (siRNAs),3 with a subsequent reduction in the expression of the corresponding protein. Introduction of chemically synthesized siRNA into cells to reduce expression of a specific gene can be used to determine gene function or for therapeutic purposes.
However, delivery of siRNA proves a constant challenge and there are specific issues which have to be addressed when delivering to neurons and the central nervous system (CNS).4 Neurons are notoriously difficult to transfect for reasons that have yet to be fully understood, although it is likely to be partly related to their postmitotic nature.5 There are many barriers to siRNA delivery to the CNS including neuronal uptake, vesicular escape, and, not least, the blood-brain barrier.6 Appropriately designed delivery systems are therefore required in order to mediate neuronal siRNA delivery and gene silencing. Viral vectors have been extensively reported and achieve efficient transfection.7 However, nonviral vectors are more recently the focus of research to achieve a safer means of transfection. To date, there are limited reports of successful neuronal delivery of siRNA with these. The most widely used are cationic lipid based vectors, in particular Lipofectamine 2000 (Invitrogen), which has been shown to mediate effective gene knockdown in primary neurons,8−10 with up to 70% reduction in gene or protein expression achieved.9 However, such vectors are also associated with considerable toxicity in primary cells, particularly neurons.11−13 Therefore, there remains a need for the development of alternative less toxic vectors for siRNA and gene delivery to neurons.
Cyclodextrins (CDs) are naturally occurring oligosaccharides and are well characterized pharmaceutical excipients with a favorable toxicological profile.14 The potential of CDs as delivery agents for therapeutic oligonucleotides is well established.15,16 Chemical modification of CDs has yielded monodisperse nonviral gene delivery vectors.17−21 Modifications of the basic β-CD structure, including introduction of cationic and/or amphiphilic moieties have resulted in a group of novel vectors which can complex pDNA and efficiently deliver it to various in vitro cell culture models, including liver (Hep G2) cells22,23 and undifferentiated and differentiated intestinal epithelial (Caco2) cells.24 A novel synthetic approach, involving the application of cuprous-catalyzed click chemistry to modify the 2-hydroxyl position of β-CD with polar groups was recently reported.25 An amphiphilic cationic β-CD with hydrocarbon chains (C12) on the primary face and polar (propylamino) groups on the secondary face was synthesized in this way. This CD achieved high levels of gene silencing in Caco2 cells.25
Given the ease with which they can now be chemically modified and their low toxicity and efficiency in non-neuronal cells, it is perhaps surprising that the utility of such CDs for neuronal delivery remains to be exploited. Therefore, in this study, we investigate the aforementioned cationic amphiphilic CD for siRNA delivery in hippocampal and hypothalamic neurons.
Results and Discussion
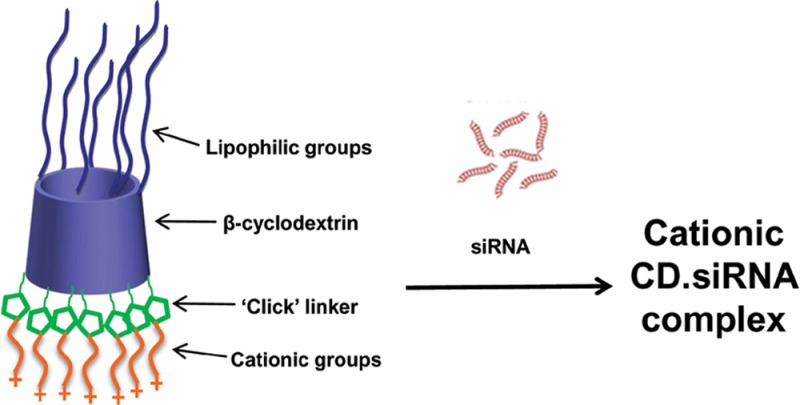
The vast potential for RNA interference technology in terms of identifying new targets in many neurological diseases, as well as offering an alternative therapeutic strategy, warrants research into the development of improved and low toxicity vectors for delivering siRNA. Developments in the chemistry of such vectors are facilitating major biological advances and, in this study, we demonstrate that a novel chemically modified CD can be used for neuronal siRNA delivery. The CD chemical structure is shown in Figure 1a. The synthesis of this CD was previously reported.25 Briefly, β-CD was substituted on the primary side with lipophilic chains (C12) by thioalkylation.18,26 Introduction of amphiphilic moieties improves the transfection efficiency of cationic CDs.18,22 Functionalization at the 2-position was by means of a copper-catalyzed “click” reaction, which represents an efficient and versatile strategy for modifying the secondary side with diverse groups, including cationic groups and polyethyleneglycol (PEG) chains. Click chemistry has been previously applied to modify the primary face of CD-based gene delivery vectors,27,28 but the selective modification at the 2-hydroxyl presented a greater challenge.19 A schematic representation of the CD is shown in Figure 1b. This CD has shown promise as a nonviral siRNA delivery vector in non-neuronal cells.25
Figure 1.
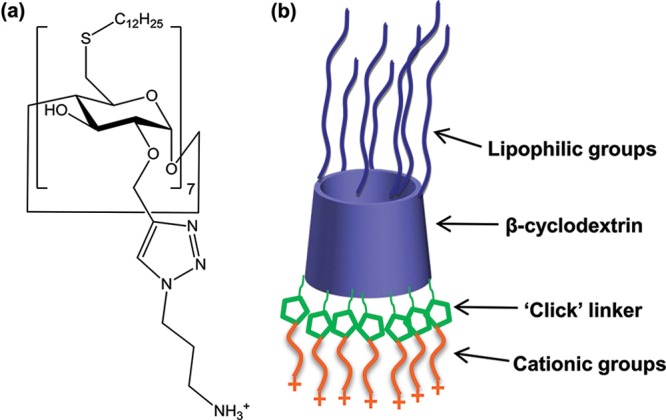
(a) Chemical structure of CD used for siRNA delivery and (b) schematic representation of CD structure.
Other nonviral methods for siRNA delivery have been reported in recent years, including cationic lipids, polymers, and cell-penetrating peptides (CPPs). Cationic lipids and polymers, although highly efficacious, exhibit considerable toxicity.29 CPPs, on the other hand, exert minimal toxicity and have been successfully used for siRNA delivery to primary hippocampal neurons,12 primary cortical neurons,30 and the hippocampus31in vivo. However, additional nonviral delivery systems would be desirable and, to this end, modified CDs offer great potential. In particular, the chemical synthesis for the CD used herein can be also applied for functionalization of CDs with PEG groups for improved stability.25 Moreover, there is potential to extend this chemistry to allow for the attachment of neuronal and CNS targeting ligands. Therefore, we believe that exciting prospects exist for the application of modified CDs to neuronal RNAi.
To determine the suitability of the modified CD as a siRNA delivery vector, several physical properties including the ability to complex siRNA, size and charge of CD.siRNA complexes, and protection of siRNA from serum degradation were assessed. Following this, in vitro experiments in neurons showed that the CD successfully mediated efficient uptake of siRNA and knockdown of reporter and housekeeping genes.
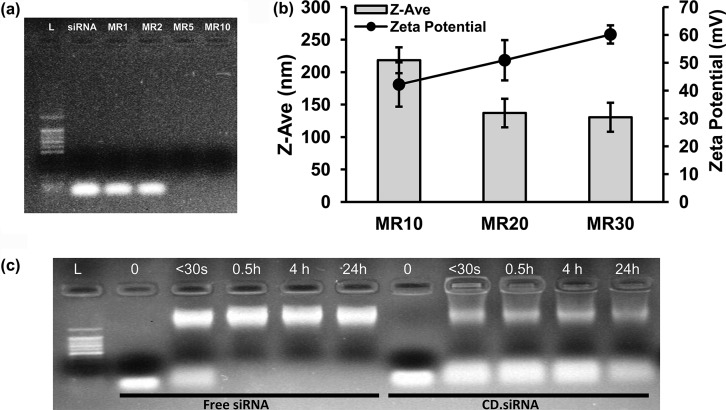
In the first instance, CD.siRNA complex formation was identified using agarose gel electrophoresis. Uncomplexed siRNA was used as a control and migrated freely through the gel. This was compared with CD.siRNA complexes, which were formed at increasing mass ratios (MRs; μg CD/μg siRNA). The positive charges from the primary amine group in the CD enable association with the negatively charged phosphate groups of the siRNA backbone.32 Complete binding of siRNA appeared to occur abruptly at MR5, with no migration of siRNA seen at MR 5 or MR10 (Figure 2a), demonstrating that the negatively charged siRNA had been effectively complexed by the CD at these MRs. Furthermore, there was no fluorescence apparent in the wells of the CD.siRNA lanes, indicating that complexed siRNA was not accessible to the intercalating agent ethidium bromide.
Figure 2.
Properties of CD.siRNA complexes. (a) CD.siRNA binding was shown by gel electrophoresis. Fluorescent bands correspond to 100 bp DNA ladder (L), free siRNA and unbound siRNA. (b) Z-Ave (size (nm)) and zeta potential (charge (mV)) of CD-siRNA complexes at increasing mass ratios (MR; μg CD/μg siRNA) were measured in DIW (final siRNA concentration 3 μg/mL) using dynamic light scattering (Malvern ZetaSizer Nano). (c) Serum degradation assay, as shown by gel electrophoresis. Uncomplexed siRNA or CD.siRNA was exposed to 50% serum for up to 24 h. CD at least partially protected siRNA from degradation.
Second, the physicochemical properties of siRNA delivery vectors, which are well-known to impact on their transfection efficiency, were examined. Key factors which can affect transfection include the size of the complexes formed with siRNA and their surface charge.33,34 The particle size of CD.siRNA complexes, measured using the Malvern ZetaSizer Nano particle size analyzer, was dependent on MR (Figure 2b). Although siRNA was shown to be fully bound at MR5, these complexes were very polydisperse and size and charge data were therefore excluded. Complexes produced at MR10 were 218 ± 19.9 nm, whereas those complexes produced at MR20 and MR30 were 136.9 ± 21.9 nm and 130.4 ± 22.3 nm, respectively. These sizes fall within the nanometer range required for cellular uptake by endocytic processes.33
The polydispersity index (PDI) was also noted for each size measurement, with PDI generally in the range 0.2–0.4, indicating some variation in the size distribution.17 The particle sizes were stable when stored for up to 6 months at 4 °C, with no aggregation or change in appearance observed (data not shown).
The surface charge of CD.siRNA complexes was also measured and was similarly dependent on MR. Complexes were cationic at all MRs, with charge (zeta potential) increasing as the MR increased from +42.2 ± 7.9 mV at MR10 to +60.2 ± 3.3 mV at MR30; that is, increasing the amount of cationic CD relative to siRNA resulted in more highly positively charged complexes. The cationic nature of the complexes formed allows association with the negatively charged proteoglycans on the surface of the cell membrane.24
Furthermore, stability of siRNA in serum is an important feature when considering future in vivo experiments. Agarose gel electrophoresis was used to characterize the protective effects of CDs on siRNA degradation by serum (Figure 2c). Uncomplexed siRNA, shown in lanes 2–5, was partially degraded after less than 30 s exposure to serum and completely degraded after 0.5–24 h exposure. In contrast, siRNA in CD complexes (lanes 6–11), which was released from the complexes by heparin displacement after serum exposure, remained intact for up to 4 h which is comparable to the protection conferred by other vectors, including liposomes.35 Even after 24 h, the CD prevented complete degradation of siRNA.
Having confirmed that the physical properties of the CD vector were suitable for siRNA delivery, in vitro testing was carried out in a mouse embryonic hypothalamic neuronal cell line, mHypoE N41,36 and in primary rat embryonic (E18) hippocampal neurons.
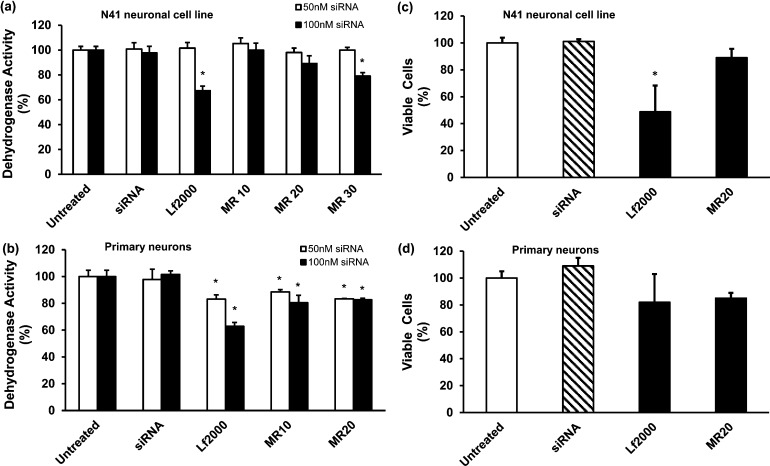
First, the cytotoxic effects of CD.siRNA complexes on neuronal cell cultures were determined by MTT assays, after cells were treated with CD.siRNA complexes for 24 h, using 50 or 100 nM siRNA. Overall, a favorable toxicity profile for this vector was observed in both cultures, although the primary neurons were more susceptible to CD.siRNA mediated toxicity when compared to the immortalized N41 cells. Mitochondrial dehydrogenase activity compared to untreated neurons was used as an indicator of toxicity.
In N41 cells, CD.siRNA complexes had no effect on dehydrogenase activity at 50 nM siRNA (Figure 3a). After 24 h treatment at 100 nM siRNA, cell viability was unaffected at MR10 and was maintained at 88 ± 1.5% at MR20 (Figure 3a). Only at MR30 was there a significant reduction, corresponding to minor cytotoxicity (80 ± 0.9%, *p < 0.05 relative to untreated controls).
Figure 3.
Toxicity was determined by MTT assay (a, b) and trypan blue exclusion assay (c, d) after 24 h incubation with CD.siRNA complexes (50 or 100 nM siRNA) in mHypoE N41 cells and primary rat (E18) hippocampal neurons. Data (n = 3) are presented as the mean ± SEM; *p < 0.05 relative to untreated controls.
Meanwhile, in primary hippocampal neuronal cultures, siRNA alone did not cause any cytotoxicity (Figure 3b) but some reduction in cell viability occurred after treatment; but after treatment with CD.siRNA complexes at 50 nM siRNA, dehydrogenase activity was reduced to 88 ± 1.6% (*p < 0.05) at MR10 and 83 ± 0.4% (*p < 0.05) at MR20. At 100 nM siRNA, however, there was no further decrease in cell viability.
Trypan blue exclusion assays were also carried out as an alternative measure of cytotoxicity (Figure 3c, d). These data confirm the favorable cytotoxicity profile of CD vectors in both immortalized and primary neurons.
Neuronal cells, in particular primary neuronal cells, are quite susceptible to the toxic effects of cationic vectors. For example, in primary cortical cultures, layered double hydroxide (LDH) nanoparticles caused a 20–30% reduction in viability37 and in another study, toxicity with Oligofectamine (∼80%) and with polyethyleneimine (PEI; ∼76%) was reported.38 In primary hippocampal neurons, stearylated octaarginine and artificial viruslike particles reduced cell viability to 73% and 75%, respectively.39 The data presented here showed that the modified CD did not induce significant toxicity in the hypothalamic neuronal cell line at MR10 or MR20 (Figure 3a) and caused limited impairment of viability of the primary hippocampal neurons (80%, Figure 3b). Under similar experimental conditions, Lipofectamine 2000 was found to be considerably more toxic to primary hippocampal neurons than the CD vector, reducing viability to 61.8 ± 3.5% (Figure 3b), in the MTT assay. This was comparable to previously reported toxicity of Lipofectamine 2000 in primary hippocampal cultures.12
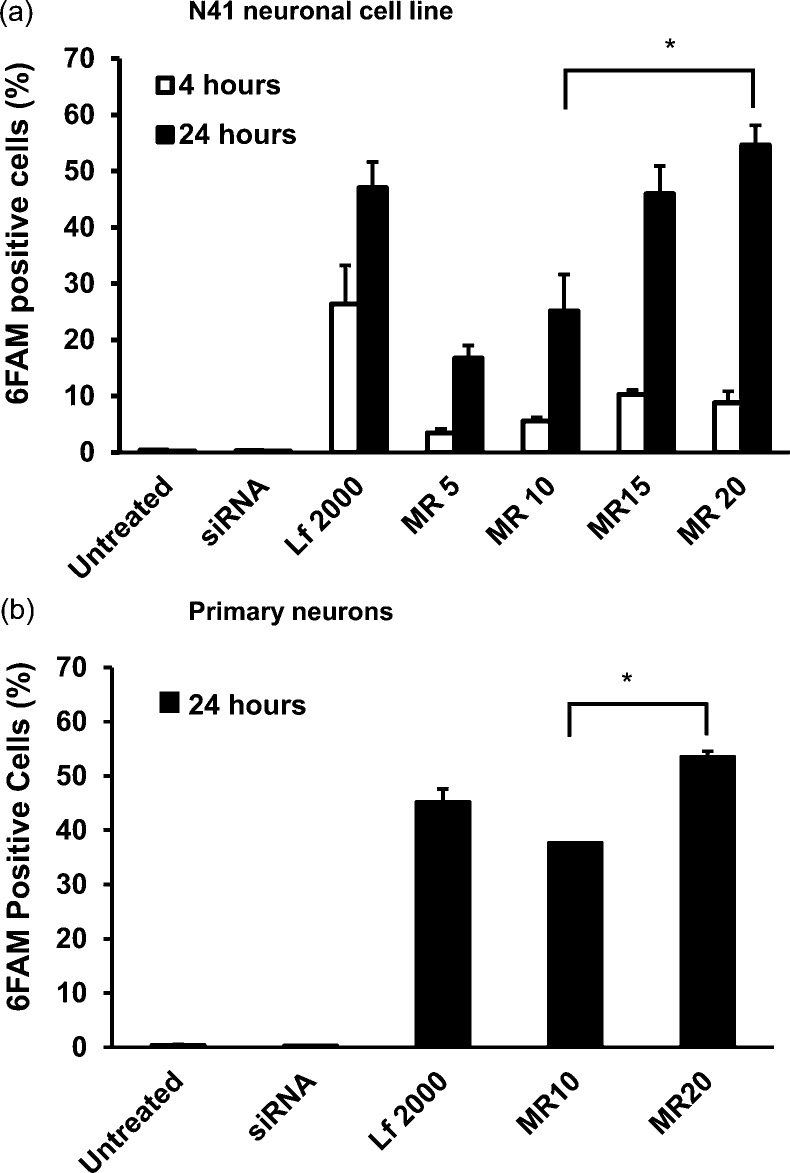
Second, cellular uptake of CD.siRNA complexes was assessed by flow cytometry. For these experiments, 6-FAM labeled siRNA was used, at a concentration of 50 nM. Figure 4a shows the percentage of 6-FAM positive cells in N41 cultures, after treatment with CD.siRNA complexes at increasing mass ratios from MR5 to MR20, for either 4 or 24 h. No uptake was observed when cells were treated with uncomplexed siRNA at either time point. After 4 h treatment, low levels of uptake were seen, ranging from 3.5 ± 1.2% at MR5 to 8.8 ± 3.6% at MR20. However, a highly significant increase in uptake was observed after 24 h treatment, with the highest level at MR20 (54.6 ± 6.1%, *p < 0.05 relative to untreated controls). One possible explanation for this delayed uptake is slow settling of the CD.siRNA complexes onto the adherent cells. The inclusion of a triazole moiety in a polycationic amphiphilic CD was previously reported to improve stability in a high salt environment.21 Aggregation can enhance transfection in vitro as the aggregates are quick to settle down onto adherent cells for association and uptake.40,41 Therefore, delayed uptake may result when aggregation occurs more slowly.
Figure 4.
Internalization of CD.siRNA complexes by (a) mHypoE N41 cells and (b) primary rat (E18) hippocampal cells. FAM-siRNA uptake, after incubation with CD.siRNA complexes (siRNA 50 nM), was measured by flow cytometry. Data (n = 3) are expressed as the mean ± SEM; *p < 0.05 relative to MR10.
Although aggregation may aid cellular association in vitro, this is not the case in vivo and methods to improve formulations to prevent aggregation are required to enable prolonged circulation and prevent opsonisation.
Significant levels of uptake were also achieved when primary hippocampal neurons were treated with CD.siRNA complexes for 24 h (Figure 4b). The percentage of 6-FAM positive cells increased from 37.7 ± 1.8% at MR10 to 53.5 ± 2.3% at MR20. In both neuronal cultures, the level of uptake achieved with MR20 was significantly greater than that at MR10 with the same concentration of siRNA. Lipofectamine 2000 mediated similar levels of uptake in both neuronal cell models (Figure 4).
After delivery into neurons, it is critical that siRNA is made available for RNAi by incorporation into the RISC complex in the cytoplasm, to enable downstream gene silencing. To this end, the ability of the CD to deliver luciferase siRNA into neuronal cells and reduce luciferase gene expression was investigated. Given that significantly greater uptake was achieved after 24 h, this was chosen as the appropriate length of time for transfection experiments.
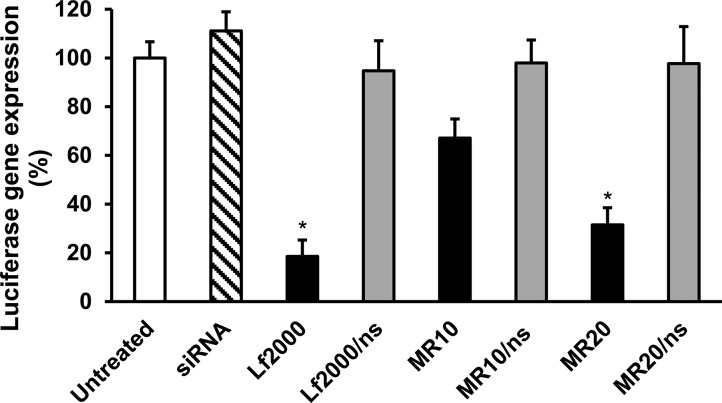
First, N41 cells were transfected for 2 h with the firefly luciferase reporter plasmid by cationic lipid mediated delivery (Lipofectamine 2000). Subsequently, cells were treated for 24 h with CD.siRNA complexes against the luciferase gene. Nonsilencing siRNA complexed to CD was used as a negative control. Luciferase expression levels decreased after treatment with MR10 and MR20 (Figure 5), with a significant reduction achieved at MR20 (68.5 ± 7%, *p < 0.05 relative to untreated control). Transfection with uncomplexed siRNA did not have any effect on luciferase expression. Similarly, CD complexes with nonsilencing (ns) siRNA did not reduce luciferase expression, demonstrating the specificity of knockdown. These data confirm that effective and specific gene silencing was achieved when siRNA was delivered to neuronal cells by the CD.siRNA delivery system.
Figure 5.
Knockdown of luciferase reporter gene in mHypoE N41 cells by CD.siRNA complexes. Cells were incubated with luciferase reporter plasmid (1 μg/well). Two hours later, after washing and changing media, luciferase siRNA (50 nM), either uncomplexed or complexed to CD, was added. After 24 h, reduction in gene expression was measured by luciferase assay. Data (n = 3) are expressed as the mean ± SEM; *p < 0.05 relative to untreated controls.
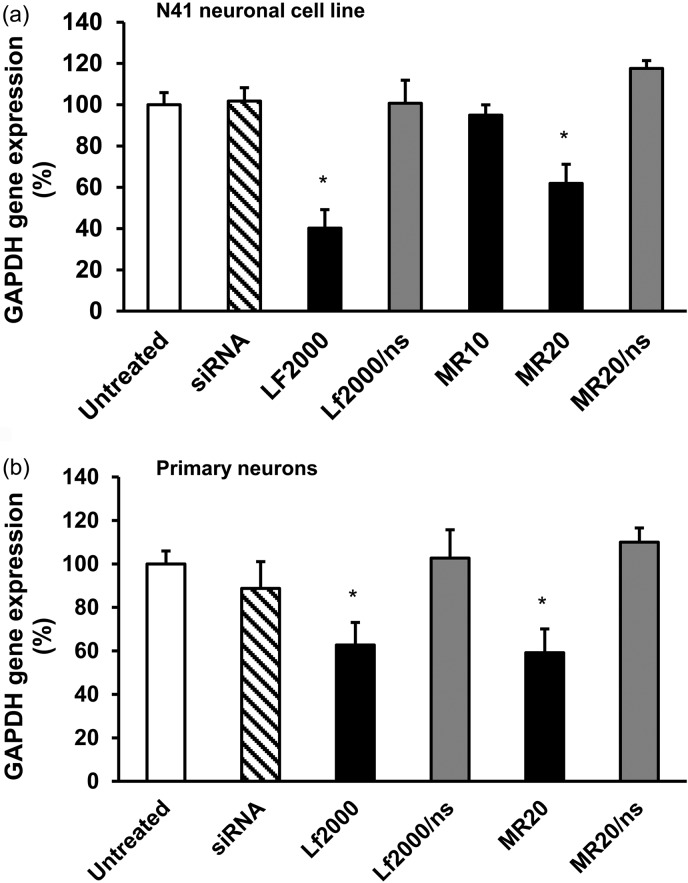
Having achieved knockdown of an exogenously transfected gene with CD.siRNA complexes, the ability of the vector to mediate knockdown of an endogenous housekeeping gene, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), was assessed. Preliminary experiments in N41 cells showed that when siRNA was used at a concentration of 50 nM, CD.siRNA complexes had no effect on GAPDH expression. Therefore, subsequent experiments were carried out using 100 nM siRNA. GAPDH siRNA and a negative control siRNA were used. Neurons were treated for 24 h, and GAPDH mRNA levels were quantified by qRT-PCR, after extraction of total RNA from the treated cells. These data are shown in Figure 6. In N41 cells (Figure 6a), at MR20, a significant reduction in GAPDH expression was achieved (38% ± 9.3%, *p < 0.05 relative to untreated control). Meanwhile, the complexes formed at MR10 did not reduce GAPDH expression under these experimental conditions. Figure 6b shows a similar level of knockdown in the primary hippocampal neurons (41% ± 5.7%, *p < 0.05 relative to untreated control). Neither nonsilencing siRNA formulated with the CD nor uncomplexed siRNA reduced GAPDH expression, confirming specificity of gene silencing and the lack of effectiveness of naked siRNA.
Figure 6.
Knockdown of endogenous GAPDH in (a) mHypoE N41 cells and (b) primary rat (E18) hippocampal cells by CD.siRNA complexes. Cells were incubated GAPDH siRNA (100 nM), either uncomplexed or complexed to CD, for 24 h. Total RNA was extracted and reverse transcribed to cDNA for qRT-PCR, to determine the reduction in GAPDH mRNA expression. Data (n = 3) are expressed as the mean ± SEM; *p < 0.05 relative to untreated controls.
The level of GAPDH knockdown reported here was moderate and although there is some possibility that this may be due to incomplete dissociation of siRNA from the CD, the reduction in GAPDH gene expression achieved with Lipofectamine 2000 was not significantly different (∼ 40%, Figure 6b). Furthermore, similar levels of knockdown have been reported for other endogenous gene targets in neuronal cells. For example, a modest reduction in HMGB-1 mRNA (<40%) was achieved in primary cortical neurons using PAMAM-arginine vector to deliver shRNA13 and a stearylated octaarginine vector mediated ∼50% reduction in reporter gene expression in primary hippocampal neurons.39 Furthermore, Lipofectamine 2000 has been reported for siRNA mediated knockdown in N-1 hypothalamic neurons, with varying levels of knockdown depending on the target. Reductions in levels of resistin mRNA of 25%42 to 55–60%42,43 were reported, depending on the siRNA sequence. However, other RNAi delivery methods have achieved more pronounced gene silencing effects. For example, >80% reduction in target protein expression was reported for Penetratin-1-siRNA conjugates in primary hippocampal neurons12 and in the hippocampus in vivo.31 More substantial levels of knockdown (∼90%) of another target, CCAAT enhancer binding protein α, were achieved using Lipofectamine 2000 in N-1 neurons.44 In this study, similar experimental conditions to those reported in this paper were used; however, “stealth” siRNA sequences, which are chemically modified RNA duplexes, with improved stability and transfection efficiency, were used which may explain the greater knockdown levels achieved.
It is also worth noting that a moderate reduction in target gene expression may be sufficient for a functional output in an animal model.45−47 For example, a 30% decrease in striatal tyrosine hydroxylase was sufficient to reduce the hyperactive locomotor response to amphetamine in mice46 and antidepressant-like behaviors were observed in mice, after icv infusion of siRNA led to a 40% reduction in expression of the serotonin transporter.47
In summary, an amphiphilic cationic modified β-cyclodextrin was shown to deliver siRNA to neurons for gene silencing. Efficient uptake into neuronal cells was shown, with a favorable cytotoxicity profile. Moreover, significant levels of knockdown were achieved for an exogenous gene (luciferase) and an endogenous gene (GAPDH). The physical properties of the complexes formed with siRNA and the ability of the CD to protect siRNA from serum degradation were key features in its efficiency as a delivery vector. Furthermore, future studies focused on optimizing the structural modifications applied to CDs can now be carried out having established the potential of this CD as a novel nonviral vector for neuronal siRNA delivery.
Methods
Cell Culture
mHypoE N41
A mouse embryonic hypothalamic cell line36 was obtained from tebu-bio (France) and was maintained in Dulbecco’s modified Eagle’s medium (DMEM, Sigma), supplemented with 10% fetal bovine serum (FBS, Sigma) in a humidified 37 °C incubator with 5% CO2. Cells were seeded in 12-well, 24-well and 96-well plates at 6.6 × 104, 3.5 × 104, and 1.5 × 104 cells/well, respectively.
Primary Neuronal Culture
Primary rat embryonic hippocampal neurons were obtained from Sprague–Dawley rats, at day 18 of gestation as previously described.11 Briefly, the animals were sacrificed and the embryos removed. Embryonic hippocampal neurons were dissected out and plated onto poly-l-lysine coated coverslips (for immunofluoresence) or poly-l-lysine treated wells (for all other experiments), in 12-well or 96-well plates and maintained in Neurobasal medium (Gibco), supplemented with 2% B-27 (Invitrogen) and 0.5 mM l-glutamine (Sigma). Cells were maintained in a humidified 37 °C incubator with 5% CO2. Seeding densities were 5 × 105 and 1.5 × 104 for 12- and 96-well plates, respectively. Characterization of the cultures by immunostaining for β III tubulin and GFAP confirmed the presence of 90–95% neurons in the culture.
siRNAs
siRNAs were synthesized by Qiagen. siRNA sequences are shown in Table 1.
Table 1. Sequences of siRNA Obtained from Qiagen.
| sense | antisense | |
|---|---|---|
| 6FAM negative control siRNA | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
| negative control siRNA | UUCUCCGAACGUGUCACGUdTdT | ACGUGACACGUUCGGAGAAdTdT |
| luciferase GL3 siRNA | CUUACGCUGAGUACUUCGAdTdT | UCGAAGUACUCAGCGUAAGdTdT |
| anti-mouse GAPDH siRNA | GGUCGGUGUGAACGGAUUUdTdT | AAAUCCGUUCACACCGACCdTdT |
| anti-rat GAPDH siRNA | GGUCGGUGUGAACGGAUUUdTdT | AAAUCCGUUCACACCGACCdTdT |
Preparation of CD.siRNA Complexes
An amphiphilic cationic CD was synthesized as previously described25 and was dissolved in chloroform to give a 1 mg/mL solution. A stream of nitrogen was then applied to remove the solvent, leaving a CD film, which was rehydrated with water (CD concentration 1 mg/mL), followed by sonication at room temperature (RT) for 1 h for vesicle size reduction. A solution of siRNA in water was then mixed with CD solution (equal volumes) and complexed for 15–20 min at RT (final CD concentration of 0.5 mg/mL). Different mass ratios (MR) of CD to siRNA were chosen (μg CD/μg siRNA). This “mixing method” for CD.siRNA complex preparation was previously optimized for CD.DNA complexes.17,22,24
Preparation of Lf2000.siRNA Complexes
Lipofectamine 2000 (Invitrogen) (Lf2000) is a cationic lipid-based transfection reagent. Lf2000.siRNA complexes were prepared as per the manufacturer’s protocol. Briefly, the required volume of Lf2000 was diluted in 50 μL of OptiMEM, mixed gently, and incubated at RT for 5 min. siRNA was diluted in 50 μL of OptiMEM and combined with the diluted Lf2000, and then mixed gently and incubated at RT for 20 min. One microliter of Lf2000 was used per 20 pmol of siRNA.
Size and Charge Measurements
Particle size and charge were measured with Malvern’s Zetasizer Nano ZS instrument, using laser-light scattering and electrophoretic mobility measurements, respectively.24,48 CD.siRNA (3 μg siRNA) complexes were prepared by the “mixing method”. The resulting mixtures were made up to 1 mL with 0.2 μm filtered deionized water (DIW). Five readings of Z-average size (nm), polydispersity (25 °C, measurement angle 170°), and zeta potential (mV) (25 °C, measurement angle 12.8°) were taken. For data analysis, the viscosity (0.8872 mPa·s) and refractive index (1.33) of water were used to determine Z-average size. The data are presented as mean ± SD.
Gel Retardation Assay
Agarose gel electrophoresis was used to determine the binding of siRNA. CD.siRNA complexes were prepared and mixed with loading buffer and DIW to a final volume of 20 μL (containing 0.3 μg siRNA). Samples were added to wells in a 1% agarose gel containing ethidium bromide (0.5 μg/mL). Electrophoresis was carried out at 90 V for 20 min, with a Tris-borate-EDTA buffer.49 Bands corresponding to the DNA ladder (100 bp) and unbound siRNA were visualized by UV, using the DNR Bioimaging Systems MiniBis Pro and Gel Capture US B2 software.
Serum Stability Studies
To evaluate protection of siRNA from serum nucleases by the CD, CD.siRNA complexes were incubated with 50% FBS for 30 s, 0.5 h, 4 h, or 24 h at 37 °C, followed by addition of excess heparin to displace the siRNA.50 Uncomplexed siRNA samples were also incubated with serum in the same way. Samples were analyzed by agarose gel electrophoresis (1.5% agarose gel) as described above.
Toxicity Assays
MTT Toxicity Assay
The MTT assay is widely used as an indicator of the toxicity caused to neurons by nonviral vectors.13,37,38,51 This assay measures the reduction of MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide) by mitochondrial dehydrogenase, in viable cells only, to give a dark blue product. However, as it only measures one end-point, namely, the change in mitochondrial integrity, it is not, therefore, a direct measure of cell viability. Cells were seeded in 96-well plates for 1 day before transfection then were treated with CD.siRNA complexes for 24 h. Medium was removed and replaced with 100 μL of fresh medium and MTT reagent (20 μL of a 5 mg/mL solution) for 2 h, after which the formazan crystals produced were dissolved in 100 μL of dimethyl sulfoxide. Absorbance was measured at 590 nm using a UV plate reader. Each experiment was carried out in triplicate. Results were expressed as % dehydrogenase activity compared to untreated controls. The data are presented as the mean ± SEM.
Trypan Blue Exclusion Assay
Cell viability was also assessed using trypan blue exclusion analysis as previously reported.37,52 Briefly, cells were seeded in 24-well plates for 1 day before transfection and then CD.siRNA complexes (100 nM siRNA), diluted in OptiMEM, were applied in serum-containing medium, for 24 h. Cells were detached with trypsin-EDTA (0.25%) and stained with 0.02% trypan blue. Viable cells were counted and results were expressed as percent of viable cells compared to untreated controls. The data are presented as the mean ± SEM.
Cellular Uptake Experiments
The level of uptake mediated by transfection complexes was assessed by flow cytometry.37,53,54 Fluorescently labeled (6FAM) siRNA (Qiagen) was used for these experiments. Cells were seeded in 24-well plates for 1 day before transfection. siRNA (50 nM) alone or in CD.siRNA complexes was diluted in OptiMEM and added to cells in serum-containing medium, for 4 or 24 h. Following this, uninternalized complexes were removed by washing cells with PBS and by incubation with 250 μL of CellScrub buffer for 15 min at room temperature.48 Cells were removed from the wells and prepared for analysis following several washing steps. The fluorescence associated with 10 000 cells was measured with a FACS Caliber instrument (BD Biosciences), and data was analyzed using Cell Quest Pro software. Each experiment was carried out in triplicate. The data are presented as the mean ± SEM.
Knockdown of Luciferase Reporter Gene
Silencing of an exogenous gene was assessed by measuring knockdown of the firefly luciferase gene.51,55,56 Cells were seeded in 24-well plates for 1 day before transfection. Cells were transfected with luciferase reporter plasmid, pGL3 luciferase (1 μg/well), complexed to Lipofectamine 2000 (2.5 μL/ μg pDNA) for 2 h. Following this, cells were washed twice with PBS prior to siRNA transfection. siRNA (50 nM) alone, or complexed to CD, was diluted in OptiMEM and added to the cells in serum-containing medium. Nonsilencing siRNA complexed to CD was used as a control. After 24 h, cells were washed with PBS and lysed. Lysate (20 μL) was assayed for expression of luciferase by adding to 100 μL of luciferin (Promega) and measuring the luminesence in a Junior LB 9059 luminometer (Promega). Total protein levels in each sample were determined via usthe BCA protein assay (Thermo Scientific) to allow normalization of luciferase activity. Results were expressed as percent of gene expression relative to control (no RNAi) samples. Data are presented as the mean ± SEM.
Knockdown of Endogenous GAPDH
Silencing of an endogenous gene was assessed by measuring knockdown of the housekeeping gene GAPDH. This gene is widely used to assess knockdown efficiency in in vitro studies.57,58 Cells were seeded in 12-well plates for 1 day before transfection. GAPDH siRNA (100 nM) alone, or complexed to CD, was diluted in OptiMEM and added to the cells in serum-containing medium. Nonsilencing siRNA complexed to CD was used as a control. After 24 h, total RNA was extracted from N41 cells using the Stratagene Absolutely RNA Miniprep Kit, according to the manufacturer’s instructions and from primary hippocampal cells using Trizol reagent (Invitrogen) for primary hippocampal cultures. The concentration of RNA was measured by UV absorbance on the NanoDrop ND-1000 UV–vis spectrophotometer, and the RNA integrity was confirmed by analysis using the Agilent 2100 bioanalyzer. A high-capacity cDNA reverse transcriptase kit (Applied Biosystems) was used for cDNA (cDNA) synthesis. Gene expression was assessed by real-time qPCR using the Applied Biosystems Real Time PCR System (model 7300). Assays were performed using appropriate primer sets for GAPDH and β-actin (TaqMan, Applied Biosystems). Amplification was carried out by 40 cycles of denaturation at 95 °C (15 s) and annealing at 60 °C (1 min). β-Actin endogenous gene was used for relative gene quantification.58 The (2–ΔCt) method was used to calculate relative changes in mRNA.59 Each experiment was carried out in triplicate. Results were expressed as percent of GAPDH gene expression relative to untreated (nontransfected) controls. The data are presented as the mean ± SEM.
Statistics
One-way analysis of variance (ANOVA) was used to compare multiple groups followed by Bonferroni’s post hoc test. Statistical significance was set at *p < 0.05.
Author Contributions
A.O.M. carried out all experiments and performed data analysis under the direction of C.O.D. and J.C. B.G. completed trypan blue exclusion assays in primary cultures. J.O. synthesized the modified cyclodextrin under direction of R.D. M.D. assisted in the chemistry of the CDs, especially the ‘click’ approach. A.O.M. wrote the manuscript, with assistance from C.O.D. and J.C.
This work was supported by Science Foundation Ireland (Grant No. 07/SRC/B1154), the Irish Drug Delivery Network and by a scholarship to A.M.O’M. from the Irish Research Council of Science, Engineering and Technology.
The authors declare no competing financial interest.
References
- Elbashir S. M.; Harborth J.; Lendeckel W.; Yalcin A.; Weber K.; Tuschl T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Davis M. E.; Zuckerman J. E.; Choi C. H. J.; Seligson D.; Tolcher A.; Alabi C. A.; Yen Y.; Heidel J. D.; Ribas A. (2010) Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–U1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S. M.; Bernstein E.; Beach D.; Hannon G. J. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Thakker D. R.; Hoyer D.; Cryan J. F. (2006) Interfering with the brain: Use of RNA interference for understanding the pathophysiology of psychiatric and neurological disorders. Pharmacol. Ther. 109, 413–438. [DOI] [PubMed] [Google Scholar]
- Ohki E. C.; Tilkins M. L.; Ciccarone V. C.; Price P. J. (2001) Improving the transfection efficiency of post-mitotic neurons. J. Neurosci. Methods 112, 95–99. [DOI] [PubMed] [Google Scholar]
- Bergen J. M.; Pun S. H. (2008) Analysis of the intracellular barriers encountered by nonviral gene carriers in a model of spatially controlled delivery to neurons. J. Gene Med. 10, 187–197. [DOI] [PubMed] [Google Scholar]
- Davidson B. L.; Breakefield X. O. (2003) Viral vectors for gene delivery to the nervous system. Nat. Rev. Neurosci. 4, 353–364. [DOI] [PubMed] [Google Scholar]
- Lu J.; Nozumi M.; Fujii H.; Igarashi M. (2008) A novel method for RNA interference in neurons using enhanced green fluorescent protein (EGFP)-transgenic rats. Neurosci. Res. 61, 219–224. [DOI] [PubMed] [Google Scholar]
- Kao S. C.; Krichevsky A. M.; Kosik K. S.; Tsai L. H. (2004) BACE1 suppression by RNA interference in primary cortical neurons. J. Biol. Chem. 279, 1942–1949. [DOI] [PubMed] [Google Scholar]
- Li L.; Prabhakaran K.; Zhang X.; Borowitz J. L.; Isom G. E. (2006) PPAR {alpha}-Mediated Upregulation of Uncoupling Protein-2 Switches Cyanide-Induced Apoptosis to Necrosis in Primary Cortical Cells. Toxicol. Sci. 93, 136. [DOI] [PubMed] [Google Scholar]
- Krichevsky A. M.; Kosik K. S. (2002) RNAi functions in cultured mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 99, 11926–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson T. J.; Harel S.; Arboleda V. A.; Prunell G. F.; Shelanski M. L.; Greene L. A.; Troy C. M. (2004) Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation. J. Neurosci. 24, 10040–10046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-B.; Choi J. S.; Nam K.; Lee M.; Park J.-S.; Lee J.-K. (2006) Enhanced transfection of primary cortical cultures using arginine-grafted PAMAM dendrimer, PAMAM-Arg. J. Controlled Release 114, 110–117. [DOI] [PubMed] [Google Scholar]
- Guo J.; Fisher K. A.; Darcy R.; Cryan J. F.; O’Driscoll C. (2010) Therapeutic targeting in the silent era: advances in non-viral siRNA delivery. Mol. BioSyst. 6, 1143–1161. [DOI] [PubMed] [Google Scholar]
- Mellet C. O.; Fernandez J. M. G.; Benito J. M. (2011) Cyclodextrin-based gene delivery systems. Chem. Soc. Rev. 40, 1586–1608. [DOI] [PubMed] [Google Scholar]
- Méndez-Ardoy A.; Urbiola K.; Aranda C.; Ortiz-Mellet C.; García-Fernández J. M.; Tros de Ilarduya C. (2011) Polycationic amphiphilic cyclodextrin-based nanoparticles for therapeutic gene delivery. Nanomedicine 6, 1697–1707. [DOI] [PubMed] [Google Scholar]
- Cryan S. A.; Holohan A.; Donohue R.; Darcy R.; O’Driscoll C. M. (2004) Cell transfection with polycationic cyclodextrin vectors. Eur. J. Pharm. Sci. 21, 625–633. [DOI] [PubMed] [Google Scholar]
- Cryan S. A.; Donohue R.; Ravo B. J.; Darcy R.; O’Driscoll C. M. (2004) Cationic cyclodextrin amphiphiles as gene delivery vectors. J. Drug Delivery Sci. Technol. 14, 57–62. [Google Scholar]
- Diaz-Moscoso A.; Le Gourrierec L.; Gomez-Garcia M.; Benito J. M.; Balbuena P.; Ortega-Caballero F.; Guilloteau N.; Di Giorgio C.; Vierling P.; Defaye J.; Mellet C. O.; Fernandez J. M. G. (2009) Polycationic Amphiphilic Cyclodextrins for Gene Delivery: Synthesis and Effect of Structural Modifications on Plasmid DNA Complex Stability, Cytotoxicity, and Gene Expression. Chem.—Eur. J. 15, 12871–12888. [DOI] [PubMed] [Google Scholar]
- Ortega-Caballero F.; Mellet C. O.; Le Gourrierec L.; Guilloteau N.; Di Giorgio C.; Vierling P.; Defaye J.; Fernandez J. M. G. (2008) Tailoring beta-Cyclodextrin for DNA Complexation and Delivery by Homogeneous Functionalization at the Secondary Face. Org. Lett. 10, 5143–5146. [DOI] [PubMed] [Google Scholar]
- Méndez-Ardoy A.; Guilloteau N.; Di Giorgio C.; Vierling P.; Santoyo-González F.; Ortiz Mellet C.; García Fernández J. M. (2011) β-Cyclodextrin-Based Polycationic Amphiphilic “Click” Clusters: Effect of Structural Modifications in Their DNA Complexing and Delivery Properties. J. Org. Chem. 76, 5882–5894. [DOI] [PubMed] [Google Scholar]
- McMahon A.; Gomez E.; Donohue R.; Forde D.; Darcy R.; O’Driscoll C. M. (2008) Cyclodextrin gene vectors: cell trafficking and the influence of lipophilic chain length. J. Drug Delivery Sci. Technol. 18, 303–307. [Google Scholar]
- McMahon A.; O’Neill M. J.; Gomez E.; Donohue R.; Forde D.; Darcy R.; O’ Driscoll C. M. (2012) Targeted gene delivery to hepatocytes with galactosylated amphiphilic cyclodextrins. J Pharm. Pharmacol. 54, 1063–1073. [DOI] [PubMed] [Google Scholar]
- O’ Neill M. J.; Guo J.; Byrne C.; Darcy R.; O’ Driscoll C. M. (2011) Mechanistic studies on the uptake and intracellular trafficking of novel cyclodextrin transfection complexes by intestinal epithelial cells. Int. J. Pharm. 413, 174–183. [DOI] [PubMed] [Google Scholar]
- O’Mahony A. M.; Ogier J.; Desgranges S.; Cryan J. F.; Darcy R.; O’ Driscoll C. M. (2012) A click chemistry route to 2-functionalised PEGylated and cationic β-cyclodextrins: co-formulation opportunities for siRNA delivery. Org. Biomol. Chem. 10, 4954–4960. [DOI] [PubMed] [Google Scholar]
- Donohue R.; Mazzaglia A.; Ravoo B. J.; Darcy R. (2002) Cationic beta-cyclodextrin bilayer vesicles. Chem. Commun. 2864–2865. [DOI] [PubMed] [Google Scholar]
- Srinivasachari S.; Fichter K. M.; Reineke T. M. (2008) Polycationic β-Cyclodextrin “Click Clusters”: Monodisperse and Versatile Scaffolds for Nucleic Acid Delivery. J. Am. Chem. Soc. 130, 4618–4627. [DOI] [PubMed] [Google Scholar]
- Srinivasachari S.; Reineke T. M. (2009) Versatile supramolecular pDNA vehicles via ″click polymerization″ of beta-cyclodextrin with oligoethyleneamines. Biomaterials 30, 928–938. [DOI] [PubMed] [Google Scholar]
- Bergen J. M.; Park I. K.; Horner P. J.; Pun S. H. (2008) Nonviral approaches for neuronal delivery of nucleic acids. Pharm. Res. 25, 983–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifediba M. A.; Medarova Z.; Ng S. W.; Yang J. Z.; Moore A. (2010) siRNA Delivery to CNS Cells using a Membrane Translocation Peptide. Bioconjugate Chem. 21, 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzzo D.; Privitera L.; Fa M.; Staniszewski A.; Hashimoto G.; Aziz F.; Sakurai M.; Ribe E. M.; Troy C. M.; Mercken M.; Jung S. S.; Palmeri A.; Arancio O. (2011) Endogenous amyloid-β is necessary for hippocampal synaptic plasticity and memory. Ann. Neurol. 69, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby B.; Cates S.; Harris A.; Ohki E. C.; Tilkins M. L.; Price P. J.; Ciccarone V. C. (2004) Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods 33, 95–103. [DOI] [PubMed] [Google Scholar]
- Rejman J.; Oberle V.; Zuhorn I. S.; Hoekstra D. (2004) Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J. 377, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resina S.; Prevot P.; Thierry A. R. (2009) Physico-Chemical Characteristics of Lipoplexes Influence Cell Uptake Mechanisms and Transfection Efficacy. Plos One 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulford B.; Reim N.; Bell A.; Veatch J.; Forster G.; Bender H.; Meyerett C.; Hafeman S.; Michel B.; Johnson T.; Wyckoff A. C.; Miele G.; Julius C.; Kranich J.; Schenkel A.; Dow S.; Zabel M. D. (2010) Liposome-siRNA-Peptide Complexes Cross the Blood-Brain Barrier and Significantly Decrease PrPC on Neuronal Cells and PrPRES in Infected Cell Cultures. PLoS One 5, e11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsham D. D.; Cai F.; Cui H.; Smukler S. R.; Salapatek A. M. F.; Shkreta L. (2004) Generation of a phenotypic array of hypothalamic neuronal cell models to study complex neuroendocrine disorders. Endocrinology 145, 393–400. [DOI] [PubMed] [Google Scholar]
- Wong Y. Y.; Markham K.; Xu Z. P.; Chen M.; Lu G. Q.; Bartlett P. F.; Cooper H. M. (2010) Efficient delivery of siRNA to cortical neurons using layered double hydroxide nanoparticles. Biomaterials 31, 8770–8779. [DOI] [PubMed] [Google Scholar]
- Kim I. D.; Lim C. M.; Kim J. B.; Nam H. Y.; Nam K.; Kim S. W.; Park J. S.; Lee J. K. (2010) Neuroprotection by biodegradable PAMAM ester (e-PAM-R)-mediated HMGB1 siRNA delivery in primary cortical cultures and in the postischemic brain. J. Controlled Release 142, 422–430. [DOI] [PubMed] [Google Scholar]
- Tonges L.; Lingor P.; Egle R.; Dietz G. P. H.; Fahr A.; Bahr M. (2006) Stearylated octaarginine and artificial virus-like particles for transfection of siRNA into primary rat neurons. RNA 12, 1431–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S.; Webster P.; Davis M. E. (2004) PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur. J. Cell Biol. 83, 97. [DOI] [PubMed] [Google Scholar]
- Ogris M.; Steinlein P.; Kursa M.; Mechtler K.; Kircheis R.; Wagner E. (1998) The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 5, 1425–1433. [DOI] [PubMed] [Google Scholar]
- Brown R.; Imran S. A.; Belsham D. D.; Ur E.; Wilkinson M. (2007) Adipokine gene expression in a novel hypothalamic neuronal cell line: Resistin-dependent regulation of fasting-induced adipose factor and SOCS-3. Neuroendocrinology 85, 232–241. [DOI] [PubMed] [Google Scholar]
- Brown R. E.; Wilkinson P. M. H.; Imran S. A.; Wilkinson M. (2009) Resistin differentially modulates neuropeptide gene expression and AMP-activated protein kinase activity in N-1 hypothalamic neurons. Brain Res. 1294, 52–60. [DOI] [PubMed] [Google Scholar]
- Brown R.; Imran S. A.; Ur E.; Wilkinson M. (2008) Valproic acid and CEBP alpha-mediated regulation of adipokine gene expression in hypothalamic neurons and 3T3-L1 adipocytes. Neuroendocrinology 88, 25–34. [DOI] [PubMed] [Google Scholar]
- Fendt M.; Schmid S.; Thakker D. R.; Jacobson L. H.; Yamamoto R.; Mitsukawa K.; Maier R.; Natt F.; Husken D.; Kelly P. H.; McAllister K. H.; Hoyer D.; van der Putten H.; Cryan J. F.; Flor P. J. (2007) mGluR7 facilitates extinction of aversive memories and controls amygdala plasticity. Mol Psychiatry 13, 970–979. [DOI] [PubMed] [Google Scholar]
- Salahpour A.; Medvedev I. O.; Beaulieu J. M.; Gainetdinov R. R.; Caron M. G. (2007) Local knockdown of genes in the brain using small interfering RNA: A phenotypic comparison with knockout animals. Biol. Psychiatry 61, 65–69. [DOI] [PubMed] [Google Scholar]
- Thakker D. R.; Natt F.; Husken D.; van der Putten H.; Maier R.; Hoyer D.; Cryan J. F. (2005) siRNA-mediated knockdown of the serotonin transporter in the adult mouse brain. Mol. Psychiatry 10, 782–789. [DOI] [PubMed] [Google Scholar]
- Guo J.; Cheng W. P.; Gu J.; Ding C.; Qu X.; Yang Z.; O’Driscoll C. (2012) Systemic delivery of therapeutic small interfering RNA using a pH-triggered amphiphilic poly-l-lysine nanocarrier to suppress prostate cancer growth in mice. Eur. J. Pharm. Sci. 45, 521–532. [DOI] [PubMed] [Google Scholar]
- Tsutsumi T.; Hirayama F.; Uekama K.; Arima H. (2008) Potential use of polyamidoamine dendrimer/alpha-cyclodextrin conjugate (generation 3, G3) as a novel carrier for short hairpin RNA-expressing plasmid DNA. J. Pharm. Sci. 97, 3022–3034. [DOI] [PubMed] [Google Scholar]
- Katas H.; Alpar H. O. (2006) Development and characterisation of chitosan nanoparticles for siRNA delivery. J. Controlled Release 115, 216–225. [DOI] [PubMed] [Google Scholar]
- Cardoso A. L. C.; Simoes S.; de Almeida L. P.; Plesnila N.; de Lima M. C. P.; Wagner E.; Culmsee C. (2008) Tf-lipoplexes for neuronal siRNA delivery: A promising system to mediate gene silencing in the CNS. J. Controlled Release 132, 113–123. [DOI] [PubMed] [Google Scholar]
- Dey D.; Inayathullah M.; Lee A. S.; LeMieux M. C.; Zhang X.; Wu Y.; Nag D.; De Almeida P. E.; Han L.; Rajadas J.; Wu J. C. (2011) Efficient gene delivery of primary human cells using peptide linked polyethylenimine polymer hybrid. Biomaterials 32, 4647–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P.; Wu H. Q.; McBride J. L.; Jung K. E.; Kim M. H.; Davidson B. L.; Lee S. K.; Shankar P.; Manjunath N. (2007) Transvascular delivery of small interfering RNA to the central nervous system. Nature 448, 39–U32. [DOI] [PubMed] [Google Scholar]
- Suk J. S.; Suh J.; Choy K.; Lai S. K.; Fu J.; Hanes J. (2006) Gene delivery to differentiated neurotypic cells with RGD and HIV Tat peptide functionalized polymeric nanoparticles. Biomaterials 27, 5143–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi K.; Tokunaga K.; Hohjoh H. (2004) Long-lasting RNAi activity in mammalian neurons. FEBS Lett. 558, 89–95. [DOI] [PubMed] [Google Scholar]
- Cardoso A. L. C.; Simoes S.; de Almeida L. P.; Pelisek J.; Culmsee C.; Wagner E.; de Lima M. C. P. (2007) siRNA delivery by a transferrin-associated lipid-based vector: a non-viral strategy to mediate gene silencing. J. Gene Med. 9, 170–183. [DOI] [PubMed] [Google Scholar]
- Crombez L.; Aldrian-Herrada G.; Konate K.; Nguyen Q. N.; McMaster G. K.; Brasseur R.; Heitz F.; Divita G. (2009) A New Potent Secondary Amphipathic Cell-penetrating Peptide for siRNA Delivery Into Mammalian Cells. Mol. Ther. 17, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y.; Kurihara R.; Tsuchida A.; Hasegawa M.; Nagashima T.; Mori T.; Niidome T.; Katayama Y.; Okitsu O. (2008) Efficient delivery of siRNA using dendritic poly(L-lysine) for loss-of-function analysis. J. Controlled Release 126, 59–66. [DOI] [PubMed] [Google Scholar]
- Simen B. B.; Duman C. H.; Simen A. A.; Duman R. S. (2006) TNF alpha signaling in depression and anxiety: Behavioral consequences of individual receptor targeting. Biol. Psychiatry 59, 775–785. [DOI] [PubMed] [Google Scholar]