Abstract
KIR2DL1 alleles were identified by DNA sequencing of the coding region from amplified genomic DNA from 100 random African Americans. The majority of individuals (97%) carried a KIR2DL1 locus. Allele KIR2DL1*00302 was found in 68% of individuals but KIR2DL1*00401, *002, *00303, *006, and *007 were also frequent. Eleven new alleles were described: KIR2DL1*00403, *01101,*01102 *012, *013N, *014, *015, *016, *017,*018, and *019. Nine of the novel alleles encoded amino acid substitutions located throughout the receptor; one allele carried a stop codon in the exon encoding the first extracellular domain.
Introduction
The natural killer (NK) cell receptor KIR2DL1 is a cell surface inhibitory receptor that recognizes a subset of HLA-C allelic products carrying Lys80 (1,2). Recognition of its ligand results in signals that prevent the NK cell from responding to normal cells. Loss of its HLA ligand on virally infected or malignant cells removes the inhibitory signal and may result in cytotoxicity or in cytokine production (3). The gene encoding KIR2DL1 is located in a cluster of similar genes in the Leukocyte Receptor Complex (4) although some KIR haplotypes lack the KIR2DL1 gene entirely (5). Fifteen alleles encoding ten variant receptors have been identified for KIR2DL1 (6,7). Studies of a similar receptor, KIR3DL1, have demonstrated that allelic variants may differ in their level of expression at the cell surface and ligand binding affinity (8). The purpose of this study was to evaluate KIR2DL1 allelic diversity in a population of random African American individuals.
Materials and Methods
Genomic DNA was isolated from 100 unique and unrelated African Americans from the human variation panel obtained from the National Institute of General Medical Sciences (NIGMS) Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/nigms/) using a QIAamp® DNA Blood Mini Kit (Qiagen, Valencia, CA). Testing for the presence or absence of KIR2DL1 used polymerase chain reaction (PCR) primers and reaction conditions previously described (9,10,11). A primer pair, described by Gomez-Lozano et al., specific for KIR2DS1 and KIR2DL1 *004, *007, and *010 was a useful control (12). For sequencing of exons 1 through 9, the strategy based on PCR amplification of genomic DNA followed by DNA sequence analysis has been described (13) with the following exception: The antisense PCR primer used to obtain the amplicon including exon 4 through exon 9 was 2DL1-E89R (TGTGAGGAAGATCGATGCCCTAAG). The latter, described by Murdoch et al (14), was useful to identify a 3’untranslated region variation distinguishing KIR2DL1*0040101 from *0040102. Allele assignments were obtained by comparison with a KIR2DL1 cDNA reference library from ImmunoPolymorphismDatabase (IPD)-KIR Release 2.1.0 (6). In this report, the numbering of nucleotides and codons is based on IPD-KIR unless noted. Novel alleles were isolated using allele specific amplification, cloning and/or by using haplotype specific extraction (2DL1-435G; HaploPrep, Qiagen, Valencia, CA). Sequences were submitted to GenBank and novel allele designations were assigned by the KIR subcommittee of the World Health Organization Nomenclature Committee for Factors of the HLA System (7). Confirmatory sequences of previously described alleles were submitted to the IPD-KIR database and include: KIR2DL1*00303 (Cell GM17132; GenBank GQ406045), KIR2DL1*0040102 (GM17116; GQ844297); KIR2DL1*006 (GM17177; GQ406047), KIR2DL1*007 (GM17178; GQ406043), and KIR2DL1*010 (GM17108, GQ406044). The first four alleles had been previously identified in AfroCaribbean or African American individuals (15,13); the last allele in an individual of European ancestry (14). A phylogenetic tree of KIR2DL1 was constructed using a maximum likelihood technique with a molecular clock (DNAMLK version 3.66). Since the 5’ sequence is unknown for some alleles, the tree was based on the nucleotide sequence from codon -17 through codon 327 (16).
HLA allele identification
To identify the HLA-A,-B,-C alleles carried by each individual, PCR primers were used to amplify each locus as previously described (17). Applied Biosystems Big Dye terminator chemistry and an Applied Biosystems Models 3730xl DNA analyzer (PE Applied Biosystems, Foster City, CA) were used to obtain the sequences of both strands of exons 2 and 3. IMGT/HLA database 2.21.0 was used for the interpretation of sequencing results. DRB1 alleles were amplified and identified using the One Lambda LABType® SSO HD Kit (version RSSOH2B1_002, One Lambda, Canoga Park, California) following manufacturer’s protocols. Alleles identical in exons 2 and 3 (class I) or exon 2 (DRB1) were not resolved. For those class I samples yielding alternative genotypes, either allele specific sequencing primers, allele specific PCR amplification, or HaploPrep (Qiagen, Valencia, CA) was used to link polymorphisms and to identify the specific allele combination. [In-house primer sequences used for all loci are available at www.dodmarrow.org.] Supplemental Table 1 provides the HLA data.
Results and Discussion
Three individuals (3%) showed a complete absence of the KIR2DL1; another study observed 2% of African Americans negative for this locus (18). Forty two individuals carried a single KIR2DL1 coding sequence and 55 were heterozygous. Of the 13 previously described alleles that differ within the coding region of the gene, eight were observed (Table 1). KIR2DL1*00302 was the most common allele being found in 68 individuals. Two other frequent alleles included KIR2DL1*00401 (23 individuals) and KIR2DL1*002 (13 individuals),. These alleles are the ones most frequently observed in studies of other populations (e.g., Japanese (8), Irish (19)). The G to A transition in the 3’ untranslated region of KIR2DL1*00401 at nucleotide 34 following the stop codon which distinguishes KIR2DL1*0040101 from *0040102 was observed in this population. We identified the G variant (KIR2DL1*0040101) in 14 individuals and the A variant (KIR2DL1*0040102) in nine individuals; one individual carried both alleles. The A variant was initially observed in an Afro-Carribean population (15). Other alleles observed in multiple individuals, KIR2DL1*006 (11 individuals), KIR2DL1*00303 (8 individuals) and KIR2DL1*007 (7 individuals), have been previously observed only in populations of African origin (15,19,13).
Table 1.
Frequency of KIR2DL1 Alleles in an African American Panel (n=100)
| KIR2DL1 Allele | Number Of Individuals Positive (%) |
KIR2DL1 Allele | Number Of Individuals Positive (%) |
|---|---|---|---|
| *001 | 2 (2) | *008 | 0 |
| *002 | 13 (13) | *009 | 0 |
| *00301 | 0 | *010 | 1 (1) |
| *00302a | 68 (68) | *01101b | 1 (1) |
| *00303 | 8 (8) | *01102b | 2 (2) |
| *0040101 | 14 (14) | *012b | 5 (5) |
| *0040102 | 9 (9) | *013Nb | 1 (1) |
| *00402 | 0 | *014b | 3 (3) |
| *00403b | 1 (1) | *015b | 1 (1) |
| *005 | 0 | *016b | 1 (1) |
| *006 | 11 (11) | *017b | 1 (1) |
| *007 | 7 (7) | *018 b | 1 (1) |
| *019b | 2 (2) |
KIR2DL1*0030201 and KIR2DL1*0030202 were not individually distinguished in this study.
Novel allele described in this study.
Eleven novel alleles were characterized. Four of the novel alleles were found in multiple individuals: KIR2DL1*01102 (2 individuals), KIR2DL1*012 (5 individuals), KIR2DL1*014 (3 individuals), and KIR2DL1*019 (2 individuals). One allele carried a synonymous substitution of the common allele KIR2DL1*00401 creating KIR2DL1*00403. Five novel alleles, KIR2DL1*01101, KIR2DL1*01102, KIR2DL1*012, KIR2DL1*013N, and KIR2DL1*019 (codon 114), carried substitutions that were found in other alleles at the locus. For example, KIR2DL1*012 carried a substitution altering the first domain at codon 16 altering a proline to an arginine. Twelve of the 15 KIR2DL1 alleles encode arginine; the other three encode proline at this position. Five novel alleles introduced polymorphism at codons which were previously conserved in all or the majority of alleles altering the amino acid: KIR2DL1*014, KIR2DL1*015, KIR2DL1*016, KIR2DL1*017, KIR2DL1*018, KIR2DL1*019 (codon 226). Four novel alleles, KIR2DL1*01101, KIR2DL1*01102, KIR2DL1*013N, and KIR2DL1*019, exhibited two substitutions. KIR2DL1*013N is interesting because the second substitution in the exon encoding the first extracellular domain creates a termination codon. Other truncated alleles previously described carry termination codons in the exon encoding the second extracellular domain: KIR2DS3*003N (substitution), KIR3DL1*024N (deletion of nucleotide), and KIR3DS1*049N (deletion of nucleotide) (20,21).
The majority of the new alleles described carried nonsynonymous substitutions. The substitutions altered the amino acid sequence of the signal peptide (altered in 2 alleles), domain 1 (1 allele), domain 2 (2 alleles), transmembrane region (2 alleles) and cytoplasmic tail (3 alleles). Only one of the substitutions in the two external domains of KIR introduced a novel amino acid; the other two are observed in other alleles. Based on the crystal structure, none of the extracellular variations appear to impact residues that contact the HLA-C/peptide complex (2). In the cytoplasmic tail, KIR2DL1*018 (CCA/Pro) has a substitution at codon 282 that was conserved in most alleles (ACA/Thr). This residue falls within the membrane proximal immunoreceptor inhibitory motif (ITIM) motif, adjacent to the tyrosine which is phosphorylated during signaling (22,23). While there is some latitude in amino acids at this position in ITIM motifs, the substitution of a cyclic amino acid (Pro) with properties that affect secondary protein stucture may impact signaling for this form of the receptor.
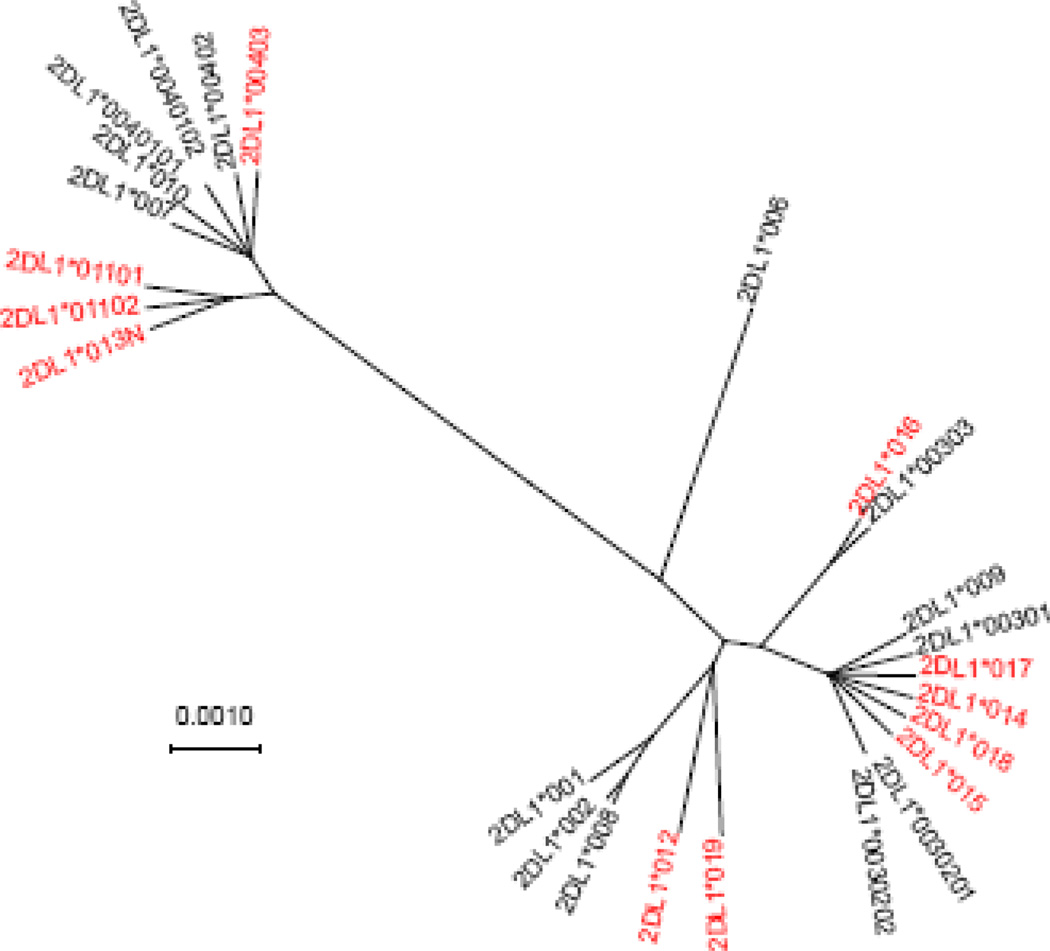
Figure 1 shows a phylogenetic tree showing the relationships among all of the reported KIR2DL1 alleles. There are two major clusters of alleles, each containing frequent alleles in the African American population. The novel alleles found in African Americans are found in both of these clusters. The number of KIR2DL1 alleles observed in this population of 100 individuals is remarkable; 19 alleles with 68% of individuals carrying KIR2DL1*00302. In contrast, studies of European American (n=75) or Northern Irish (n=140) populations observed 7 alleles with KIR2DL1*00302 being found in 63–66% of individuals (13,19).
Figure 1.
Phylogenetic tree of KIR2DL1 alleles. Alleles shown in red were described in this study. Previously described KIR2DL1*005 was not included because it is missing a portion of the nucleotide sequence encoding the signal peptide.
Identification of HLA-C alleles in the 97 KIR2DL1 positive African Americans showed that 25 individuals (26%) did not carry an HLA-C allele that would serve as a ligand for KIR2DL1, that is, an HLA-C allelic product with Lys80 (Group 2). In these individuals without the cognate ligand, NK cells expressing, as their only inhibitory KIR receptor, KIR2DL1, would not be “licensed” and are likely to be hyporeactive (24).
Supplementary Material
Table 2.
Novel KIR2DL1 alleles observed in African Americans
|
KIR2DL1 Novel Allele |
Most Similar Allele |
Alteration Nucleotide Codon (Amino Acid)a |
Protein Region Altered |
GenBank Accession Number |
Cell Line |
|---|---|---|---|---|---|
| 2DL1*00403 | 2DL1*00401 | 254 GCG (A ) => GCT (A) | -- | EU930369 | GM17109 |
| 2DL1*01101 | 2DL1*00401 | -17 GTC (V) => TTC (F), 221 CGA (R) => AGA (R) | Signal peptide | EU930368 | GM17159 |
| 2DL1*01102 | 2DL1*00401 | -17 GTC (V) => TTC (F), 186 TAC (Y) => TAT (Y) | Signal peptide | FJ655851 | GM17200 b |
| 2DL1*012 | 2DL1*001 | 16 CCC (P) => CGC (R) | Domain 1 | EU930370 | GM17119b |
| 2DL1*013N | 2DL1*00401 | -17 GTC (V) => TTC (F), 35 GAA (E) => TAA (Stop) | Truncated | EU930371 | GM17120 |
| 2DL1*014 | 2DL1*00302 | 179 GGC (G) => AGC (S) | Domain 2 | EU930372 | GM17123 b |
| 2DL1*015 | 2DL1*00302 | 275 GAC (D) => GAA (E) | Cytoplasmic | EU930373 | GM17124 |
| 2DL1*016 | 2DL1*00303 | 296 CGC (R) => TGC (C) | Cytoplasmic | FJ655852 | GM17126 |
| 2DL1*017 | 2DL1*00302 | 221 CGA (R) => CAA (Q) | Transmembrane | FJ655853 | GM17131 |
| 2DL1*018 | 2DL1*00302 | 282 ACA (T) => CCA (P) | Cytoplasmic | FJ655854 | GM17153 |
| 2DL1*019 | 2DL1*00303 | 114 CTG (L) => CCG (P), 226 CTG (L) => GTG (V) | Domain 2 Transmembrane |
FJ655855 | GM17160 b |
Most similar known allele => novel allele
This allele was found in more than one individual: KIR2DL1*01102, GM17198; KIR2DL1*012, GM17141, GM17142, GM17166, GM17185; KIR2DL1*014, GM17140, GM17174; KIR2LD1*019, GM17102.
Acknowledgments
This research is supported by funding from the Office of Naval Research N00014-07-2108. The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Navy, the Department of Defense, or the U.S. government. The phylogetic tree was created by Dr. Dongying Wu. We would like to thank him.
References
- 1.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp.Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DLI-HLA-Cw4 complex. Nature Immunology. 2001;2:452–460. doi: 10.1038/87766. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK cell recognition. Annu.Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 5.Hsu KC, Chida S, Dupont B, Geraghty DE. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunological Reviews. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 6.Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2005;33:D523–D526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh SGE, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Tissue Antigens. 2003;62:79–86. doi: 10.1034/j.1399-0039.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 8.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp.Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou L, Steiner NK, Chen M, Belle I, Kubit AL, Ng J, Hurley CK. Limited Allelic Diversity of Stimulatory Two Domain Killer Immunoglobulin-Like Receptors. Human Immunology. 2008;69:174–178. doi: 10.1016/j.humimm.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 11.Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: An update. Tissue Antigens. 2002;59:184–193. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 13.Hou LH, Steiner NK, Chen M, Belle I, Ng J, Hurley CK. KIR2DL1 allelic diversity: four new alleles characterized in a bone marrow transplant population and three families. Tissue Antigens. 2007;69:250–254. doi: 10.1111/j.1399-0039.2006.00793.x. [DOI] [PubMed] [Google Scholar]
- 14.Murdoch S, Seoud M, Kircheisen R, Mazhar B, Slim R. Detailed gene and allele content analysis of three homozygous KIR haplotypes. Tissue Antigens. 2006;68:72–77. doi: 10.1111/j.1399-0039.2006.00606.x. [DOI] [PubMed] [Google Scholar]
- 15.Artavanis-Tsakonas K, Eleme K, McQueen KL, Cheng NW, Parham P, Davis DM, Riley EM. Activation of a subset of human NK cells upon contact with Plasmodium falciparum-infected erythrocytes. J Immunol. 2003;171:5396–5405. doi: 10.4049/jimmunol.171.10.5396. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 17.Tu B, Mack SJ, Lazaro A, Lancaster A, Thomson G, Cao K, Chen M, Ling G, Hartzman R, Ng J, Hurley CK. HLA-A, -B, -C, -DRB1 allele and haplotype frequencies in an African American population. Tissue Antigens. 2007;69:73–85. doi: 10.1111/j.1399-0039.2006.00728.x. [DOI] [PubMed] [Google Scholar]
- 18.Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007;59:1–15. doi: 10.1007/s00251-006-0168-4. [DOI] [PubMed] [Google Scholar]
- 19.Meenagh A, Gonzalez A, Sleator C, McQuaid S, Middleton D. Investigation of killer cell immunoglobulin-like receptor gene diversity, KIR2DL1 and KIR2DS1. Tissue Antigens. 2008;72:383–391. doi: 10.1111/j.1399-0039.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- 20.Luo L, Du Z, Sharma SK, Cullen R, Spellman S, Reed EF, Rajalingam R. Chainterminating natural mutations affect the function of activating KIR receptors 3DS1 and 2DS3. Immunogenetics. 2007;59:779–792. doi: 10.1007/s00251-007-0239-1. [DOI] [PubMed] [Google Scholar]
- 21.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 22.Burshtyn DN, Scharenberg AM, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet JP, Long EO. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravetch JV, Lanier LL. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 24.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.