Abstract
Equine influenza A (H3N8) virus is a leading cause of infectious respiratory disease in horses causing widespread morbidity and economic losses. As with influenza in other species, equine influenza strains continuously mutate, requiring constant re-evaluation of current vaccines and development of new vaccines. Current inactivated (killed) vaccines, while efficacious, only offer limited protection against multiple strains and require frequent boosts. Ongoing research into new vaccine technologies, including gene-based vaccines, aims to increase the neutralization potency, breadth, and duration of protective immunity of new or existing vaccines. In these hypothesis-generating experiments, we demonstrate that a DNA vaccine expressing the hemagglutinin protein of equine H3N8 influenza virus generates homologous and heterologous immune responses, and protects against clinical disease and viral replication following homologous H3N8 infection in horses. Furthermore, we demonstrate that a needle-free delivery device is as efficient and effective as conventional parenteral injection using a needle and syringe. The observed trends in this study drive the hypothesis that DNA vaccines offer a safe, effective, and promising alternative approach for veterinary vaccines against influenza, and applicable to combat equine influenza.
Keywords: Influenza, Equine, DNA Vaccine, Immunogenicity, Protection
1. Introduction
Equine influenza (EI) is considered one of the most important and common infectious respiratory disease of equids (11–13, 60). As in other species, EI is a difficult target for vaccination due to continuous and frequent viral mutations, requiring constant evaluation and development of new vaccines. Yet despite intensive vaccination efforts, transmission and outbreaks of equine influenza still occur (44), causing major economic losses to the horse industry and posing a serious threat to equine welfare by causing high morbidity and periodically, mortality in infected horses.
While current inactivated whole virus vaccines are efficacious in inducing robust antibody responses, they are limited in their ability to induce broad-based immune responses and do not always fully protect against infection (37, 41, 53). In addition, these vaccines require egg-based production methods which are inefficient and require large bio containment facilities. Research has focused on exploring alternative vaccine technologies that can confer broad, long-lived protection against infection (44), with more efficient production methods (64). Recent studies have utilized modified live viruses (MLV)(8,9, 35, 57), pox virus vectors (5, 45, 54), DNA technology (35), and adjuvants (21, 55) to develop improved vaccines, some of which are now in commercial use.
MLV vaccines have been shown to be protective and immunogenic, but carry the risk of reversion to virulence and other complications, especially in animals that are immune-compromised or pregnant (53). Alternatively, licensed canarypox-vectored vaccines may be a valuable alternative to older MLV technologies, as previous studies have demonstrated full and durable protection from viral shedding (17, 36, 54). However, each of these require the use of live viruses which require advanced bio containment facilities and bio safety measures. DNA vaccines are a logical alternative since they are safer to produce, and may provide more complete clinical protection than pox-vectored vaccines (53). Also, DNA vaccines can be tailored quickly and specifically to respond to outbreaks of novel strains, while encoding multiple immunogens to elicit broad-based protection. While there are currently no licensed DNA vaccines for use against EI in the horse, a DNA vaccine has been licensed for use against West Nile Virus (58). In other species, DNA vaccines have been licensed for use against infectious hematopoietic necrosis virus in salmon, as well as melanoma in dogs (28).
In the current study, our primary goal was to conduct a series of hypothesis-generating experiments to develop and test novel EI DNA vaccines encoding the viral HA in the equine influenza challenge model. We have previously demonstrated the safety and immunogenicity of DNA vaccines in nonhuman primates and human clinical trials (29, 52, 62). We have also established efficacy, safety, and protection of DNA vaccines against H5N1 influenza in mice and chickens (48), as well as classic and pandemic H1N1 influenza in pigs (20). Here, we test the immunogenicity of monovalent and trivalent CMV/R-HA DNA vaccine constructs for EI in mice, and assess immune responses and protection against H3N8 challenge in ponies. The experimental schema and animal numbers used in this experiment are consistent with previous publications (9, 21, 59).
We have previously shown efficacy and efficiency of novel vaccination methods in various animal models, particularly needle-free delivery (20, 48, 56). Needle-free delivery offers the potential to enhance immune responses to DNA vaccination (16) and could facilitate wide-scale administration of vaccines by eliminating the need for handling and disposal of sharps. Here, we evaluate the efficiency of needle-free vaccine delivery in horses, as this method is ideally suited to administer multiple doses to large animals in farm settings.
2. Materials and Methods
2.1 Immunogen and plasmid construction
Plasmids encoding HA from influenza A/equine/Ohio/1/2003 (H3N8) (GenBank #ABA39846), influenza A/equine/Bari/2005 (H3N8) (GenBank #ABM47075), and influenza A/equine/Newmarket/2/93 (H3N8) (GenBank #X85088) were synthesized by GeneArt (Regensburg, Germany). Amino acids 1–345 (HA1 subunit) of influenza A/equine/Aboyne/1/2005 (H3N8) HA were fused with amino acids 346–565 (HA2 subunit) of influenza A/equine/Hong Kong/1/1992 (H3N8) to generate full-length HA gene as the conserved portion of HA2 of A/equine/Aboyne/1/2005 is not available in the NCBI protein database. All HA genes were synthesized using mammalian preferred codons as described (27) and cloned into CMV/R expression vector (3) for efficient expression in mammalian cells. Hereafter the virus strains are referred to as Ohio/03, Bari/05, and Aboyne/05.
2.2 Serum neutralization test (SNT)
We utilized a 50% Tissue Culture Infectious Dose (TCID50) reduction assay to determine serum neutralizing antibody titers. Confluent Madin-Darby canine kidney (MDCK) cells, grown in 96-well tissue culture plates, were used for all serum neutralization test (SNT) assays. Sera were serially diluted 2-fold using infection medium (Medium 199 (GIBCO) + 1 µg/ml TPCK Trypsin) and virus added to the diluted sera to yield final concentrations of 200 TCID50 units of virus per well. Serum-virus mixtures were incubated at room temperature for 60 min and then were added to washed MDCK cells, with each serum dilution plated in quadruplicate. Media controls (no virus), and virus controls (no serum) were included on each plate. Plates were incubated at 37°C for 2 hr, then washed twice with PBS, filled with infection medium, and incubated for a further 48 hr. Results were read by both HA assay of supernatant from each well and by staining wells with crystal violet to visualize lysis of the monolayers. SNT titers were calculated as 50% endpoints (50) for the greatest serum dilution giving complete inhibition of virus growth.
2.3 Mouse Immunogenicity Studies
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH (Bethesda, MD), in accordance with the Guide and for the Care and Use of Laboratory Animals. Groups of five mice were immunized intramuscularly with a total of 20µg CMV/R DNA plasmids expressing Ohio/03 HA, Bari/05 HA, and/or Newmarket/2/93 HA in a monovalent or multivalent vaccine. Mice were immunized three times at 2 week intervals. Pooled sera (5 mice/group) collected two weeks after the last immunization were evaluated using the serum neutralization test described above. Serum endpoint dilutions were calculated as the greatest dilution sufficient to completely neutralize 200 TCID50 units of three H3N8 viruses (Ohio/03, Newmarket/2/93 and Canine/KY/06) in 50% of wells. Canine/KY/06 is a related, heterologous H3N8 strain that causes flu in dogs, and was selected to evaluate the potential for cross-protective immune responses. Convalescent serum from an Ohio/03 H3N8 influenza-infected horse was used as the positive control.
2.4 Influenza virus preparation
Ohio/03 challenge virus was originally isolated in embryonated hens’ eggs from a nasopharyngeal swab taken from a clinical case. The allantoic fluids from the second egg passage of this isolate were used for the experimental challenges. Virus content was titrated by 50% Egg Infectious Dose (EID50) or TCID50 assays.
2.5 Experimental Ponies
All animal procedures were approved by the Institutional Animal Care and Use Committees of the University of Kentucky (Lexington, KY) and the Vaccine Research Center, NIAID, NIH (Bethesda, MD). All animal work was carried out in accordance with the EC Directive 86/609/EEC. Animals were maintained from birth at the Veterinary Science Research Farm of the University of Kentucky. Prior to the start of the study, health data and immunologic parameters were determined and ponies of both sexes (1–2 years of age at time of first vaccination) were randomly assigned to experimental groups. Ponies consisted of the following mixed breeds: Shetland blood, Welsh blood, Florida swamp pony blood. All ponies were seronegative for detectable antibody to equine influenza virus and had no history of signs of influenza infection prior to the start of the study. Ponies were housed at 2/stall, in directly adjacent stalls with free airflow between, and pairings were based on which ponies were compatible with each other, with no regard to the vaccine group which was unknown to the veterinarian or staff. During immunizations, ponies were held in individual isolation stalls and were pastured between immunizations. Animals were identified by microchip. At the end of the study, all animals were returned to the university farm for other use.
2.6 Experimental Design
Ponies were assigned to one of 4 groups, consisting of 4 ponies per group. In accordance with the experimental schedule indicated in Figure 1, animals in the first two groups were immunized with 4 mg per dose of monovalent DNA expressing the HA gene of A/Equine/Ohio/03 in 1 ml PBS, either by intramuscular injection using a needle and syringe (Group 1) or needle-free delivery system (PharmaJet®, PharmaJet, Inc., Golden, CO)(Group 2), using spring-powered jet technology to effectively deliver vaccines sub-dermally. Group 3 ponies were immunized with a trivalent DNA mix of three plasmids expressing the HAs of Ohio/03, Bari/05, and Aboyne/05 via the NF device. 1.33mg of each of the 3 DNA components was delivered for a total of 4mg per dose of DNA in 1ml PBS. Group 4 control animals received sham DNA (CMV/R plasmid with no insert) via the PharmaJet® at the same total dose and volume as the experimental groups. All injections were given in the brachiocephalicus/serratus cervicus muscles. For all groups, 0.5mL DNA was administered at two separate injection sites for a total volume of 1mL. Each of the two injection sites were on the same side of the shaved lateral neck approximately 8 cm apart. Each site was monitored for adverse reactions to the vaccine at 24 and 48 hours following immunization.
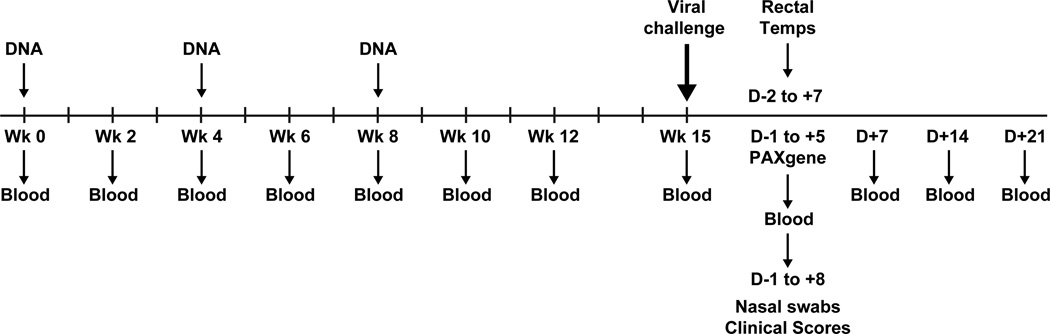
Figure 1. Experimental schema.
Ponies were immunized at weeks 0, 4 and 8 with blood draws in between vaccinations. All animals were challenged at week 15 with Ohio/03 virus. Pre- and post-challenge rectal temperatures, clinical scores, blood, and nasal swabs were collected for assays.
2.7 Serological analysis
As indicated by the schedule in Figure 1, venous blood samples were collected via jugular venipuncture prior to the first vaccination and weekly or bi-weekly thereafter. Other blood samples were drawn on Days -1, 0, 7, 14, and 21 post-challenge for serological analyses. All sera were tested for the presence of antibody to influenza A H3N8 strains Ohio/03 (American lineage/Florida clade 1), Richmond/07 (American lineage/Florida clade 2), and Aboyne/05 (Eurasian lineage) (6, 7) using the single radial hemolysis (SRH) (43, 63) and hemagglutination-inhibition (HI) (43) assays. Richmond/07 was used as the third antigen in these assays since it is antigenically and genetically very similar to the third component of the multivalent DNA vaccine – Influenza/A/eq/Bari/2005 – which was not obtainable at the time of assay. A positive control serum was included in each assay. For SRH analysis, sheep erythrocytes were used and zones of hemolysis were measured after a 20 hr incubation period. All sera were tested for non-specific lysis. SRH antibody levels were expressed as the area of hemolysis (mm2). Seroconversion was defined as an increase in SRH value of >25 mm2 or 50% (42). For HI analysis, all sera were pre-treated with trypsin-periodate as described (4) and assays were done using 2-fold dilutions starting from a 1:10 dilution with viruses at 8 HA units per well and 0.5% chicken erythrocytes.
2.8 Challenge
Seven weeks after the third immunization, vaccinated and control ponies were challenged with wild-type Ohio/03 H3N8 virus as previously described (8) in a large-animal isolation facility. Challenge virus (2.1 × 108 EID50 units in 7 ml allantoic fluid plus 25 ml PBS) was nebulized using a DeVillbis Ultra-Neb 99 nebulizer, and pumped into a 21.5m3 tented stall where it was inhaled by a group of 4–6 ponies for 45 minutes (39, 57). Each such group of ponies included both vaccinates and controls. The dose administered was equivalent to approximately 107 EID50 per cubic meter of tented stall volume. This dose reliably induces typical clinical signs of disease and active virus shedding for 5–8 days post-infection in influenza-naïve horses (9, 39, 57).
2.9 Clinical monitoring
For assessment of clinical protection following experimental challenge with virus, thorough physical examinations and assessments were performed by two experienced, certified veterinarians on all horses daily for 8 days after virus administration. Adverse reactions to immunization were monitored by daily visual inspection of the vaccination site which was circled with ink. Clinical examinations included measurements of rectal temperature, respiratory rate, auscultation of lung sounds, quality/quantity of nasal discharge, palpation of submandibular and parotid lymph nodes, general demeanor, and presentation of nasal discharge or spontaneous coughing. Examinations and clinical scoring were done as previously described by a licensed veterinarian blinded to the vaccination status of the horses (Table 1).
Table 1.
Clinical signs scoring index
| Clinical Sign | Degree | Score |
|---|---|---|
| Coughing | No coughing | 0 |
| Coughed once | 1 | |
| Coughed twice or more | 2 | |
| Nasal discharge | No discharge | 0 |
| Abnormal serous | 1 | |
| Abnormal mucopurulent | 2 | |
| Abnormal profuse | 3 | |
| Respiration | Normal <36/min | 0 |
| Abnormal (dyspnea/tachypnea) >36/min | 1 | |
| Demeanor | No depression | 0 |
| Depression present (lethargy, inappetence) | 1 | |
2.10 Viral shedding and quantitation via RT-PCR and EID50
Nasopharyngeal swabs were collected on the day prior to challenge, and daily for 8 days post challenge. Dacron swabs were inserted 10–15 cm up the nasal meatus, immersed in 1 ml of PBS/5% glycerol/1% antibiotic solution and stored at 4°C until testing. Virus RNA content was also measured by quantitative real-time RT-PCR (qRT-PCR)(33). One step qRT-PCR was performed using in vitro-transcribed (IVT) RNA as a standard. The IVT-RNA concentration was determined mathematically and the genome copy number was calculated (33). Viral shedding was detected by inoculation of 0.1ml undiluted nasal swab sample into each of three embryonated 10-day-old hen eggs followed by 3 days incubation at 35°C. Allantoic fluids were then harvested and tested for detectable virus by hemagglutination (HA) assay, as previously described (43). EID50 was performed in the same manner, except that serial ten-fold dilutions of the nasal swab samples were done using PBS + 1% bovine serum albumin. Each dilution was then injected into four embryonated hens’ eggs (0.1ml/egg) and incubated as before. Allantoic fluids were then tested by HA assay for virus growth and the EID50 titer was calculated as previously described (50).
2.11 Inflammatory cytokine response post-challenge
Venous blood samples were taken on the day prior to challenge, and daily for 6 days post-challenge. These were collected into PAXgene (Qiagen, Valencia, CA) tubes, which contain a stabilizing additive to preserve the RNA expression profile (47). RNA was isolated as per manufacturer’s recommendations and quantified by OD260 and later reverse transcribed into cDNA. Reverse transcription conditions and FAM-labeled primer probes for Granzyme B (GrzB), IFNγ, IL-1β, IL-6, TNF-α and β-GUS were as previously described (9). Equine β-glucuronidase (β-GUS) was used as an endogenous control to normalize for differences in RNA isolation and cDNA synthesis (1, 15, 51). Relative quantitative RT-PCR was carried out on the Applied Biosystems 7900 HT Fast Real-Time PCR System (31) as previously described (9). The 2−ΔΔCT method for analyzing relative gene expression from real-time quantitative PCR experiments was employed for analysis of data (9, 31). The average of the samples taken prior to challenge (Day -1) was chosen as the calibrator sample and results are expressed as relative quantities (RQ). Each sample was tested in duplicate for each of the cytokine targets and the endogenous control.
3. Results
3.1 Mouse Immunogenicity Studies
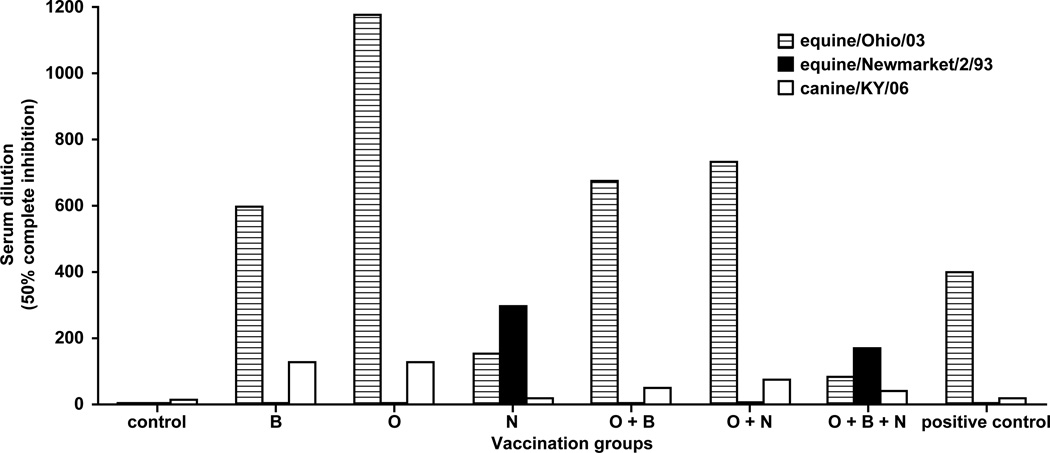
Studies were initially performed in mice to verify the immunogenicity of DNA plasmids against different equine influenza A H3N8 viruses. Microneutralization data showed that mice immunized with DNA expressing Bari/05 or Ohio/03 HA elicited high titer neutralizing antibodies against homologous and in some cases heterologous viruses (Bari/05 HA-immunized tested against Ohio/03), suggesting cross-neutralization (Fig. 2). However, immunization with Newmarket/2/93 HA (either monovalent or trivalent) only elicited detectable neutralization titers against Newmarket/2/93, with the exception of the bivalent group combined with Ohio/03 HA. Since no Bari/05 viruses were available for microneutralization, specific anti-Bari/05 activity was not determined. Interestingly, although Canine/KY/06 HA was not encoded in any vaccine, low levels of cross-reactive neutralizing antibodies against Canine/KY/06 were detected in most groups. These preliminary mouse data indicated that the Ohio/03 and Bari/05 HAs elicited optimal neutralizing activity in mice, prompting their use as the basis for further investigation in the equine influenza model.
Figure 2. Microneutralization titers against H3N8 viruses from mouse studies.
Serum endpoint dilutions calculated to completely neutralize 200 TCID50 units of Ohio/03 (striped bar), Newmarket/2/93 (solid bar) and canine/KY/06 (open bar) viruses in 50% of wells are shown. The x-axis indicates the different vaccination groups. B = Bari/05, O = Ohio/03, N = Newmarket/93. Horse convalescent serum challenged with Ohio/03 was used as the positive control.
3.2.1 Hemagglutination inhibition (HI) responses
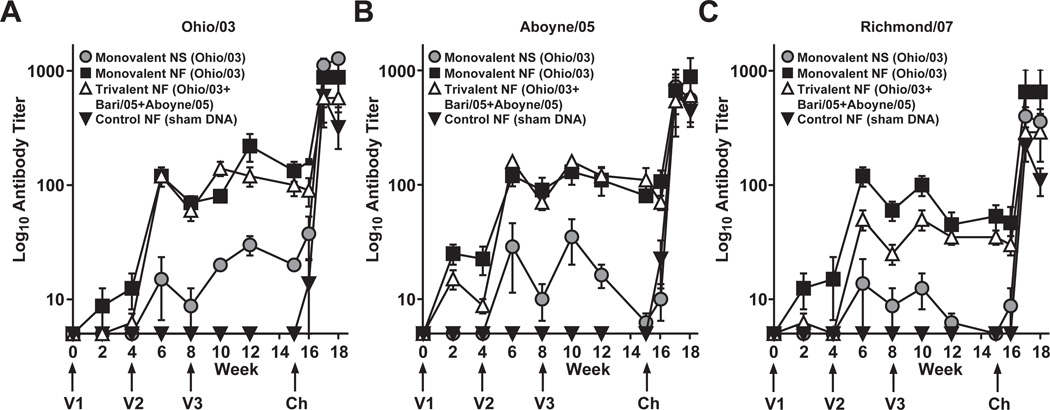
Control ponies remained seronegative for influenza virus by HI throughout the vaccination period. Both monovalent and trivalent needle-free vaccine groups showed robust HI titers against Ohio/03, Aboyne/05, and Richmond/07 antigens after the second immunization, while the monovalent NS group showed moderate titers (Fig. 3 A–C). Additionally, these titers were more stable in the needle-free groups than in the needle/syringe group in the period up to the challenge, and interestingly, appeared to be minimally affected by the second boost. All vaccinated animals showed antibody responses.
Figure 3. HI antibody responses following vaccination and challenge.
Mean HI titers to Ohio/03 (a), Aboyne/05 (b), and Richmond/07 (c) antigen. For graphing purposes, a titer of <10 was assumed to be 5 and is denoted as a value of log10(5), which also serves as the y origin. Error bars represent Standard Error of the Mean (SEM). Ponies received three doses of vaccine (V1, V2 and V3) and were challenged with live equine influenza virus (Ch). Monovalent = Ohio/03 HA, Trivalent = Ohio/03 HA + Aboyne/05 HA + Bari/05 HA, Control = empty CMV/R vector, NS = needle and syringe, NF = needle-free device.
3.2.2 Single radial hemolysis (SRH) responses
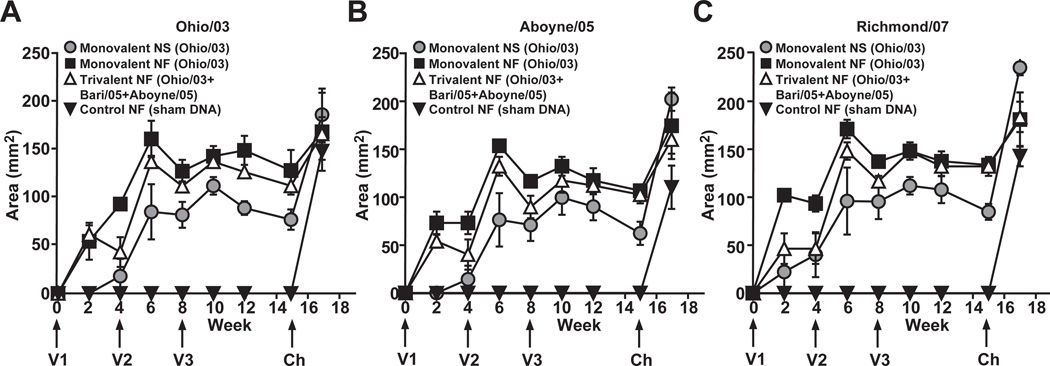
As another method to confirm antibody responses, SRH assays were performed on horse serum. All animals were seronegative by SRH before immunization and controls remained seronegative throughout the vaccination period. Trends similar to the HI assay results were observed in response to Ohio/03, Aboyne/05, and Richmond/07 (Fig. 4 A–C). Homologous and heterologous neutralizing antibody responses were observed following the first immunization, which were amplified by the first boost, and less by the second boost. Like the HI results, all vaccinated animals showed responses, as well as large anamnestic responses following challenge with Ohio/03 virus (Fig. 4).
Figure 4. SRH antibody responses following vaccination and challenge.
Mean SRH antibody in vaccinated horses measured against (a) Ohio/03, (b) Aboyne/05, and (c) Richmond/07. Error bars represent SEM. Ponies received three doses of vaccine (V1, V2 and V3) and were challenged with live equine influenza virus (Ch). Monovalent = Ohio/03 HA, Trivalent = Ohio/03 HA + Aboyne/05 HA + Bari/05 HA, Control = empty CMV/R vector, NS = needle and syringe, NF = needle-free device.
3.3 Clinical responses of ponies to vaccination and challenge
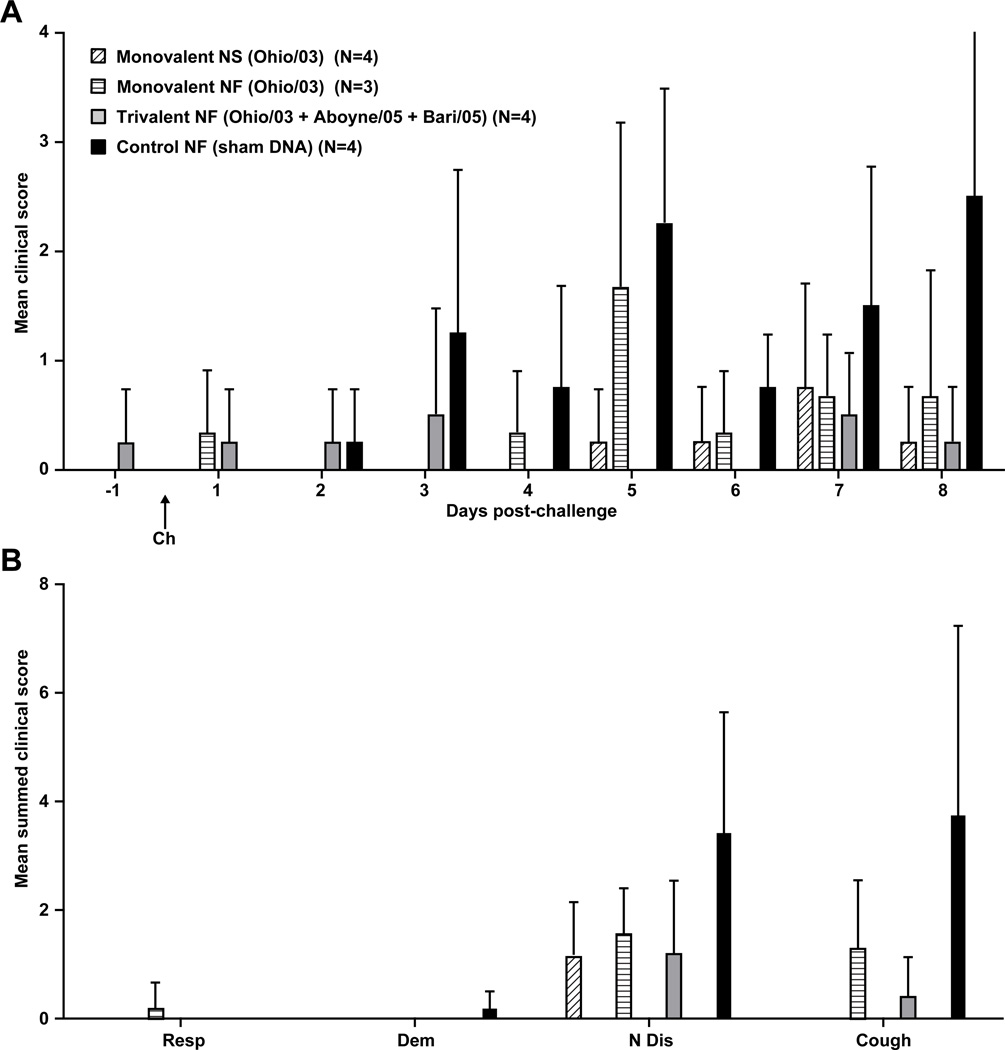
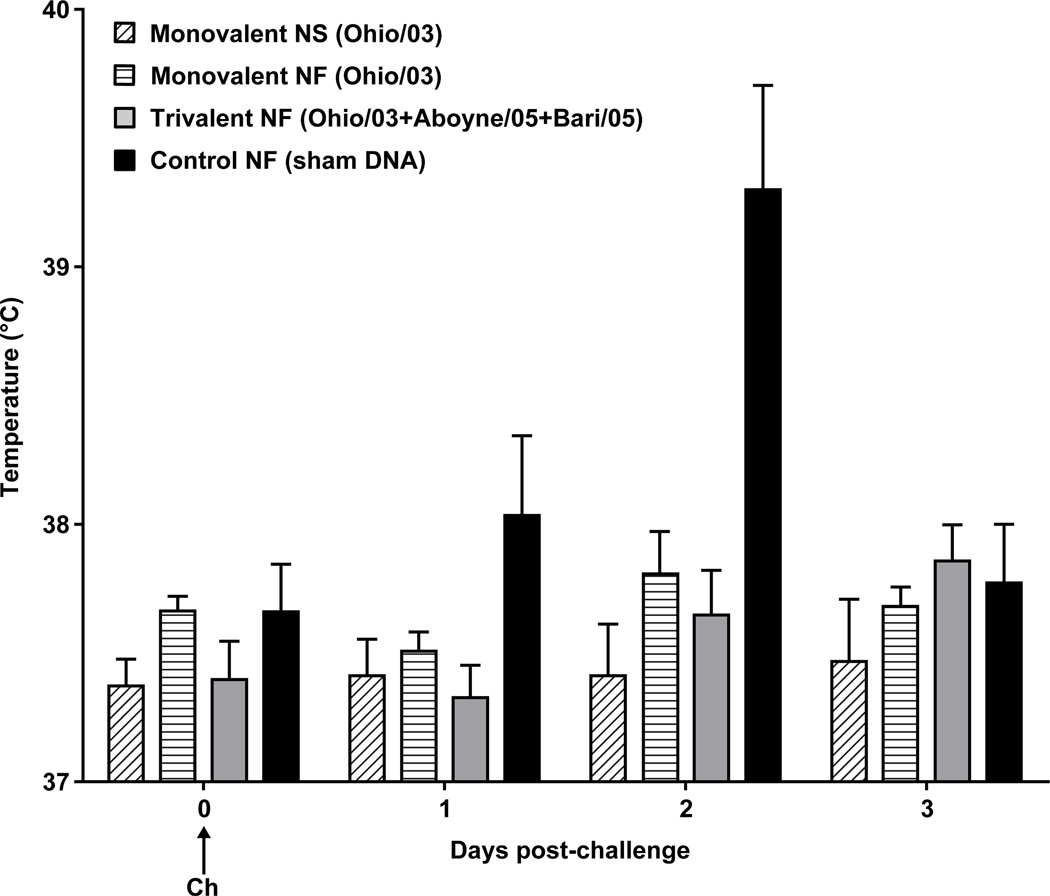
Vaccination was followed by daily monitoring of the injection site (skin above the brachiocephalicus/serratus cervicus muscles), and evaluation of clinical responses by a two equine clinical veterinarians using the scoring index outlined in Table 1. Apart from rare, transient swelling at the injection site (1 cm diameter, 1 mm raised, <24 hrs after injection), there were no untoward effects of vaccination in both control and vaccine groups. One pony sustained a cecocolic intussusception with peritonitis not attributed to vaccination (confirmed by detailed pathologic evaluation) and was humanely euthanized prior to challenge. Following challenge, vaccinated animals showed a trend of lower mean clinical scores compared to the control animals (Fig. 5A), particularly using coughing and nasal discharge as an index (Fig. 5B). Similar trends applied to rectal temperatures, as control animals appeared to show higher temperatures on day 2 post-challenge (Fig. 6). Control animals suffered mild to moderate pyrexia (T ≥ 38.9°C), with the mean rectal temperature peaking at an average of 39.3°C on day 2 post-challenge. No pyrexia was observed in vaccinated ponies.
Figure 5. Mean clinical scores post-challenge.
(a) Mean daily clinical scores for 8 days post-challenge. (b) Mean summed clinical scores for days 1 through 8 broken down by clinical sign. Error bars represent SEM. See Table 1 for scoring index. (Resp = respiration, Dem = demeanor, N Dis = nasal discharge, Cough = coughing) Ponies received three doses of vaccine (V1, V2 and V3) and were challenged with live equine influenza virus (Ch). Monovalent = Ohio/03 HA, Trivalent = Ohio/03 HA + Aboyne/05 HA + Bari/05 HA, Control = empty CMV/R vector, NS = needle and syringe, NF = needle-free device. One pony in the monovalent NF group was euthanized at wk 10 due to unrelated cecocolic intusseption with secondary peritonitis. Necropsy investigation revealed that this was not a vaccine-related event.
Figure 6. Mean daily rectal temperature post-challenge.
The mean rectal temperatures for the first three days following challenge. After day +3, all temperatures returned to normal range. Error bars represent SEM. Ponies received three doses of vaccine (V1, V2 and V3) and were challenged with live equine influenza virus (Ch). Monovalent = Ohio/03 HA, Trivalent = Ohio/03 HA + Aboyne/05 HA + Bari/05 HA, Control = empty CMV/R vector, NS = needle and syringe, NF = needle-free device.
3.4 Virus shedding
Viral shedding was assessed in frozen nasal swab samples by virus growth in embryonated hens’ eggs, PCR, and EID50 assays. The frequency of virus isolation was lower and the duration in days of virus shedding was shorter in all vaccinated animals compared to the controls. All four of the unvaccinated control ponies shed detectable virus for 5 consecutive days. One horse in the needle/syringe vaccine group shed detectable virus for 2 days. Of the seven ponies that were vaccinated by needle-free device, only one shed detectable virus, for a group average of 0.3 days (Table 2).
Table 2.
Viral isolation from nasal swabs taken from vaccinated and unvaccinated ponies after challenge with A/eq/Ohio/03 virus.
| Group | N | Egga (No. ponies with positive results) b |
Duration of shedding d (egg) a |
EID50 (per mL)e |
PCRf | Duration of shedding d (PCR) |
|---|---|---|---|---|---|---|
| Monovalent NS | 4 | 1 | 2 | - | 3256 | 2.5 |
| Monovalent NF | 4 | 1 | 0.67 | - | 2491 | 1.67 |
| Trivalent NF | 3 | 0 | 0 | - | 2525 | 0.5 |
| Control NF | 4 | 4c | 5 | + (0 – 1×104) | 3.07×106 | 5.25 |
virus growth in embryonated eggs. Nasal swab samples (0.1ml, undiluted) were injected into allantoic cavities of each of three 10-day-old eggs. Following incubation, allantoic fluids were harvested and tested for virus by hemagglutination assay. If any one egg tested positive, the swab sample was called virus-positive and the pony from which it originated was counted as shedding virus on that day.
results (positive or negative) are cumulative from days 2–5 post-challenge
on day 2 post-challenge, only 2 of the 4 ponies were positive by HA assay. On all other days, all 4 ponies were positive.
mean number of days of virus shedding
value on day 2 post-challenge
mean value on day 3 post-challenge (day of peak shedding in control group)
+ = influenza detected, − = influenza not detected
3.5 Viral load reduction via RT- PCR and EID50
Viral RNA from nasal swabs was quantified by RT-PCR for eight days post-challenge (Table 2). All four control animals shed detectable virus for at least 5 days post-challenge, whereas only four out of the eleven vaccinated animals shed virus between 1 to 4 days total post-challenge. The mean control viral load peaked at day 3, with control animals shedding an average of 3.07 × 106 copies of influenza virus RNA per 50µl of nasal secretion. Vaccinated animals shed less virus as compared to the controls on day 3 (vaccinated range = 0 – 1.3 × 104 viral RNA copies) (Table 2). Viral shedding was also evaluated by EID50. Several vaccinated animals were positive by PCR but negative by EID50. Only two of the controls were positive by EID50 with titers of 103 – 104 EID/ml on day 2 post-challenge (Table 2).
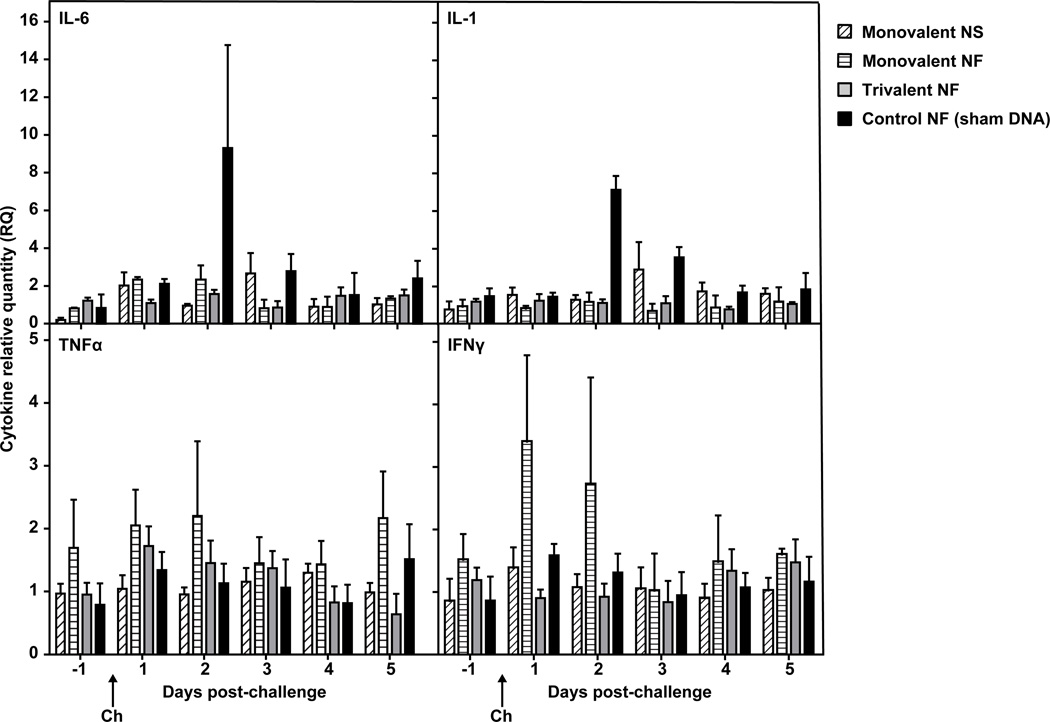
3.6 Cytokine response post-challenge
Expression of specific cytokine mRNAs in peripheral blood samples was used to assess the pro-inflammatory cytokine response post-challenge (Fig. 7). In controls, mean post-challenge IL-6 mRNA expression peaked on day 2. Like IL-6, mean IL-1 expression also increased in the controls from pre-challenge to day 2 post-challenge, but to a lesser degree (less than five-fold). This increase was not observed in any of the vaccine groups between days −1 and +2. No noticeable changes in TNFα or IFNγ mRNA expression occurred post-challenge most vaccinated and control ponies, although the monovalent NF group showed increased values of IFN-γ expression 1 and 2 days post-challenge.
Figure 7. Pro-inflammatory cytokine mRNA induction post-challenge.
Interleukin 6 (IL-6) (a), interleukin 1 (IL-1) (b), tumor necrosis factor-alpha (TNFα) (c), and interferon-gamma (IFNγ) (d) expression in whole blood collected in PAXgene tubes was determined by quantitative RT-PCR. RQ is the ratio of each daily sample’s normalized measurement to that of the Day -1 average (calibrator). Error bars represent SEM. Ponies received three doses of vaccine (V1, V2 and V3) and were challenged with live equine influenza virus (Ch). Monovalent = Ohio/03 HA, Trivalent = Ohio/03 HA + Aboyne/05 HA + Bari/05 HA, Control = empty CMV/R vector, NS = needle and syringe, NF = needle-free device.
4. Discussion
Current inactivated, whole-virus vaccines have been shown to protect horses from equine influenza by reducing clinical signs and virus excretion (13, 14, 38, 40). However, outbreaks of equine flu continue to occur despite vaccination, due to mismatching between vaccine strains and circulating viruses (12, 13). Gene-based vaccines have been indicated as a promising alternative with the potential to induce humoral and cellular responses, as well as the benefit of more efficient cell-based production methods.
The results of our pilot studies of DNA vaccination against equine H3N8 influenza in horses are consistent with our previous observations in the chicken and pig influenza challenge models (20, 48). Our primary focus was to test the immunogenicity and protective efficacy of equine influenza DNA vaccines and alternative vaccine delivery methods in an established equine influenza infection model. The immunogenicity of these DNA vaccines was first verified in mice, in which Ohio/03 and Bari/05 encoded antigens elicited neutralizing antibodies and evidence of cross-reactive neutralizing activity. Based on these data and on previous studies using HA gene-based vaccines (35, 53), we hypothesized that vaccinating ponies against EI virus with a DNA vaccine encoding these antigens could elicit a robust immune response and confer clinical protection from infection. Consistent with previous studies (20, 48), this experiment demonstrates that DNA vaccination elicits strong immune responses and provides clinical and virologic protection against homologous challenge. This broadens the potential of DNA vaccine enhancement including multivalency and an alternative needle-free delivery method. Due to logistical limitations and animal availability, low sample numbers in this experiment do not allow for robust statistical analysis. Therefore, while the findings in this study may not be statistically conclusive, the observed hypothesis-generating trends can be used for the design of more robust follow-up experiments.
Serological data in the current study were compared to previous studies of modified live virus (8, 9, 34, 57), inactivated (10, 14, 21, 22, 32, 46), and pox-vectored (17, 36, 45, 54) equine influenza vaccines. Review of these studies revealed that both our monovalent and multivalent DNA vaccines are potentially comparable in terms of immunogenicity and protection, although DNA vaccines showed a slightly delayed onset of immunity compared to modified live and canarypox-vectored vaccines (26). Specifically, SRH responses were similar to those elicited by licensed inactivated vaccines (Duvaxyn IE-T Plus and EQUIP F) (10, 14). Based on other studies, an SRH titer of roughly 150mm2 is expected to be adequate for short-term protection against a homologous EI challenge (13). It is important to note that SRH benchmarks for protection were developed using traditional vaccines, and may not be relevant to DNA vaccines which may induce alternative specific antibody isotype profiles (19, 23, 24).
Our vaccines induced SRH titers of close to 150mm2 following the second immunization, most notably in the needle-free groups and clinical protection was conferred by both the monovalent and multivalent vaccines when challenged with a homologous virus. HI and SRH data also suggest that the multivalent vaccine consisting of HA genes of Ohio/03, Aboyne/05, and Bari/05 EI viruses is capable of eliciting a broad antibody response. Based on previous studies in mice, chickens, and swine (20, 48), these broad responses to different strains have been associated with heterologous protection. Interestingly, the monovalent NF group generated a higher titers of cross-reactive antibodies compared to the monovalent NS group, and was also similar to the trivalent NF group. It is possible that needle-free delivery enhances responses and that broad trivalent NF responses may be due to cross-reactivity between antigens, common in SRH assays (30). Future studies should examine this and assess whether the multivalent approach confers heterologous protection against multiple equine influenza strains.
The monovalent and trivalent DNA vaccines tested here did not completely eliminate viral shedding (as measured by PCR) following challenge, but as determined by the conventional method of EID50 assay, no detectable live virus was shed from vaccinated animals. In addition to enhancing the pre-challenge immune responses, it is possible that immune modulators such as interleukin-23 (IL-23) or CCL5 (RANTES) may offer superior protection by completely eliminating any viral shedding following challenge, and could be a focus for future studies.
Since cytokines produced during antigen exposure are the primary regulators of the immune responses, analyzing levels in the blood allows us to better understand the immune response elicited by each of the different vaccines and delivery routes. In the control sera, mean levels of expression of interleukin-1 (IL-1) and interleukin-6 (IL-6) peaked at day 2 (at which mean febrile responses also peaked), while expression in vaccinates remained relatively constant. These findings are consistent with previous studies (9, 61). The trend toward increased expression of IFNγ and TNFα mRNA in vivo in the monovalent needle-free group was also consistent with previous studies, indicating a stimulation of the Th-1 cell-mediated immune response (9, 46, 53), similar to that of natural infection (25). This was further supported by the anamnestic in vitro IFNγ response of equine influenza virus-stimulated PBMC from the vaccinates.
Assessment of serum antibody titers and cytokine responses shows that DNA vaccination is capable of eliciting both humoral and cellular immune responses. Future studies should focus on developing pathways increase DNA vaccine feasibility, and overcome limitations that include high cost of production. Towards this, a dosing-down study or the incorporation of adjuvants will contribute to the goals of improving cost-efficacy and increasing potency.
It is encouraging that needle-free delivery of DNA elicited similar and comparable immunogenicity and protection as conventional injection with needle and syringe, as consistent with another previous equine study utilizing a gene gun (35). Needle-free delivery can improve the administration of vaccines by increasing the speed of distribution and the reduction of safety risks and logistical problems associated with the handling of needles suited for farm animals (2, 18). Furthermore, previous studies show that needle-free delivery of DNA vaccines may enhance vaccine efficacy partly by exposing the dermal layer to the immunogens (48, 49), whereas intramuscular needle/syringe injections bypass the dermis entirely. The advantages of needle-free delivery with this particular device have also been demonstrated against H1N1 influenza in the swine model (20) and this method should continue to be developed as a practical alternative to parenteral injection. In fact, needle-free delivery enhances cost-efficacy since the device is re-usable, does not carry the risk of handling sharps, and a growing competitive industry is making these devices more affordable.
4.1 Conclusions
We have provided evidence that gene-based vaccination is a potentially effective method for immunizing horses against H3N8 EI infection. DNA may be a viable alternative to both viral-vectored vaccines (54) and older vaccine technology due to its advantages in safety, efficiency of production, and potential for broad-based protection. To the best of our knowledge, this is the first multivalent gene-based equine influenza vaccine to be tested. Our data also suggests that delivery via needle-free device may enhance immune responses compared to traditional needle/syringe delivery, however does not impact the degree of protection. Future studies will be scaled up and focused to determine the potential for DNA vaccines to provide heterologous protection against multiple strains and subtypes, closely analyze the effects of monovalency vs. multivalency, and to delineate more clearly any enhancements offered by needle-free delivery in terms of immunogenicity and clinical protection.
Highlights.
DNA vaccines expressing the HA gene of equine H3N8 influenza virus were generated
DNA vaccines elicit homologous & heterologous immune responses after 3 vaccinations
DNA vaccines protect against disease and viral replication following H3N8 challenge
Needle-free delivery is as efficient and effective as conventional needle/syringe
DNA vaccines are a safe, effective alternative for veterinary vaccines against flu
Acknowledgments
We thank the University of Kentucky Veterinary Science farm crew for their expert animal care and handling. We also gratefully acknowledge the contributions of Ms. Judy Stein for material transfer and contractual requirements; Ms. Brenda Hartman for figure formatting; Dr. Mythreyi Shastri for manuscript preparation; and Ms. Martha Nason for assistance with statistical analysis. This research was supported by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, US National Institutes of Health and by the Kentucky Agricultural Experiment Station (project no. KY014041).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
To fully disclose any potential conflicts of interest, authors Gary J. Nabel, Srinivas S. Rao, Wing-Pui Kong, and Chih-Jen Wei are each listed on a patent filing for our DNA vaccine technology, entitled “U.S. Continuation in Part Patent Application No. 12/838,292”, which is an adjunct to an existing patent entitled “Influenza DNA Vaccination and Methods of Use thereof”, Serial #61/023,341.
Reference List
- 1.Aerts J, Gonzalez M, Topalian S. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques. 2004;36:84–86. doi: 10.2144/04361ST04. 88,90. [DOI] [PubMed] [Google Scholar]
- 2.Amorij JP, Hinrichs WL, Frijlink HW, Wilschut JC, Huckriede A. Needle-free influenza vaccination. Lancet Infect. Dis. 2010;10:699–711. doi: 10.1016/S1473-3099(10)70157-2. [DOI] [PubMed] [Google Scholar]
- 3.Barouch DH, Yang Zy, Kong Wp, Korioth-Schmitz B, Sumida SM, Truitt DM, Kishko MG, Arthur JC, Miura A, Mascola JR, Letvin NL, Nabel GJ. A Human T-Cell Leukemia Virus Type 1 Regulatory Element Enhances the Immunogenicity of Human Immunodeficiency Virus Type 1 DNA Vaccines in Mice and Nonhuman Primates. J. Viral. 2005;79:8828–8834. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boliar S, Stanislawek W, Chambers TM. Inability of kaolin treatment to remove nonspecific inhibitors from equine serum for the hemagglutination inhibition test against equine H7N7 influenza virus. J Vet Diagn Invest. 2006;18:264–267. doi: 10.1177/104063870601800305. [DOI] [PubMed] [Google Scholar]
- 5.Breathnach CC, Clark HJ, Clark RC, Olsen CW, Townsend HG, Lunn DP. Immunization with recombinant modified vaccinia Ankara (rMVA) constructs encoding the HA or NP gene protects ponies from equine influenza virus challenge. Vaccine. 2006;24:1180–1190. doi: 10.1016/j.vaccine.2005.08.091. [DOI] [PubMed] [Google Scholar]
- 6.Bryant NA, Rash AS, Russell CA, Ross J, Cooke A, Bowman S, MacRae S, Lewis NS, Paillot R, Zanoni R, Meier H, Griffiths LA, Daly JM, Tiwari A, Chambers TM, Newton JR, Elton DM. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2009. Vet Microbiol. 2007;138:41–52. doi: 10.1016/j.vetmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Bryant NA, Rash AS, Woodward AL, Medcalf E, Helwegen M, Wohlfender F, Cruz F, Herrmann C, Borchers K, Tiwari A, Chambers TM, Newton JR, Mumford JA, Elton DM. Isolation and characterisation of equine influenza viruses (H3N8) from Europe and North America from 2008 to 2011. Vet Microbiol. 2009;147:19–27. doi: 10.1016/j.vetmic.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 8.Chambers TM, Holland RE, Tudor LR, Townsend HG, Cook A, Bogdan J, Lunn DP, Hussey S, Whitaker-Dowling P, Youngner JS, Sebring RW, Penner SJ, Stiegler GL. A new modified live equine influenza virus vaccine: phenotypic stability, restricted spread and efficacy against heterologous virus challenge. Equine Vet. 2001;33:630–636. doi: 10.2746/042516401776249291. [DOI] [PubMed] [Google Scholar]
- 9.Chambers TM, Quinlivan M, Sturgill T, Cullinane A, Horohov DW, Zamarin D, Arkins S, Garcia-Sastre A, Palese P. Influenza A viruses with truncated NS1 as modified live virus vaccines: pilot studies of safety and efficacy in horses. Equine Vet. J. 2009;41:87–92. doi: 10.2746/042516408x371937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crouch CF, Daly J, Henley W, Hannant D, Wilkins J, Francis MJ. The use of a systemic prime/mucosal boost strategy with an equine influenza ISCOM vaccine to induce protective immunity in horses. Vet Immunol Immunop. 2005;108:345–355. doi: 10.1016/j.vetimm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Cullinane A, Elton D, Mumford J. Equine influenza - surveillance and control. Influenza. Other Respi. Viruses. 2010;4:339–344. doi: 10.1111/j.1750-2659.2010.00176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly JM, MacRae S, Newton JR, Wattrang E, Elton DM. Equine influenza: a review of an unpredictable virus. Vet. J. 2011;189:7–14. doi: 10.1016/j.tvjl.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Daly JM, Newton JR, Mumford JA. Current perspectives on control of equine influenza. Vet. Res. 2004;35:411–423. doi: 10.1051/vetres:2004023. [DOI] [PubMed] [Google Scholar]
- 14.Daly JM, Sindle T, Tearle J, Barquero N, Newton JR, Corning S. Equine influenza vaccine containing older H3N8 strains offers protection against A/eq/South Africa/4/03 (H3N8) strain in a short-term vaccine efficacy study. Equine Vet J. 2007;39:446–450. doi: 10.2746/042516407x180327. [DOI] [PubMed] [Google Scholar]
- 15.Dheda KHJBSJMRGZA. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–114. doi: 10.2144/04371RR03. 116,118. [DOI] [PubMed] [Google Scholar]
- 16.Drunen Littel-van den Hurk S, Babiuk S, Babiuk LA. Needle-Free Delivery of Veterinary DNA Vaccines. Ann NY Acad Sci. 2006:91–105. doi: 10.1385/1-59745-168-1:91. [DOI] [PubMed] [Google Scholar]
- 17.Edlund TC, Daly J, Sindle T, Guigal PM, Audonnet JC, Minke JM. Efficacy of a recombinant equine influenza vaccine against challenge with an American lineage H3N8 influenza virus responsible for the 2003 outbreak in the United Kingdom. Vet. Rec. 2005;156:367–371. doi: 10.1136/vr.156.12.367. [DOI] [PubMed] [Google Scholar]
- 18.Ekwueme DU, Weniger BG, Chen RT. Model-based estimates of risks of disease transmission and economic costs of seven injection devices in sub-Saharan Africa. Bull. World Health Organ. 2002;80:859–870. [PMC free article] [PubMed] [Google Scholar]
- 19.Feltquate DM, Heaney S, Webster RG, Robinson HL. Different T helper cell types and antibody isotypes generated by saline and gene gun DNA immunization. J. Immunol. 1997;158:2278–2284. [PubMed] [Google Scholar]
- 20.Gorres JP, Lager KM, Kong WP, Royals M, Todd JP, Vincent AL, Wei CJ, Loving CL, Zanella EL, Janke B, Kehrli ME, Jr, Nabel GJ, Rao SS. DNA Vaccination Elicits Protective Immune Responses against Pandemic and Classic Swine Influenza Viruses in Pigs. Clin. Vaccine Immunol. 2011 doi: 10.1128/CVI.05171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heldens JG, Pouwels HG, Derks CG, Van de Zande SM, Hoeijmakers MJ. The first safe inactivated equine influenza vaccine formulation adjuvanted with ISCOM-Matrix that closes the immunity gap. Vaccine. 2009;27:5530–5537. doi: 10.1016/j.vaccine.2009.06.085. [DOI] [PubMed] [Google Scholar]
- 22.Heldens JGM, Pouwels HGW, Derks CGG, Van de Zande SMA, Hoeijmakers MJH. Duration of immunity induced by an equine influenza and tetanus combination vaccine formulation adjuvanted with ISCOM-Matrix. Vaccine. 2010;28:6989–6996. doi: 10.1016/j.vaccine.2010.08.043. [DOI] [PubMed] [Google Scholar]
- 23.Huber VC, McKeon RM, Brackin MN, Miller LA, Keating R, Brown SA, Makarova N, Perez DR, Macdonald GH, McCullers JA. Distinct contributions of vaccine-induced immunoglobulin G1 (IgG1) and IgG2a antibodies to protective immunity against influenza. Clin. Vaccine Immunol. 2006;13:981–990. doi: 10.1128/CVI.00156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber VC, Thomas PG, McCullers JA. A multi-valent vaccine approach that elicits broad immunity within an influenza subtype. Vaccine. 2009;27:1192–1200. doi: 10.1016/j.vaccine.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson PA, Conway M, Daly J, Nicolson C, Robertson JS, Mills KHG. Influenza HA DNA induces Th1 cells and protection despite limited antibody responses. Int Congr Ser. 2001;1219:911–915. [Google Scholar]
- 26.Kannegieter NJ, Frogley A, Crispe E, Kirkland PD. Clinical outcomes and virology of equine influenza in a naive population and in horses infected soon after receiving one dose of vaccine. Aust. Vet. J. 2011;89(Suppl 1):139–142. doi: 10.1111/j.1751-0813.2011.00768.x. [DOI] [PubMed] [Google Scholar]
- 27.Kong W-P, Hood C, Yang Z-Y, Wei C-J, Xu L, Garcia-Sastre A, Tumpey TM, Nabel GJ. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. P Natl Acad Sci. 2006;103:15987–15991. doi: 10.1073/pnas.0607564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat. Rev. Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, Yeh WW, Asmal M, Gelman RS, Shen L, Whitney JB, Seoighe C, Lacerda M, Keating S, Norris PJ, Hudgens MG, Gilbert PB, Buzby AP, Mach LV, Zhang J, Balachandran H, Shaw GM, Schmidt SD, Todd JP, Dodson A, Mascola JR, Nabel GJ. Immune and Genetic Correlates of Vaccine Protection Against Mucosal Infection by SIV in Monkeys. Sci. Transl. Med. 2011;3:81ra36. doi: 10.1126/scitranslmed.3002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis NS, Daly JM, Russell CA, Horton DL, Skepner E, Bryant NA, Burke DF, Rash AS, Wood JL, Chambers TM, Fouchier RA, Mumford JA, Elton DM, Smith DJ. Antigenic and genetic evolution of equine influenza A (H3N8) virus from 1968 to 2007. J. Virol. 2011;85:12742–12749. doi: 10.1128/JVI.05319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-[Delta][Delta]CT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Lopez AM, Hecker R, Mutwiri, van Drunen Littel-van den Hurk G, Babiuk LA, Townsend HGG. Formulation with CpG ODN enhances antibody responses to an equine influenza virus vaccine. Vet Immunol Immunop. 2006;114:103–110. doi: 10.1016/j.vetimm.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Lu Z, Chambers TM, Boliar S, Branscum AJ, Sturgill TL, Timoney PJ, Reedy SE, Tudor LR, Dubovi EJ, Vickers ML, Sells S, Balasuriya UBR. Development and Evaluation of One-Step TaqMan Real-Time Reverse Transcription-PCR Assays Targeting Nucleoprotein, Matrix, and Hemagglutinin Genes of Equine Influenza Virus. J. Clin. Microbiol. 2009;47:3907–3913. doi: 10.1128/JCM.00598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunn DP, Hussey S, Sebing R, Rushlow KE, Radecki SV, Whitaker-Dowling P, Youngner JS, Chambers TM, Holland RE, Jr, Horohov DW. Safety, efficacy, and immunogenicity of a modified-live equine influenza virus vaccine in ponies after induction of exercise-induced immunosuppression. J. Am. Vet. Med. Assoc. 2001;218:900–906. doi: 10.2460/javma.2001.218.900. [DOI] [PubMed] [Google Scholar]
- 35.Lunn DP, Soboll G, Schram BR, Quass J, McGregor MW, Drape RJ, Macklin MD, McCabe DE, Swain WF, Olsen CW. Antibody responses to DNA vaccination of horses using the influenza virus hemagglutinin gene. Vaccine. 1999;17:2245–2258. doi: 10.1016/s0264-410x(98)00496-4. [DOI] [PubMed] [Google Scholar]
- 36.Minke JM, Toulemonde CE, Coupier H, Guigal PM, Dinic S, Sindle T, Jessett D, Black L, Bublot M, Pardo MC, Audonnet JC. Efficacy of a canarypox-vectored recombinant vaccine expressing the hemagglutinin gene of equine influenza H3N8 virus in the protection of ponies from viral challenge. Am. J. Vet. Res. 2007;68:213–219. doi: 10.2460/ajvr.68.2.213. [DOI] [PubMed] [Google Scholar]
- 37.Morley PS, Townsend HG, Bogdan JR, Haines DM. Efficacy of a commercial vaccine for preventing disease caused by influenza virus infection in horses. J. Am. Vet. Med. Assoc. 1999;215:61–66. [PubMed] [Google Scholar]
- 38.Mumford J, Wood JM, Scott AM, Folkers C, Schild GC. Studies with inactivated equine influenza vaccine. 2. Protection against experimental infection with influenza virus A/equine/Newmarket/79 (H3N8) J. Hyg. (Lond) 1983;90:385–395. doi: 10.1017/s0022172400029016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mumford JA, Hannant D, Jessett DM. Experimental infection of ponies with equine influenza (H3N8) viruses by intranasal inoculation or exposure to aerosols. Equine Vet J. 1990;22:93–98. doi: 10.1111/j.2042-3306.1990.tb04217.x. [DOI] [PubMed] [Google Scholar]
- 40.Mumford JA, Wood JM, Folkers C, Schild GC. Protection against experimental infection with influenza virus A/equine/Miami/63 (H3N8) provided by inactivated whole virus vaccines containing homologous virus. Epidemiol. Infect. 1988;100:501–510. doi: 10.1017/s0950268800067236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson KM, Schram BR, McGregor MW, Sheoran AS, Olsen CW, Lunn DP. Local and systemic isotype-specific antibody responses to equine influenza virus infection versus conventional vaccination. Vaccine. 1998;16:1306–1313. doi: 10.1016/s0264-410x(98)00009-7. [DOI] [PubMed] [Google Scholar]
- 42.Newton JR, Townsend HG, Wood JL, Sinclair R, Hannant D, Mumford JA. Immunity to equine influenza: relationship of vaccine-induced antibody in young Thoroughbred racehorses to protection against field infection with influenza A/equine-2 viruses (H3N8) Equine Vet. J. 2000;32:65–74. doi: 10.2746/042516400777612116. [DOI] [PubMed] [Google Scholar]
- 43.OIE. OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris: Office International des Epizooties; 2004. Equine Influenza; pp. 686–697. [Google Scholar]
- 44.Paillot R, Hannant D, Kydd JH, Daly JM. Vaccination against equine influenza: quid novi? Vaccine. 2006;24:4047–4061. doi: 10.1016/j.vaccine.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 45.Paillot R, Kydd JH, Sindle T, Hannant D, Edlund Toulemonde C, Audonnet JC, Minke JM, Daly JM. Antibody and IFN-[gamma] responses induced by a recombinant canarypox vaccine and challenge infection with equine influenza virus. Vet Immunol Immunop. 2006;112:225–233. doi: 10.1016/j.vetimm.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 46.Quinlivan M, Nelly M, Prendergast M, Breathnach C, Horohov D, Arkins S, Chiang YW, Chu HJ, Ng T, Cullinane A. Pro-inflammatory and antiviral cytokine expression in vaccinated and unvaccinated horses exposed to equine influenza virus. Vaccine. 2007;25:7056–7064. doi: 10.1016/j.vaccine.2007.07.059. [DOI] [PubMed] [Google Scholar]
- 47.Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, Tryon V. Stabilization of mRNA Expression in Whole Blood Samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- 48.Rao SS, Kong WP, Wei CJ, Yang ZY, Nason M, Styles D, DeTolla LJ, Panda A, Sorrell EM, Song Haichen, Wan H, Ramirez-Nieto GC, Perez D, Nabel GJ. Multivalent HA DNA Vaccination Protects against Highly Pathogenic H5N1 Avian Influenza Infection in Chickens and Mice. PLoS one. 2008;3 doi: 10.1371/journal.pone.0002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao SS, Styles D, Kong W, Andrews C, Gorres JP, Nabel GJ. A gene-based avian influenza vaccine in poultry. Poult Sci. 2009;88:860–866. doi: 10.3382/ps.2008-00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- 51.Shipley JM, Miller RD, Wu BM, Grubb JH, Christensen SG, Kyle JW, Sly WS. Analysis of the 5' flanking region of the human beta-glucuronidase gene. Genomics. 1991;10:1009–1018. doi: 10.1016/0888-7543(91)90192-h. [DOI] [PubMed] [Google Scholar]
- 52.Shu Y, Winfrey S, Yang ZY, Xu L, Rao S, Srivastava I, Barnett S, Nabel GJ, Mascola J. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007;25:1398–1408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soboll G, Horohov DW, Aldridge BM, Olsen CW, McGregor MW, Drape RJ, Macklin MD, Swain WF, Lunn DP. Regional antibody and cellular immune responses to equine influenza virus infection, and particle mediated DNA vaccination. Vet. Immunol. Immunopathol. 2003;94:47–62. doi: 10.1016/s0165-2427(03)00060-6. [DOI] [PubMed] [Google Scholar]
- 54.Soboll G, Hussey SB, Minke JM, Landolt GA, Hunter JS, Jagannatha S, Lunn DP. Onset and duration of immunity to equine influenza virus resulting from canarypox-vectored (ALVAC) vaccination. Vet. Immunol. Immunopathol. 2010;135:100–107. doi: 10.1016/j.vetimm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Soboll G, Nelson KM, Leuthner ES, Clark RJ, Drape R, Macklin MD, Swain WF, Olsen CW, Lunn DP. Mucosal co-administration of cholera toxin and influenza virus hemagglutinin-DNA in ponies generates a local IgA response. Vaccine. 2003;21:3081–3092. doi: 10.1016/s0264-410x(03)00161-0. [DOI] [PubMed] [Google Scholar]
- 56.Song K, Bolton DL, Wei CJ, Wilson RL, Camp JV, Bao S, Mattapallil JJ, Herzenberg LA, Herzenberg LA, Andrews CA, Sadoff JC, Goudsmit J, Pau MG, Seder RA, Kozlowski PA, Nabel GJ, Roederer M, Rao SS. Genetic immunization in the lung induces potent local and systemic immune responses. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22213–22218. doi: 10.1073/pnas.1015536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Townsend HG, Penner SJ, Watts TC, Cook A, Bogdan J, Haines DM, Griffin S, Chambers T, Holland RE, Whitaker-Dowling P, Youngner JS, Sebring RW. Efficacy of a cold-adapted, intranasal, equine influenza vaccine: challenge trials. Equine Vet. J. 2001;33:637–643. doi: 10.2746/042516401776249354. [DOI] [PubMed] [Google Scholar]
- 58.United States Department of Agriculture. USDA issues license for West Nile Virus DNA vaccine for horses. 2005 http://www. aphis. usda. gov/lpa/news/2005/07/wnvdna_vs. html. [Online.].
- 59.Van de Walle GR, May MA, Peters ST, Metzger SM, Rosas CT, Osterrieder N. A vectored equine herpesvirus type 1 (EHV-1) vaccine elicits protective immune responses against EHV-1 and H3N8 equine influenza virus. Vaccine. 2010;28:1048–1055. doi: 10.1016/j.vaccine.2009.10.123. [DOI] [PubMed] [Google Scholar]
- 60.van MC, Cullinane A. Equine influenza virus infections: an update. Vet. Q. 2002;24:79–94. doi: 10.1080/01652176.2002.9695127. [DOI] [PubMed] [Google Scholar]
- 61.Wattrang E, Jessett DM, Yates P, Fuxler L, Hannant D. Experimental infection of ponies with equine influenza A2 (H3N8) virus strains of different pathogenicity elicits varying interferon and interleukin-6 responses. Viral Immunol. 2003;16:57–67. doi: 10.1089/088282403763635456. [DOI] [PubMed] [Google Scholar]
- 62.Wei CJ, Boyington JC, McTamney PM, Kong WP, Pearce MB, Xu L, Andersen H, Rao S, Tumpey TM, Yang ZY, Nabel GJ. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science. 2010;329:1060–1064. doi: 10.1126/science.1192517. [DOI] [PubMed] [Google Scholar]
- 63.Wood JM, Mumford J, Folkers C, Scott AM, Schild GC. Studies with inactivated equine influenza vaccine. 1. Serological responses of ponies to graded doses of vaccine. J Hyg (Lond) 1983;90:371–384. doi: 10.1017/s0022172400029004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright PF. Vaccine preparedness--are we ready for the next influenza pandemic? N. Engl. J. Med. 2008;358:2540–2543. doi: 10.1056/NEJMp0803650. [DOI] [PubMed] [Google Scholar]