Abstract
It was reported previously that enolase enzyme activity and ENO1 transcript levels are induced by anaerobic stress in maize (Zea mays). Here we show that not all isoforms of maize enolase are anaerobically induced. We cloned and sequenced a second enolase cDNA clone (pENO2) from maize. Sequence analysis showed that pENO2 shares 75.6% nucleotide and 89.5% deduced amino acid sequence identity with pENO1 and is encoded by a distinct gene. Expression of ENO2 is constitutive under aerobic conditions, whereas ENO1 levels are induced 10-fold in maize roots after 24 h of anaerobic treatment. Western-blot analysis and N-terminal sequencing of in vivo-labeled maize roots identified two major proteins selectively synthesized upon anaerobic stress as isozymes of enolase. We describe the expression of enolase in maize roots under anaerobic stress.
Anaerobic treatment of maize (Zea mays) seedlings drastically alters the profile of total protein synthesis. In an anaerobic environment, 20 proteins that account for more than 70% of the total translation are selectively synthesized (Sachs et al., 1980). Most of the ANPs identified have been found to be enzymes used in glycolysis or sugar-phosphate metabolism (Freeling, 1973; Kelley and Freeling, 1984; Kelley and Tolan, 1986; Springer et al., 1986; Russell and Sachs, 1991). In addition, the induction of transcription and enzyme activity of enolase (Lal et al., 1991) and PDC (Kelley, 1989) in maize has been reported during anaerobic stress, indicating that they may represent other ANPs. Synthesis of ANPs is regulated at the level of transcription and translation (Sachs et al., 1980; Hake et al., 1985).
Enolase (2-phospho-d-glycerate hydratase; EC 4.2.1.11) is an integral enzyme in glycolysis. It catalyzes the interconversion of 2-phosphoglycerate to PEP. The enzyme is extensively characterized in yeast (Lebioda et al., 1989) and in vertebrates (Giallongo et al., 1986). Genes encoding plant enolases have been cloned from Arabidopsis, tomato (Van Der Straeten et al., 1991), castor bean (Blakeley et al., 1994), Mesembryanthemum crystallinum L. (Forsthoefel et al., 1995), and Echinochloa phyllopogon (Fox et al., 1995), and the purification of the enzyme to apparent homogeneity has been reported from potato tubers (Boser, 1959), spinach (Sinha and Brewer, 1984), E. phyllopogon, and Echinochloa crus-pavonis (Mujer et al., 1995). Two enolase isozymes, one found in the cytosol and the other compartmentalized in the plastid, have been reported in castor bean (Miernyk and Dennis, 1984, 1992). Lal et al. (1991) previously identified a cDNA clone (pZM245 or pENO1) encoding maize enolase by functional genetic complementation of an enolase-deficient mutant of Escherichia coli. The gene encoding this cDNA is designated eno1.
The transcript levels detected by the ENO1 probe were induced in maize roots during anaerobic treatment. In contrast, no apparent increase in the level of enolase protein was observed during 24 h of anaerobic stress (Lal et al., 1994). Enolase was also purified from maize seeds, and this isolated protein resolved as a doublet by SDS-PAGE, with apparent molecular masses of 55 and 56 kD (Lal et al., 1994). This protein doublet was further resolved into three isoforms upon two-dimensional IEF-SDS-PAGE. However, Southern-blot analysis at high stringency indicated that eno1 is a single-copy gene in maize (Lal et al., 1991). Based on these observations, we decided to determine if the multiple enolase isozymes are encoded by two or more different genes or by a single gene in maize.
We report the cloning of another cDNA encoding maize enolase, pENO2. The pENO2 nucleotide sequence, its deduced amino acid sequence, and its expression during the anaerobic-stress response are compared with those of the previously reported pENO1 enolase clone. Genomic Southern-blot analyses and sequence analyses confirmed that these two enolase cDNAs are the products of two different genes. We also identified two previously described major anaerobic proteins, ANP45A and ANP45B (Sachs et al., 1980), as isozymes of enolase in maize.
MATERIALS AND METHODS
Plant Material and Anaerobic Treatment
Maize (Zea mays L. cv B73) seeds were germinated for 4 d on moist filter paper (3MM, Whatman3) in the dark until the primary roots were 6 to 9 cm long. Anaerobic treatment of the seedlings was either in an anaerobic chamber (model 1025, Forma Scientific) maintained with a gas mixture of 90% (v/v) nitrogen and 10% (v/v) hydrogen, or the seedlings were placed in a sealed container through which argon was continuously bubbled during the time of incubation (Sachs et al., 1980). Maize seedlings subjected to these two forms of anaerobic treatment gave a similar induction profile of anaerobic proteins (data not presented). In both cases whole seedlings were submerged in 5 mm Tris-HCl buffer, pH 7.5, supplemented with Augmentin (250 mg of amoxicillin and 125 mg of potassium clavulanate per liter; SmithKline Beecham, Philadelphia, PA; Subbaiah et al., 1994). Sterility was monitored by streaking seedlings on a nutrient agar Petri dish and incubating overnight at 37°C.
cDNA Library Construction and Screening
A cDNA library (Lal and Sachs, 1995) was constructed from RNA extracted from 6-h anaerobically treated preemergent (leaves in coleoptile) maize seedling roots, using a cDNA-synthesis kit (ZAP, Stratagene) following the instructions provided by the manufacturer. Poly(A+) mRNA was isolated from the total RNA using the Poly-ATract I RNA-isolation system (Promega). This library was screened with a 1.6-kb EcoRI insert of pTENO1, a cDNA clone encoding tomato enolase kindly provided by Marc Van Montagu (Van Der Straeten et al., 1991), under low-stringency conditions (5× Denhardt's solution, 6× SSC, 0.1% [w/v] SDS, and 100 μg/mg denatured salmon-sperm DNA at 60°C). Fifteen positive plaques were identified after screening approximately 20,000. The positive plaques were purified and excised in vivo to produce pBSKS using the Exassist/SOLR system (Stratagene).
Three clones that did not hybridize during Southern-blot analysis to a 32P-labeled, 244-bp XhoI fragment of the pENO1 (3′-untranslated region) probe had identical 3′-untranslated sequences with each other, but differed from that in pENO1. The clone with the longest insert, designated pENO2, was characterized by restriction and DNA-sequence analysis. DNA sequencing was done for both strands by the University of Illinois Genetic Engineering Facility with a DNA sequencer (model 373A, Applied Biosystems) using dye-terminator chemistry. HIBIO MacDNASIS Pro software (Hitachi, Tokyo, Japan) was used for the analysis of DNA and for protein sequencing. Sequence homology of the clones to previously reported enolase sequences was analyzed using the Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990).
RNA Isolation and Northern-Blot Analysis
After anaerobic treatment for different times, roots were excised 5 cm proximal from the tip and immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation. RNA was extracted (Russell and Sachs, 1989) and 20 μg of total RNA per lane was resolved on a 1.3% (w/v) agarose-formaldehyde gel. The RNA was then capillary transferred to nylon membranes (Nytran, Schleicher & Schuell), as described previously (Russell and Sachs, 1989). A 244-bp XhoI fragment of pENO1 containing the nucleotide sequence from the 3′-untranslated region and a PCR-amplified, 252-bp fragment from the 3′-untranslated region of pENO2 were used as gene-specific probes for ENO1 and ENO2, respectively, during RNA analysis. Before radiolabeling the DNA probes were isolated by resolving on a 1% (w/v) agarose gel, sliced out of the gel, and purified using a gel-extraction kit (Qiaex, Qiagen, Chatsworth, CA).
These probes were labeled with [32P]dCTP using a DNA-labeling kit (PrimeIt II, Stratagene). Prehybridization was in a solution containing 50% (v/v) formamide, 5× Denhardt's solution, 5× SSPE (1× SSPE = 0.15 m NaCl, 10 mm NaH2PO4, and 1 mm EDTA, pH 7.4), 0.1% (w/v) SDS, and 100 μg/mL denatured salmon-sperm DNA at 37°C for 3 h. Hybridization was carried out under similar conditions for 12 to 18 h. The blots were washed twice for 10 min each at room temperature in 2× SSC and 0.5% (w/v) SDS, followed by a final wash for 1 h at 60°C with 0.5× SSC and 0.1% (w/v) SDS, and exposed to Kodak XAR-5 film at −80°C using an intensifying screen. Blots were routinely reprobed after being washed twice with 0.1× SSC and 0.1% SDS for 15 min at 90°C, followed by overnight exposure on film to ensure complete removal of the probe. The transcript levels were quantified by scanning autoradiographs on a scanner (ScanJet 4C, Hewlett-Packard) equipped with a transparency adapter, and densitometrically analyzed using imaging software (NIH Image, National Institutes of Health, Bethesda, MD) on a Macintosh computer. Transcript levels were determined relative to levels of a constitutive mRNA designated 1055, and averaged from three experiments.
In Vivo Labeling and Autoradiography
Maize seedlings were subjected to anaerobic treatment for different time intervals in an anaerobic chamber. In vivo labeling was performed by immersing root tips in 1 mL of drowning buffer (5 mm Tris-HCl, pH 7.5) containing 0.1 mCi of Escherichia coli hydrolysate labeling reagent (Trans35S-Label, ICN). After exposure to labeled amino acids, 10 primary roots at specific time intervals were excised and ground in 250 μL of extraction buffer (62.5 mm Tris-HCl, pH 6.8, 1 mm PMSF, and 1 mm DTT). The resulting slurry was centrifuged at 10,000g for 5 min (Speedfuge HSC 15R, Savant Instruments, Holbrook, NY). The supernatant was resolved by native two-dimensional SDS-PAGE, as described previously (Sachs et al., 1980). After electrophoresis, gels were either electroblotted for western-blot analysis, or dried and exposed to film for autoradiography.
N-Terminal Microsequencing of Proteins
Proteins were extracted from seedling roots treated anaerobically for 64 h. After separation by native two-dimensional SDS-PAGE, proteins were blotted onto a PVDF membrane (ProBlott, Applied Biosystems) according to the procedure described by Matsudaira (1987). Protein microsequencing was performed at the Genetic Engineering Facility of the University of Illinois at Urbana-Champaign. The spots of interest were excised from the PVDF membrane, and N-terminal sequencing was done using a sequencer (model 477A, Applied Biosystems) coupled to an on-line phenylthiohydantoin analyzer (model 120A, Applied Biosystems) using Edman chemistry.
Western-Blot Analysis
Maize seedling roots were pulse-labeled either for 1 h aerobically or after different times of anaerobic treatment. The protein was then extracted and resolved on native two-dimensional SDS-PAGE gels. The gels were then electroblotted onto a nitrocellulose membrane (Schleicher & Schuell) using a semidry blotting apparatus according to the manufacturer's directions (Poly Blot, American Bionetics, Hayward, CA; Towbin et al., 1979). The membrane was then processed using a procedure similar to that of Lending et al. (1988). The membrane was initially incubated for 30 min in 1% (w/v) gelatin to block the nonspecific binding of proteins. The blot was then incubated for 90 min with polyclonal antibodies raised against the overexpressed fusion protein of pENO1 (a generous gift from David T. Dennis, Queens University, Kingston, Ontario, Canada) at 1:3000 dilution in TBST (100 mm Tris, pH 7.5, 1.4 m NaCl, 1.5% [v/v] Tween 20). The blot was then given three washes for 10 min each in TBST before subjecting it to a final incubation in 1:2000 TBST-diluted goat anti-rabbit serum coupled to peroxidase (Sigma) for 90 min. After three additional washes in TBST and a 5-min wash in TBS (100 mm Tris, pH 7.5, 1.4 m NaCl), the antigen-antibody reaction was visualized using the color development reagent diamino-benzidine (Bio-Rad). The blot was then exposed to film at −80°C for autoradiography.
Southern-Blot Analysis
Genomic DNA, isolated according to the procedure described by Saghai-Maroof et al. (1984), was digested with an excess of a restriction endonuclease (New England Biolabs) in a 100-μL reaction volume supplemented with 4 mm spermidine (Dellaporta et al., 1983). The digested DNA was separated on a 0.8% (w/v) agarose gel and capillary blotted onto nitrocellulose. The blot was then hybridized with the labeled PstI fragment of pENO1 and the XhoI-EcoRI fragment of pENO2 following the protocol described previously by Russell and Sachs (1991).
RESULTS
Sequence Analysis
We have cloned a full-length enolase cDNA (pENO2) by screening an anaerobically induced maize root cDNA library under low-stringency conditions using a tomato enolase cDNA (pTENO1) as a heterologous hybridization probe. The tomato enolase cDNA was chosen over the maize pENO1 as a probe because enolases from different plant species are conserved (Van Der Straeten et al., 1989; Lal et al., 1994), and because pTENO1 shares only 75% sequence identity with maize pENO1, so the screening would less likely be biased in favor of clones representing ENO1. Figure 1 shows the comparison of nucleotide and predicted amino acid sequence between ENO1 and ENO2. The initiation Met of ENO1 perfectly aligns with Met at the same relative position in ENO2. This indicates that pENO2 contains the full protein-coding region and belongs to the same gene family as ENO1. Both proteins are predicted to be 446 amino acids in length, with predicted masses of 48.1 kD for ENO1 and 48.2 kD for ENO2. The sequence identity between the initiation Met and the putative stop codons of ENO1 and ENO2 is 75.6%. The regions flanking the translated region at the 3′ and 5′ ends are totally divergent, however, showing that these two cDNAs represent the products of two different genes.
Figure 1.
Sequence alignment of maize enolase pENO1 and pENO2. A, The complete nucleotide sequence of ENO2 is aligned with that of ENO1. The nontranslated regions are in lowercase and the coding region is in uppercase. The initiation and termination codons are underlined. The putative consensus AATAAA sequence, which in animals serves as a signal for poly(A+) tail addition, also exists in the 3′-untranslated region of ENO1 and in a slightly modified form (AATAAT) in ENO2, and is double-underlined. B, The deduced polypeptide sequence of pENO1 is compared with that of pENO2. The asterisks indicate the termination codons. In both A and B, identical sequences in the coding regions are indicated by dots. {/ANNT;84480n;69696n;92928n;118272n}
ENO1 and ENO2 share 89% sequence identity at the amino acid level (Fig. 1B). The amino acid residues found in the active site of yeast enolase (Lebioda et al., 1989) are also present in predicted sequences for both ENO1 and ENO2, suggesting that the cDNAs encode functional enzymes.
Northern-Blot Analysis
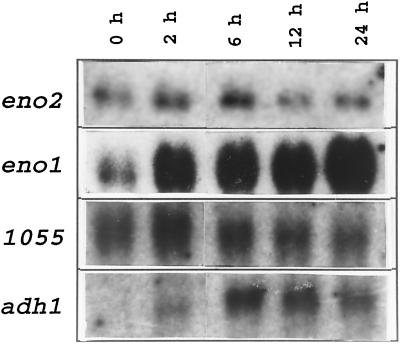
Because there is considerable variation in the induction kinetics observed in the accumulation of mRNAs encoding ANPs (Hake et al., 1985; Peschke and Sachs, 1994), we examined the levels of ENO1 and ENO2 mRNAs during anaerobic treatment. Total RNAs from aerobic and anaerobically treated (2, 6, 12, and 24 h) maize roots were separated on a formaldehyde-agarose gel and subjected to RNA analysis using ENO1 and ENO2 gene-specific probes. The ENO2 probe hybridized to a transcript of a similar size to ENO1 (approximately 1.8 kb). The transcript levels of ENO2 appeared to be significantly lower compared with the induced levels of ENO1 transcript, as judged by comparably labeled probes during multiple RNA analyses. As shown in Figure 2, ENO2 transcript levels remained relatively unchanged during anaerobic treatment, whereas the transcript levels of ENO1 reached a 5.2-fold induction level compared with the aerobic control by 24 h of anaerobic treatment. The same blot was also probed with pADH1 to monitor the efficacy of anaerobic treatment. ADH1 transcript levels were induced, reaching a maximum level between 6 and 12 h of anaerobic treatment. The RNA loading was normalized by probing with a cDNA designated 1055, the level of which has been reported to remain constant under anaerobic stress in maize (Subbaiah et al., 1994), and enolase mRNA levels were quantified relative to 1055.
Figure 2.
RNA hybridization showing the differential regulation of the two enolase genes during anaerobic treatment. Total RNA extracted from maize roots at different anaerobic time intervals (as indicated above each lane) was subjected to electrophoresis, blotted, and hybridized to the cDNA probe indicated to the left of each panel.
Southern-Blot Analysis
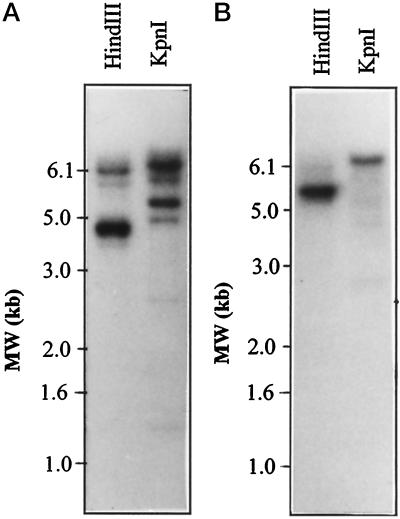
Total genomic DNA from an inbred maize line (B73) was digested with the restriction enzymes HindIII and KpnI and analyzed by Southern blotting. The nucleotide sequences of ENO1 and ENO2 transcripts are less than 80% identical and are not expected to cross-hybridize during Southern-blot analysis under high-stringency conditions. A 1.7-kb PstI insert of pENO1 and an XhoI-EcoRI insert of pENO2 were radiolabeled as probes for Southern-blot analysis. The hybridization-banding patterns observed for ENO1 and ENO2 differed from each other in the DNA digests (Fig. 3). For example, 4.5- and 6.2-kb HindIII genomic fragments hybridized to ENO1, whereas only one major fragment of 5.2 kb was detected with the ENO2 probe. Similarly, only the ENO1 probe detected a major 5.2-kb fragment in the KpnI genomic digest. These data also indicate that the two cDNAs are encoded by two different genes in the maize genome, eno1 and eno2. In addition, several other bands weakly hybridized with the ENO1 and ENO2 cDNA probes (Fig. 3). Further investigation is required to determine if these bands represent genes encoding other enolase isozymes or pseudogenes. The eno2 gene was mapped by restriction-fragment-length polymorphism analysis to the short arm of chromosome 1 in maize (B. Burr, personal communication). The eno1 gene was previously mapped to the short arm of chromosome 9 (Peschke and Sachs, 1994).
Figure 3.
Southern-blot analysis of eno1 and eno2 in the maize genome. Genomic DNA from an inbred maize line (B73) was digested to completion with the restriction enzymes indicated above the lanes and analyzed as described in Methods. A, Genomic hybridization pattern unique to pENO1. B, Genomic hybridization pattern unique to pENO2. Size markers (MW; 1-kb DNA ladder, Promega) are shown at the left of each panel.
N-Terminal Protein Sequence Analysis
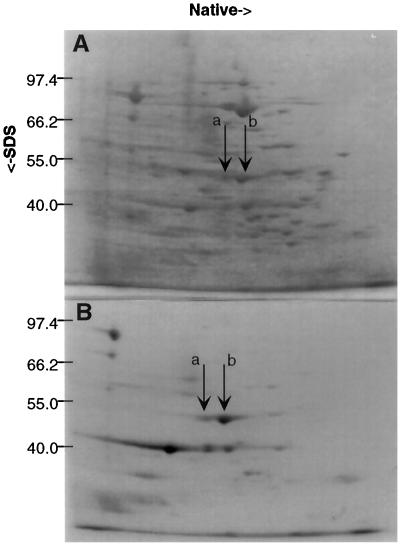
To identify some of the unknown ANPs (Sachs et al., 1980), protein spots from a two-dimensional gel (Fig. 4) were isolated and subjected to N-terminal sequencing. Two major anaerobic protein spots identified in previous work as ANP45A and ANP45B (Sachs et al., 1980) were first visualized with Coomassie blue staining (Fig. 4), and were then excised separately and subjected to N-terminal sequencing as described in Methods. The sequence of the first 12 N-terminal amino acid residues of both protein spots was MAVTITWVKARQ. Using BLAST, this sequence was also found to be identical to the N-terminal deduced amino acid sequence of maize ENO1 (Lal et al., 1991).
Figure 4.
Native two-dimensional SDS-PAGE and autoradiography. Extracts from maize seedling roots treated anaerobically for 70 h and labeled with 35S. A, Coomassie blue-stained native two-dimensional SDS-PAGE gel. ANP45A (a) and ANP45B (b) spots, indicated by arrows, were excised from similar gels and subjected to N-terminal sequencing. B, Autoradiograph of the gel shown in A.
Western-Blot Analysis
To confirm that ANP45A and ANP45B are enolase isozymes, proteins from maize seedling roots subjected to aerobic conditions and labeled with 35S for 1 h aerobically or to anaerobic stress of 15, 45, and 70 h for the last 15 h of treatment were resolved by native two-dimensional SDS-PAGE (Sachs et al., 1980). The gels were then subjected to western-blot analysis using antibodies against maize enolase. The blot was subsequently exposed to radiographic film to visualize proteins synthesized during the treatments.
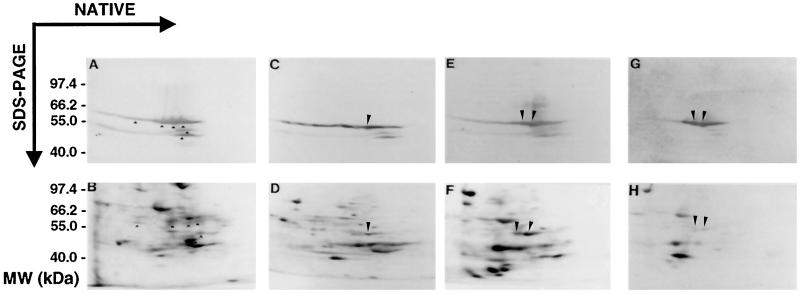
This antisera, apart from recognizing ANP45A and ANP45B, clearly cross-reacted with a set of polypeptides of the same apparent molecular mass, which are separable from one another in the native dimension. Furthermore, the enolase antibodies also detected two additional size classes of proteins. One, with an apparent mass of approximately 43.5 kD, was detectable in the aerobic sample and rapidly decreased during early anaerobic treatment. The other, with an apparent mass of approximately 42 kD, decreased gradually during the time course of anaerobic treatment, and was barely detectable by 70 h. Comparison of the autoradiograms with their corresponding western blots indicated that, along with certain other 45-kD proteins, both ANP45A and ANP45B are detected by the enolase antibody (Fig. 5, A and B). However, the only polypeptides detected by enolase antibodies that also appear to be synthesized under anaerobic treatment are ANP45A and ANP45B (Fig. 5, C–G). After 45 h of anaerobic treatment, most of the 45-kD proteins appeared to be degraded, with the exception of ANP45A and ANP45B, which continued to be synthesized during this period. During long-term anoxia (70 h; Fig. 5, G and H), the synthesis of ANP45A and ANP45B was significantly reduced, and the additional 42- and 45-kD proteins recognized by enolase antibodies were reduced to very low or nondetectable levels. Only ANP45A and ANP45B remained detectable by anti-enolase antibodies at this time point.
Figure 5.
Two-dimensional SDS-PAGE and western-blot analysis of in vivo-labeled maize root proteins. Maize root proteins, labeled in vivo either under aerobic (A and B) or anaerobic treatment (C–H), were separated on two-dimensional SDS-PAGE gels and electroblotted onto nitrocellulose filters. This blot was first subjected to western-blot analysis using enolase antibody, and then was subjected to autoradiography. The bottom panels are autoradiograms representing aerobic control (B) and plants anaerobically treated for 15 h (D), 45 h (F), and 70 h (H). The top panels (A, C, E, and G) are the western blots used for the autoradiograms B, D, F, and H, respectively. The proteins recognized by the antibody that corresponds to ANP45A and ANP45B are marked with long arrowheads. The shorter arrowheads in A and B point to cross-reacting proteins that appear to be synthesized under aerobic conditions but not during anoxia. Size markers (MW) are shown on the left (in kilodaltons).
DISCUSSION
In the absence of oxygen, there is an energy crisis caused by the limitation of oxidative phosphorylation of ADP. To survive, most plants must shift their metabolism from the oxidative pathway to a fermentative pathway. This metabolic transition is marked by a dramatic induction at the levels of transcription and translation of a few key fermentation enzymes, such as ADH (Freeling, 1973) and PDC (Kelley, 1989), in maize. In contrast, the expression of many intervening glycolytic enzymes either remains constitutive or is induced only slightly (2-fold or less; Kelley and Freeling, 1984; Bailey-Serres et al., 1988). We previously reported a 5-fold induction of enolase transcript levels and a 2-fold induction of enzyme activity in maize roots during anoxia (Lal et al., 1991). Later studies showed that such induction occurs with no apparent change in enolase protein levels, as is seen during one-dimensional SDS-PAGE western-blot analysis (Lal et al., 1994).
Enolase molecular mass, as judged by SDS-PAGE, differs from that obtained by sequence analysis. Previously, Lal et al. (1991) showed that maize enolase behaved as a doublet of 55 and 56 kD on SDS gels. This differs from the 48-kD mass predicted from the ENO cDNA sequences. Similar discrepancies were also noted for Arabidopsis and tomato enolases (Van Der Straeten et al., 1991). In addition, E. coli strain DF261 (an enolase-deficient mutant) transformed with maize pENO1 produces an enolase migrating with an apparent molecular mass of 56 kD (Lal et al., 1991). These data suggest that the difference between the apparent molecular mass of maize enolase obtained via SDS-PAGE and that based on sequence cannot be attributed to glycosylation.
In vitro phosphorylation of enolase has been demonstrated in vertebrates (Cooper et al., 1984) and in E. phyllopogon both in vitro (Mujer et al., 1995) and in vivo (M.E. Rumpho, personal communication). Although protein folding or posttranslational modification via phosphorylation can affect the mobility of some proteins during electrophoresis, the magnitude of the discrepancy observed with maize enolase appears to be greater than can be accounted for by these factors alone. The migration of the ANPs (now known to be enolase isozymes) as an apparent 45-kD protein during native two-dimensional SDS-PAGE in previous studies (Sachs et al., 1980) and in the present study remains unclear. It is possible that the migration of enolase in the native dimension somehow affects mobility during SDS-PAGE.
In this study western-blot analysis of in vivo-labeled anaerobic root proteins suggests that the other 45-kD proteins recognized by enolase antibodies may represent isozymes of enolase. Upon anoxic treatment the enolase isozymes previously synthesized in an aerobic environment appear to be selectively degraded and replaced with anaerobic-specific enolase isozymes (ANP45A and ANP45B). This could explain why Lal et al. (1994) did not detect any change in the enolase protein levels during one-dimensional SDS-PAGE western-blot analysis. The lower-molecular-mass proteins recognized by enolase antibodies (42 and 43.5 kD) in the present study may represent proteolytic derivatives of enolase protein (Lal et al., 1994), the product(s) of additional constitutive eno genes, or nonenolase determinants present in the antiserum.
The two enolase cDNA clones are clearly encoded by two separate genes. Although the coding regions of pENO1 and pENO2 are highly homologous, they are completely divergent at the 5′- and 3′-untranslated regions. In addition, the cDNAs are derived from RNA of the same inbred maize line, in which the possibility of polymorphism is negligible. The homology between ENO1 and ENO2 in maize is analogous to that of the ADH gene family members adh1 and adh2 (Dennis et al., 1985), which show 82% sequence identity, and that of gpc2 and gpc3 of the cytosolic GAPDH gene family (Russell and Sachs, 1991), which show 80% sequence identity in the coding region. However, the members of these gene families are all completely divergent in the untranslated sequences. The banding patterns observed upon genomic Southern-blot analysis are unique for ENO1 and ENO2 cDNA probes, and confirm that the cDNAs represent different genes.
Restriction-fragment-length polymorphism analysis localizes eno1 to chromosome 9S (Peschke and Sachs, 1994) and eno2 to chromosome 1S (B. Burr, personal communication). Furthermore, the pattern of transcript accumulation during anoxia is different for each gene (Fig. 2). Enolase is encoded by more than one gene in yeast (Holland et al., 1981) and in vertebrates (Giallongo et al., 1986). On the basis of genomic Southern-blot analyses, it was reported that enolase is represented by small gene families in all plants examined except Arabidopsis (Van Der Straeten et al., 1981; Blakeley et al., 1994), E. phyllopogon, and E. crus-pavonis (Fox et al., 1995), in which a single gene encodes enolase. In maize several other genes encoding the enzymes involved in Glc metabolism, such as ADH (Dennis et al., 1985), GAPDH (Russell and Sachs, 1991), and PDC (Peschke and Sachs, 1993), are represented by small gene families.
Fothergill-Gilmore (1986) postulated that the isozyme forms of glycolytic enzymes in different organisms evolved as a result of gene duplication. It is possible that the selection process might result in an isozyme that is more adapted to meet the physiological demands of an organism in a particular environment. In accordance with this postulation, ANP45A and ANP45B might represent enolase isozymes more adapted than the constitutive forms of enolase to serve the anaerobic metabolic requirements of maize, and therefore are selectively synthesized during anaerobic treatment. The other cross-reacting 45-kD proteins may represent enolase isozymes more adapted for normoxic metabolism, and appear to be degraded during prolonged anaerobic treatment.
Recently, Van Der Straeten et al. (1991) reported that enolase is not induced during the anaerobic stress response of Arabidopsis and tomato, although an ARE sequence (the putative anaerobic responsive element reported to be essential for anaerobic induction of adh1; Walker et al. [1987]) was present in the first intron of the gene encoding Arabidopsis enolase. Our data clearly demonstrate that in maize, anaerobic stress induces the expression of ENO1, whereas ENO2 is constitutive. The anaerobic expression of eno1 and eno2 is similar to that observed for gpc3/gpc4 versus gpc1/gpc2 of the maize GAPDH gene family. The anaerobic expression of gpc1 and gpc2 is constitutive, whereas gpc3 and gpc4 are induced by oxygen deprivation (Russell and Sachs, 1991). In contrast, both genes encoding ADH are anaerobically induced (Freeling, 1973), although only adh1 is essential for anaerobic tolerance in maize (Schwartz, 1969; Dlouhy, 1980; Lemke-Keyes and Sachs, 1989). Differential anaerobic expression at the level of transcription has also been reported for the two genes encoding Suc synthase (Springer et al., 1986; McElfresh and Chourey, 1988). In addition, the three genes encoding PDC are induced with varied kinetics during anoxia in maize (Peschke and Sachs, 1994).
Differential regulation of two enolase genes in response to the carbon source in the medium and growth phase has been well documented in yeast (McAlister and Holland, 1982). Uemera et al. (1986) observed that differential expression of two yeast enolase genes in response to the carbon source in the medium is regulated at the level of transcription. In vertebrates three genes encoding isozymes of enolase exhibit tissue-specific and developmental regulation (Giallongo et al., 1986; Wistaw et al., 1988). The lens-crystalline structural protein in vertebrates has been found to be α-enolase, which serves a protective function (Wistaw et al., 1988). In yeast a major heat-shock protein, HSP48, has been identified as an isozyme of enolase (Iada and Yahara, 1985), suggesting that enolase may play an important role in the thermal tolerance of this organism. A major anaerobic stress protein (ASP55) in the flood-tolerant E. crus-pavonis was also identified as enolase, and has been suggested to play a significant role in the acquisition of flooding tolerance in this plant (Zhang et al., 1994; Fox et al., 1995).
Protein-sequencing data suggest that ANP45A and ANP45B may represent the products of two different genes with identical N-terminal amino acid sequences. Alternatively, these two polypeptides might result from posttranslational modification of a single enolase protein. The regulation of enolase in plants has been proposed to be complex and, apart from transcription and translation, may involve posttranslational modifications (Van Der Straeten et al., 1991; Lal et al., 1994; Fox et al., 1995; Mujer et al., 1995). The Tyr-46 residue of vertebrate α-enolase has been shown to be phosphorylated in vitro (Eigenbrodt et al., 1983; Cooper et al., 1984). This residue and the surrounding regions are highly conserved among enolases from different species, including maize enolase (Lal et al., 1991; Van Der Straeten et al., 1991). In addition, a novel protein kinase (APK1) isolated from Arabidopsis is capable of phosphorylating several different proteins, including enolase (Hirayama and Oka, 1992).
Recently, enolase from E. phyllopogon was shown to be phosphorylated in vitro (Mujer et al., 1995) and in vivo (M.E. Rumpho, personal communication). Phosphorylation of the protein occurs under aerobic conditions and dephosphorylation occurs after exposure to anaerobic conditions. This corresponds to an increase in enolase activity during anoxia in E. phyllopogon (M.E. Rumpho, personal communication). Further investigation is required to determine if the anaerobic expression of maize enolase involves phosphorylation or other posttranslational modifications. We also anticipate that analysis of the promoter regions of eno1 and eno2 will help us understand what factors may be involved in the selective induction of genes involved in Glc-phosphate metabolism by anoxia.
ACKNOWLEDGMENTS
We thank Dr. D. Bhattramakki for his efforts and help with the Southern-blot analysis shown in Figure 3; Drs. D.T. Dennis, and K.P. Cole (Queen's University, Kingston, Ontario, Canada) for generously providing pZM245 (pENO1) antibodies; Dr. M. Van Montagu (Laboratorium voor Genetica, Gent, Belgium) for generously providing the tomato enolase cDNA clone; Dr. K.-L. Ngai (University of Illinois, Genetic Engineering Facility) for helping with the N-terminal sequencing of ANP45A and ANP45B and with cDNA sequencing of pENO2; Dr. B. Burr (Brookhaven National Laboratory) for restriction fragment-length polymorphism mapping analysis of eno2; and Drs. C.C. Subbaiah, I.N. Saab, P. Chourey, and T.E. Elthon for critical reading of the manuscript.
Abbreviations:
- ADH
alcohol dehydrogenase
- ANP
anaerobic polypeptide
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- PDC
pyruvate decarboxylase
Footnotes
This work was supported by a grant from the U.S. Public Health Service (National Institutes of Health no. 5 R01 GM34740), and by U.S. Department of Agriculture/Agricultural Research Service funds awarded to M.M.S.
Names are necessary to report factually on available data; however, the U.S. Department of Agriculture neither guarantees nor warrants the standard of the product, and the use of the name by the U.S. Department of Agriculture implies no approval of the product to the exclusion of others that may also be suitable.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Kloeckener-Gruissem B, Freeling M. Genetic and molecular approaches to the study of the anaerobic response and tissue specific gene expression in maize. Plant Cell Environ. 1988;11:351–357. [Google Scholar]
- Blakeley SD, Dekroon C, Cole KP, Kraml M, Dennis DT. Isolation of a full-length cDNA encoding cytosolic enolase from Ricinus communis. Plant Physiol. 1994;105:455–456. doi: 10.1104/pp.105.1.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boser H. Zur heterogenitat von enzymen. I. uber enolase aus kartoffelknollen. Z Physiol Chem. 1959;315:163–170. doi: 10.1515/bchm2.1959.315.1.163. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Esch FS, Taylor SS, Hunter T. Phosphorylation sites in enolase and lactate dehydrogenase utilized by tyrosine protein kinase in vivo and in vitro. J Biol Chem. 1984;259:7835–7841. [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version 2. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Dennis ES, Sachs MM, Gerlach WL, Finnegan EJ, Peacock WJ. Molecular analysis of the Alcohol dehydrogenase 2 (Adh2) gene of maize. Nucleic Acids Res. 1985;13:727–743. doi: 10.1093/nar/13.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlouhy SR (1980) Biochemical and physiological analyses involving the Adh2 locus of Zea mays. PhD thesis. Indiana University, Bloomington
- Eigenbrodt E, Fister P, Rubsamen H, Friis R. Influence of transformation by Rous sarcoma virus on the amount, phosphorylation and enzyme kinetic properties of enolase. EMBO J. 1983;2:1565–1570. doi: 10.1002/j.1460-2075.1983.tb01625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel NR, Cushman MAF, Cushman JC. Posttranscriptional and posttranslational control of enolase expression in the facultative Crassulacean acid metabolism plant Mesembryanthemum crystallinum L. Plant Physiol. 1995;108:1185–1195. doi: 10.1104/pp.108.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill-Gilmore LA. The evolution of the glycolytic pathway. Trends Biochem Sci. 1986;11:47–51. [Google Scholar]
- Fox TC, Mujer CV, Andrews DL, Williams AS, Cobb BG, Kennedy RA, Rumpho ME. Identification and gene expression of anaerobically induced enolase in Echinochloa phyllopogon and Echinochloa crus-pavonis. Plant Physiol. 1995;109:433–443. doi: 10.1104/pp.109.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. Simultaneous induction by anaerobiosis or 2,4-D of multiple enzymes specified by two unlinked genes: differential Adh1-Adh2 expression in maize. Mol Gen Genet. 1973;127:215–227. doi: 10.1007/BF00333761. [DOI] [PubMed] [Google Scholar]
- Giallongo A, Feo S, Moore R, Croce CM, Showe LC. Molecular cloning and nucleotide sequence of a full length cDNA for human α-enolase. Proc Natl Acad Sci USA. 1986;83:6741–6745. doi: 10.1073/pnas.83.18.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Kelley PM, Taylor WC, Freeling M. Coordinate induction of alcohol dehydrogenase 1, aldolase, and other anaerobic RNAs in maize. J Biol Chem. 1985;260:5050–5054. [PubMed] [Google Scholar]
- Hirayama T, Oka A. Novel protein kinase of Arabidopsis thaliana (APK1) that phosphorylates tyrosine, serine and threonine. Plant Mol Biol. 1992;20:653–662. doi: 10.1007/BF00046450. [DOI] [PubMed] [Google Scholar]
- Holland MJ, Holland JP, Thill GP, Jackson KA. The primary structure of two yeast enolase genes: homology between the 5′ noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981;256:1385–1395. [PubMed] [Google Scholar]
- Iada H, Yahara I. Yeast heat-shock protein of MR 48,000 is an isoprotein of enolase. Nature. 1985;315:688–690. [Google Scholar]
- Kelley PM. Maize pyruvate decarboxylase mRNA is induced anaerobically. Plant Mol Biol. 1989;13:213–222. doi: 10.1007/BF00016139. [DOI] [PubMed] [Google Scholar]
- Kelley PM, Freeling M. Anaerobic expression of maize glucose phosphate isomerase. J Biol Chem. 1984;259:673–677. [PubMed] [Google Scholar]
- Kelley PM, Tolan DR. The complete amino acid sequence for the anaerobically induced aldolase from maize derived from cDNA clones. Plant Physiol. 1986;82:1076–1080. doi: 10.1104/pp.82.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal SK, Elthon TE, Kelley PM. Purification and differential expression of enolase from maize. Physiol Plant. 1994;91:587–592. [Google Scholar]
- Lal SK, Johnson S, Conway T, Kelley PM. Characterization of a maize cDNA that complements an enolase-deficient mutant of Escherichia coli. Plant Mol Biol. 1991;16:787–795. doi: 10.1007/BF00015071. [DOI] [PubMed] [Google Scholar]
- Lal SK, Sachs MM. Cloning and characterization of an anaerobically induced cDNA encoding glucose-6-phosphate isomerase in maize (Zea mays L.) Plant Physiol. 1995;108:1295–1296. doi: 10.1104/pp.108.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebioda LB, Stec B, Brewer JM. The structure of yeast enolase at 2.25 angstrom resolution: an 8-fold β + α barrel with a novel ββαα(βα)6 topology. J Biol Chem. 1989;264:3685–3693. doi: 10.2210/pdb2enl/pdb. [DOI] [PubMed] [Google Scholar]
- Lemke-Keyes CA, Sachs MM. Genetic variation for seedling tolerance to anaerobic stress in maize germplasm. Maydica. 1989;34:329–337. [Google Scholar]
- Lending CR, Kriz AL, Larkins BA, Bracker CE. Structure of maize protein bodies and immunocytochemical localization of zeins. Protoplasma. 1988;143:51–62. [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membrane. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- McAlister L, Holland MJ. Targeted deletion of a yeast enolase structural gene: identification and isolation of a yeast enolase isozyme. J Biol Chem. 1982;257:7181–7188. [PubMed] [Google Scholar]
- McElfresh KC, Chourey PS. Anaerobiosis induces transcription but not translation of sucrose synthase in maize. Plant Physiol. 1988;87:542–546. doi: 10.1104/pp.87.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA, Dennis DT. Enolase isozymes from Ricinus communis: partial purification and characterization of the isozymes. Arch Biochem Biophys. 1984;233:643–651. doi: 10.1016/0003-9861(84)90490-9. [DOI] [PubMed] [Google Scholar]
- Miernyk JA, Dennis DT. A developmental analysis of the enolase isozymes from Ricinus communis. Plant Physiol. 1992;99:748–750. doi: 10.1104/pp.99.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujer CV, Fox TC, Williams AS, Andrews DL, Kennedy RA. Purification, properties and phosphorylation of anaerobically induced enolase in Echinochloa phyllopogon and E. crus-pavonis. Plant Cell Physiol. 1995;36:1459–1470. doi: 10.1104/pp.109.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke VM, Sachs MM. Multiple maize pyruvate decarboxylase genes are induced by hypoxia. Mol Gen Genet. 1993;240:206–212. doi: 10.1007/BF00277058. [DOI] [PubMed] [Google Scholar]
- Peschke VM, Sachs MM. Characterization and expression of transcripts induced by oxygen deprivation in maize (Zea mays L.) Plant Physiol. 1994;104:387–394. doi: 10.1104/pp.104.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Sachs MM. Differential expression and sequence analysis of the maize glyceraldehyde-3-phosphate dehydrogenase gene family. Plant Cell. 1989;1:793–803. doi: 10.1105/tpc.1.8.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Sachs MM. The maize cytosolic glyceraldehyde-3-phosphate dehydrogenase gene family: organ-specific expression and genetic analysis. Mol Gen Genet. 1991;229:219–228. doi: 10.1007/BF00272159. [DOI] [PubMed] [Google Scholar]
- Sachs MM, Freeling M, Okimoto R. The anaerobic proteins of maize. Cell. 1980;20:761–767. doi: 10.1016/0092-8674(80)90322-0. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. An example of gene fixation resulting from selective advantage in suboptimal conditions. Am Nat. 1969;103:479–481. [Google Scholar]
- Sinha S, Brewer JM. Purification and comparative characterization of an enolase from spinach. Plant Physiol. 1984;74:834–840. doi: 10.1104/pp.74.4.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B, Werr W, Starlinger P, Bennett DC, Zokolica M, Freeling M. The Shrunken gene on chromosome 9 of Zea mays L. is expressed in various plant tissues and encodes an anaerobic protein. Mol Gen Genet. 1986;205:461–468. doi: 10.1007/BF00338083. [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Zhang JK, Sachs MM. Involvement of intracellular calcium in anoxic gene expression and survival in maize. Plant Physiol. 1994;105:369–376. doi: 10.1104/pp.105.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon G. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemera J, Shiba T, Paterson M, Jigami Y, Tanaka H. Identification of a sequence containing the positive regulatory region of Saccharomyces cerevisiae gene ENO1. Gene. 1986;45:67–75. doi: 10.1016/0378-1119(86)90133-2. [DOI] [PubMed] [Google Scholar]
- Van Der Straeten D, Rodrigues-Pousada RA, Goodman HM, Van Montagu M. Plant enolase: gene structure, expression, and evolution. Plant Cell. 1991;3:719–735. doi: 10.1105/tpc.3.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC, Haward EA, Dennis ES, Peacock J. DNA sequences required for anaerobic expression of maize alcohol dehydrogenase 1 gene. Proc Natl Acad Sci USA. 1987;84:6623–6628. doi: 10.1073/pnas.84.19.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wistaw GJ, Lietman T, Williams LA, Stapel SO, de Jong WW, Horwitz J, Piatigorsky J. t-Crystallin/α-enolase: one gene encodes both an enzyme and a lens structural protein. J Cell Biol. 1988;107:2729–2736. doi: 10.1083/jcb.107.6.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Lin J-J, Fox TC, Mujer CV, Rumpho ME, Kennedy RA. Effect of aerobic priming on the response of Echinochloa crus-pavonis to anaerobic stress. Plant Physiol. 1994;105:1149–1157. doi: 10.1104/pp.105.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]