Abstract
Protective antigen (PA), lethal factor, and edema factor, the protein toxins of Bacillus anthracis, are among its most important virulence factors and play a key role in infection. We performed a virtual ligand screen of a library of 10,000 members to identify compounds predicted to bind to PA and prevent its oligomerization. Four of these compounds slowed PA association in a FRET-based oligomerization assay, and two of those protected cells from intoxication at concentrations of 1–10 μM. Exploration of the protective mechanism by Western blot showed decreased SDS-resistant PA oligomer on cells, and surprisingly, decreased amounts of activated PA. In vitro assays showed that one of the inhibitors blocked furin-mediated cleavage of PA, apparently through its binding to the PA substrate. Thus, we have identified inhibitors that can independently block both PA’s cleavage by furin and its subsequent oligomerization. Lead optimization on these two backbones may yield compounds with high activity and specificity for the anthrax toxins.
INTRODUCTION
Bacillus anthracis is a spore-forming, Gram-positive bacterium that causes anthrax in livestock and humans 1. The bacterium has two main virulence factors, a poly-γ-linked-D-glutamic acid capsule that protects the bacterium from phagocytosis, and the exotoxin protein complex consisting of Protective Antigen (PA), Lethal Factor (LF), and Edema Factor (EF), which act collectively to suppress the host’s innate immune system 2–5. The triad of PA, LF, and EF act as an A2B-type toxin, where only AB combinations are toxic 6, 7. The B-part of the toxin is PA, an 83-kDa cellular-binding protein (PA83), and the alternative A-part catalytic moieties are LF and EF. LF is a 91-kDa zinc-metalloprotease that cleaves rat Nlrp1 and the N-terminal substrate docking site of the mitogen-activated protein kinase kinases (MAP2K) 1, 2, 3, 4, 6, and 7, preventing passage of signals in the ERK1/2, p38, and c-Jun N-terminal kinase pathways 8–11, while EF is an 89-kDa calmodulin-dependent adenylate cyclase that raises cytosolic levels of cAMP, activating protein kinase A 12,13. The AB combinations of LF/PA or EF/PA are known as lethal toxin (LT) and edema toxin (ET), respectively, and are responsible for the symptoms of anthrax. Injection of purified toxins has been shown to produce many of the symptoms seen in infected mice 3. Infection leads to a toxemia in humans and experimental animals, rendering antibiotic therapies of limited value in later stages of infection.
Intoxication of a cell begins with PA83 binding to one of the two receptors, capillary morphogenesis protein 2 (CMG2) or tumor endothelial marker 8 (TEM8) 14–16. Once bound, furin, or a related protease, cleaves a 20-kDa fragment from the N-terminus of PA83, giving the active, 63-kDa protein PA63. Following activation, PA63 forms an oligomer and binds 3–4 molecules of EF or LF 2,17,18. Additionally, EF and LF have been shown to drive oligomerization of PA63 in vitro and in vivo 2. The toxin is taken in by clathrin-mediated endocytosis and a decrease in endosomal pH causes a conformational change in PA that induces formation of an endosomal membrane channel through which LF or EF translocate to the cytosol19.
In the intoxication pathways for LT and ET, there are at least seven steps where inhibitors can block the toxins from affecting cellular processes. Potential therapeutic agents have been described that block intoxication at almost every step. Cellular binding by PA (1) can be prevented by the monoclonal antibody 14B7, which binds to the receptor-binding domain of PA and removes PA from circulation, and soluble TEM8 and CMG2 decoy proteins that compete with the membrane-bound proteins for binding 20, 21. Furin cleavage of PA (2) can be prevented by small molecule or peptide-based furin inhibitors 22–25, and small molecule inhibition of PA oligomerization (3) was explored in a recent report, but the mechanism of action of the compounds was not well characterized 26. Cisplatin has been shown to covalently modify and inactivate PA 27. Heptavalent small molecules that fit into the pore of oligomerized PA prevent cellular intoxication by blocking effector protein binding (4) 28–31 and endocytosis (5) of the PA oligomer complex can be prevented by using genistein to block the activation of src-like kinases, which play a role in toxin uptake 32. Translocation of the toxins from endosomes (6) can be prevented in vitro using ammonium chloride or proton pump inhibitors, including bafilomycin A1, but this approach is unlikely to be used in in vivo because such agents are toxic to cells 33. While inhibition of the first six steps is general to both toxins, inhibition of the catalytic activities (7) is specific to LT or ET. Both LF-specific protease inhibitors 34–38 and EF-specific inhibitors 39, 40 have been developed. The majority of work on inhibition of catalytic activity has concentrated on LF; however, the role of EF in patient morbidity became clear during the outbreak of anthrax among IV-drug users in the UK in 2010, and blockage of EF by monoclonal antibodies significantly delayed time to death in mice in a spore model of anthrax infection 41, 42.
Targeting the oligomerization step of intoxication has several benefits, including the fact that it would be effective against both LF and EF. Oligomerization blockage may also offer better specificity of effect than some approaches since the molecules are designed to interact specifically with PA and would not interact with any host proteins. Another benefit of this approach is that small molecules can be orally bioavailable, streamlining their administration. For these reasons, we identified several small molecules that were predicted in silico to bind to a pocket on an oligomerization face of PA and showed that they have efficacy in preventing pore formation and cell death.
RESULTS
In silico screening, and mapping of lead compounds to the PA structure
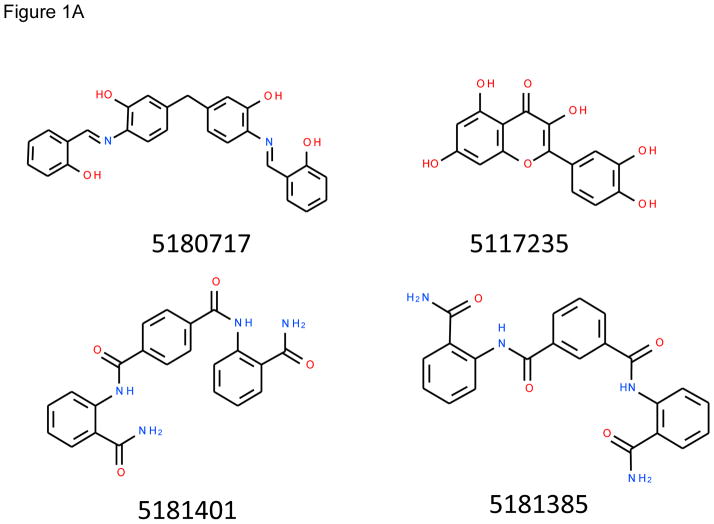
This work sought to identify small molecule inhibitors that prevent assembly of proteolytically-activated PA monomers into the oligomeric, functional PA channel. Application of the ICMPocketFinder method 43, 44 in the ICM-Pro software (MolSoft, San Diego, CA) to the PA monomer (1T6B) and heptamer (1TZO) structures available when this work began identified three potentially targetable pockets on the monomer interfaces. Screenings of a Chembridge 10,000-member library against each of the three pockets of the monomer and oligomer structures produced six rankings of the library members. Comparisons of the predicted binding strengths of high-scoring members of the separate rankings led us to focus on the pocket located closest to the furin loop (Figure 1B). Forty-two compounds ranking highly against this pocket in the monomer, oligomer, or both screens were selected for further analysis. Of these, the fifteen that were soluble at 100 mM in DMSO were tested experimentally, leading to identification of four compounds that showed activity in at least one subsequent assay. These compounds, Chembridge library members 5180717, 5181401, 5181385, and 5117235 (Figure 1A), are referred to in the text and figures below by the last two numbers of the full designations, 17, 01, 85 and 35 respectively. These compounds had rankings in the in silico screen against the monomer structure of 53, 4, 10, and 16, respectively (see Supplementary Data for additional details).
Figure 1.
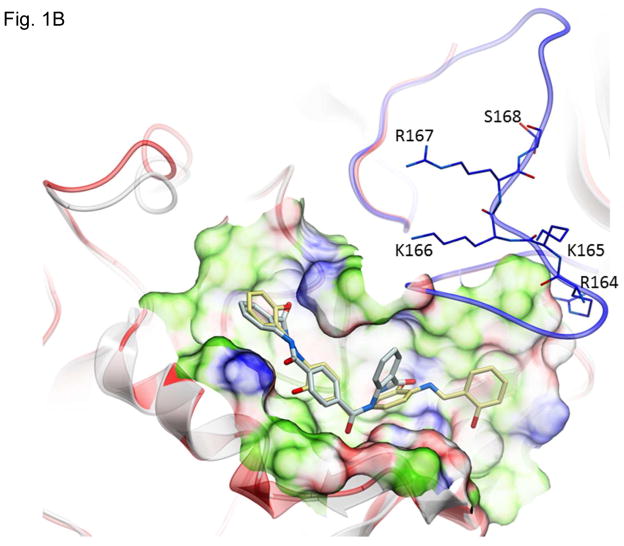
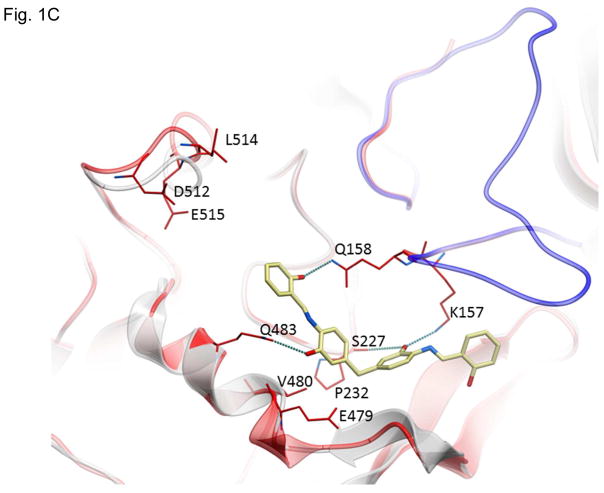
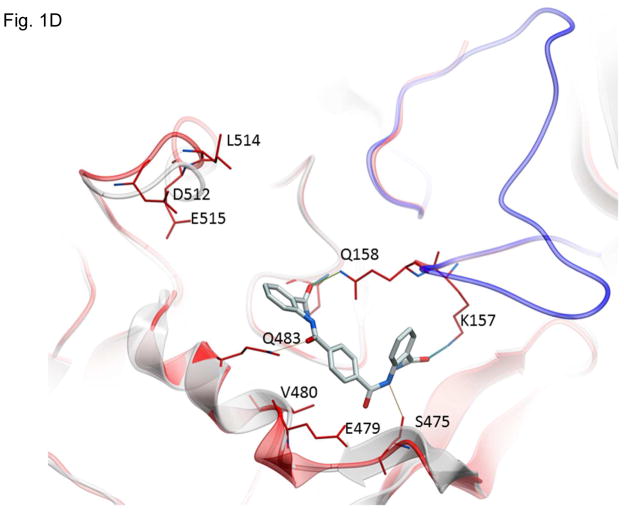
Structures of inhibitors and modes of binding to PA. (A) The compounds characterized in this study. (B) PA crystal structure 1T6B (red ribbon) superimposed on the crystal structure 3TEW (grey ribbon) with the ordered furin loop in 3TEW highlighted in blue. The furin-type protease cleaves after the sequence 164RKKR which is shown in stick representation with carbon atoms colored blue. The predicted binding poses for inhibitors 17 and 01 are displayed in stick representation with carbon atoms colored yellow and grey, respectively. The binding pocket surface for 1T6B used for virtual screening is displayed (White = neutral surface, Green = hydrophobic surface, Red = hydrogen bonding acceptor potential, Blue = hydrogen bond donor potential). (C) and (D) Predicted interactions of 17 (yellow stick) and 01 (grey stick), respectively, with PA. Hydrogen bonds are displayed as small colored spheres and both ligands make common hydrogen bonds with Q158, Q483, and K157.
The pocket targeted by these compounds is located in the vicinity of the furin loop at a site where a loop from one monomer (residues 462–469) protrudes into domains 2 and 3 of a second monomer (Figure 1B). The predicted binding poses of inhibitor 17 and 01, which are most active in cell culture assays (see below), are shown in Figure 1B. Inhibitor 17 (backbone in yellow) is predicted to extend further out of the pocket towards the furin loop compared to inhibitor 01 (backbone in grey). Both ligands make a similar hydrogen bond network with side-chains of PA residues Q158, Q483, and K157, while inhibitor 17 makes an additional side-chain hydrogen bond with S475 (Figure 1C and D). The pocket is lined by residues E479-Q483 and D512-E515, which have been shown experimentally to be important for stabilizing heptamerization 45, 46.
Inhibition of PA oligomerization in vitro
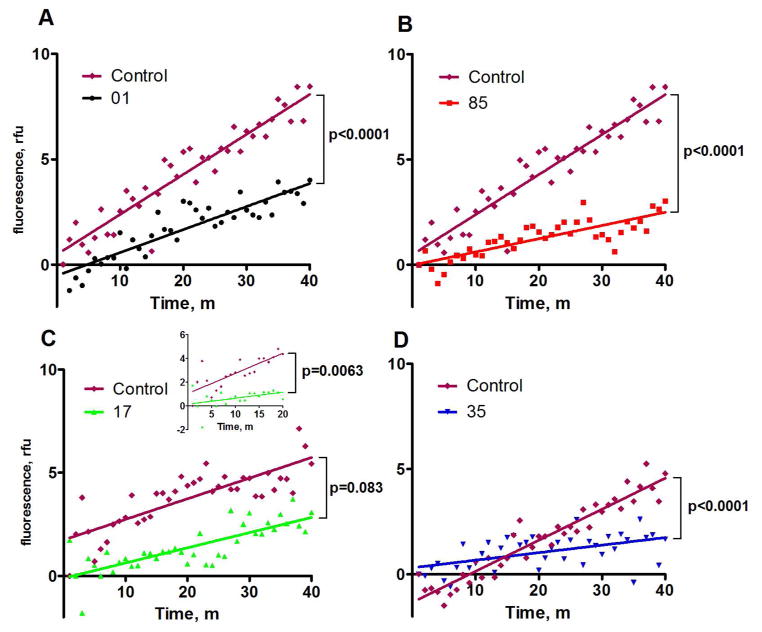
To validate hits identified by virtual ligand screening, we developed a FRET-based assay which was a modification of a method used previously to measure kinetic characteristics of assembly of anthrax toxin complexes 45, 47 . Assembly of oligomers was monitored using FRET between Alexa Fluors 488 and 594 conjugated to PA-N645C, which is a functional toxin mutant 48. The structure of PA shows that N645 is on the surface and is not buried by oligomerization. The progress of oligomerization was monitored using the FRET emission at 610 nM, indicating association of PA into higher-order species. Fifteen compounds selected as described above were tested experimentally using the FRET assay. Four of the compounds inhibited PA oligomerization, which was defined as FRET-quenching compared to a no-drug control (Figure 2). Over a 40-min period, compounds 01, 85, and 35 were able to slow the association of PA significantly compared to a no-inhibitor control. Inhibitor 17 was able to slow oligomerization significantly over a 20-min period, but not a 40-min timeframe (Figure 2C).
Figure 2.
FRET assays demonstrating inhibition of PA oligomerization. The dye-conjugated, nicked PA proteins nPA83 N645C*488 and nPA83 N645C*594 were added to a 96-well plate at 250 nM each. Inhibitors were added to a concentration of 100 μM to all wells except the control and oligomerization was driven by the addition of LF at a final concentration of 50 nM. Fluorescence at 610 nm was monitored in wells containing nPA83 and tested compounds in the presence of 50 nM LF. Plotted fluorescence is the difference in fluorescence of wells containing identical mixtures with and without inhibitors.
Inhibition of PA-induced cytotoxicity
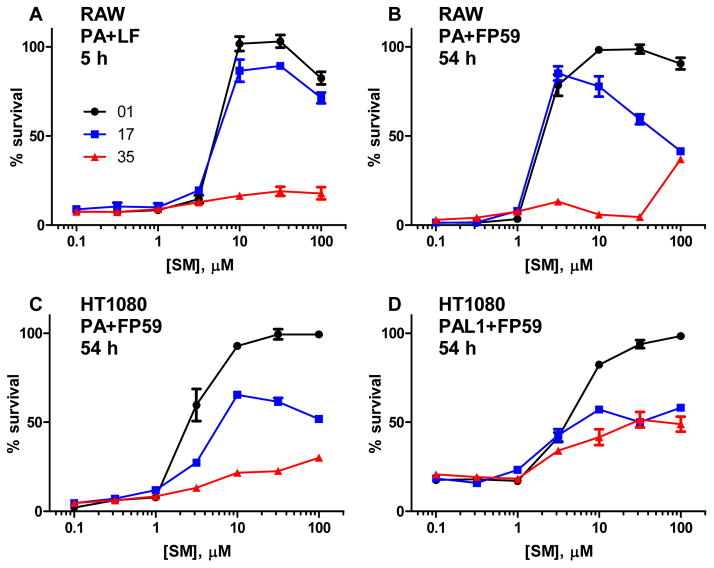
To determine whether the inhibition of PA oligomerization observed by FRET also occurred on cell-bound PA, leading to decreased toxicity, RAW264.7 macrophages were treated with PA along with either LF or the fusion protein FP59, which consists of the N-terminal PA-binding domain of LF fused to the catalytic domain of Pseudomonas exotoxin A, and varying concentrations of inhibitors 01, 17, and 35. Inhibitor 85 was included in initial testing, but became unavailable during the course of the project. HT1080 fibroblasts were treated similarly with PA or the matrix metalloprotease-activated PA-L1 and FP59 to test the generality of the protection afforded by these inhibitors. Inhibitors 01 and 17 were able to protect in all four cytotoxicity assays, with EC50 values in the range of 1–10 μM, while inhibitor 35 showed only modest protection at the highest concentrations (Figure 3). RAW264.7 cells were protected from LT almost completely by inhibitors 01 and 17 at concentrations as low as 10 μM, with inhibitor EC50 values in the range of 3–10 μM. Inhibitor 85 showed similar activity to 01 in initial testing (data not shown). The RAW264.7 cells were protected against treatment with PA + FP59 by concentrations of compounds 01 and 17 as low as 3 μM. HT1080 fibroblasts are not sensitive to lethal toxin, but can be killed by PA or PA-L1 in combination with FP59. Inhibitors 01 and 17 were able to protect the cells from PA at concentrations in the range of 3–10 μM, in the same range as was seen with the RAW macrophages. In both cell lines, inhibitor 17 had some toxicity at high concentrations (10–100 μM) and extended incubation times, whereas inhibitor 01 had little toxicity. None of the compounds were protective in the presence of fetal bovine serum (data not shown), possibly because the serum albumin bound these rather hydrophobic chemicals.
Figure 3.
Protection of RAW264.7 and HT1080 cells by small molecule toxin inhibitors. RAW264.7 cells were exposed to 0.9 nM PA with 5.6 nM LF (A) or 0.06 nM PA with 1.88 nM FP59 (B) and HT1080 cells were exposed to 0.6 nM PA (C) or PA-L1 (D) with 1.88 nM FP59 for 5 h. The PA + LF cells were stained with MTT immediately (A) while the PA + FP59 and PA-L1 + FP59 cells (B-D) were treated with 10 mM ammonium chloride after 5 h to stop intoxication and MTT stained 48 h later. Data are plotted as mean +/- standard error of four independent experiments. Percent survival is compared to a non-intoxicated control.
Inhibition of PA binding and processing
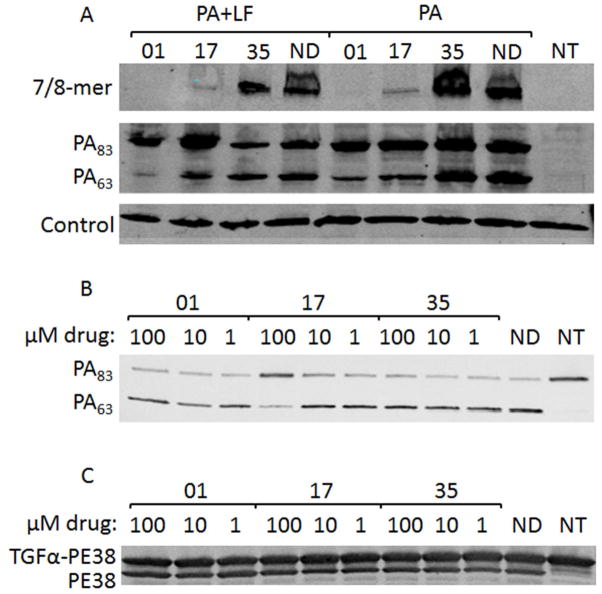
To characterize the mechanism of protection, Chinese hamster ovary cell line C4, which overexpresses the CMG2 receptor, was treated with PA alone, PA + LF or PA-U7 + LF (PA-U7 is not cleaved by any protease) and inhibitor or vehicle for 1 h and the binding, processing and oligomerization of PA was examined by Western blot. In this process, the conformational change associated with insertion of the PA oligomer into the membrane of the acidified endosome makes it resistant to SDS and boiling 33. As expected from the cytotoxicity inhibition results, inhibitor 35 showed no difference in relative levels of PA83, PA63, and SDS-resistant oligomer compared to a no-drug treatment (Figure 4). The marked decrease in formation of oligomer seen with 01 and 17 compared to both 35 and no drug correlates to the protection seen in RAW264.7 and HT1080 assays. Inhibitor 01 completely blocked oligomer formation while 17 greatly decreased oligomer formation. Both of these findings are in agreement with the cytotoxicity data in indicating that 01 shows slightly higher levels of protection than 17. There were no differences in the amounts of PA83 present in the PA, PA + LF, or PA-U7 + LF treatments, indicating that the inhibitors do not affect binding of the toxin to C4 cells (Figure 4 and data not shown). Surprisingly, treatment with 01 and 17 showed reduced levels of PA63 as compared to the other treatments. This was suggestive of an inhibition of furin action on PA.
Figure 4.
Assessment of PA binding and oligomerization to cells, and in vitro furin activity. (A) CHO C4 cells were treated with PA or PA + LF for 1 h in the presence or absence of 100 μM inhibitor and the cells were lysed and analyzed by Western blotting for PA species. (B, C) Purified PA (B) or TGFα-PE38 (C) were mixed with furin at a 100:1 molar ratio and incubated for 1 h with the indicated concentrations of inhibitors. Wells labeled ND contained no drug, and wells labeled NT were not treated with toxins in (A) or not treated with furin in (B and C).
Cell-free inhibition of PA83 cleavage by furin
To begin to elucidate the mechanism of decreased PA cleavage in the presence of inhibitors, purified PA and TGFα-PE38 (TGFα fused to PE38 by a furin-cleavable RKKR linker) were combined with purified furin in the presence and absence of varying amounts of the inhibitors. The highest concentration of inhibitor 17 reduced the extent of furin cleavage of PA83 (Figure 4B). This concentration represents a 1:1000 ratio of PA83 to drug. No differences in PA cleavage were evident in any of the other treatments. To evaluate whether the inhibitors acted directly on furin rather than through binding to PA, the cleavage of a different substrate was measured (Figure 4C). In this case, no inhibition of furin cleavage of TGFα-PE38 was observed.
DISCUSSION
To identify small molecule inhibitors of anthrax toxin oligomerization, a virtual ligand screen was performed and 15 compounds were selected for characterization. The VLS hits were screened by a FRET assay using PA63 conjugated to fluorophores, in which PA63 oligomerization was driven by addition of LF. In this assay, compounds 5180717 (17), 5117235 (35), 5181385 (85), and 5181401 (01) were found to quench the FRET signal. These four lead compounds were all screening hits to the pocket located next to the furin-loop in the monomer crystal structure (PDB: 1T6B). Compounds 17 and 01 were able to protect RAW264.7 macrophages from PA + LF and PA + FP59 treatments and HT1080 fibroblasts from both PA + FP59 and PA-L1 + FP59 treatments. Compound 85 was excluded from the majority of this study because it was unavailable in sufficient quantities for testing. However, compound 85 may merit further study because 85 and 01 are isomers (isophthalamide and terephthalamide), showed similar activity in the RAW264.7 PA + LF assay in initial testing, and their poses in the PA pocket are similar (data not shown). The two lead compounds identified (01 and 17) are able to prevent anthrax protective antigen from being cleaved by furin or forming an oligomer following cleavage. Though the difference in EC50 values between the two compounds is negligible in all cytotoxicity assays, compound 01 showed higher protection at saturating concentrations. Inhibitor 17 was mildly toxic or growth inhibitory to RAW264.7 cells in a 5-h exposure and moderately toxic or growth inhibitory to both RAW264.7 cells and HT1080 fibroblasts over a 54-h incubation, even when the inhibitor was removed from the medium following the 5-h exposure. Unfortunately, the MTT assay used here cannot distinguish between growth inhibition and toxicity. By Western blot analysis, compounds 01 and 17 prevented furin cleavage of PA on the surface of intoxicated cells and also decreased the amount of SDS-resistant oligomer present in the cell. Finally, compound 17 prevented cleavage of purified PA by purified furin in a cell-free assay. Compounds 01 and 17 are not general inhibitors of furin since they did not prevent cleavage of TGFα-PE38 by furin.
The most surprising result of this work is the ability of compounds 01 and 17 to greatly reduce the rate of furin cleavage of PA. While the compounds were not designed to inhibit this step of intoxication, retrospective analysis of the crystal structure revealed the proximity of the druggable pocket to the furin loop. Several possible mechanisms could explain this effect. The compounds may sterically prevent furin from interacting with PA, in the same way that they inhibit PA-PA interaction. Thus, inhibition would be seen if solvent-accessible side chains on furin must interact with some of the same PA residues to which the drugs bind, i.e., E479-Q483 and D512-E515. Another possible mechanism is that furin binds to PA, but is unable to achieve a conformation suitable for catalytic activity due to the presence of the drugs. An additional mechanism may be at play for inhibitor 17 since it slows cleavage in the absence of cells, unlike 01. While the increased size of inhibitor 17 means that it extends further towards the furin loop and would therefore be expected to be a better inhibitor of the furin cleavage, 17 is actually less effective than 01 in the cell intoxication experiments. This contradiction may be due to the phenyl group on 17 that is in close proximity to a hydrophobic region within 2–3 residues of the furin loop (Figure 1C). Interaction between the inhibitor and the furin loop could cause a conformational change that slows furin cleavage in vitro. Efforts are currently underway to co-crystalize inhibitors 01 and 17 with PA in order to elucidate the mechanism of furin cleavage inhibition.
These compounds are the most effective PA oligomerization-inhibiting compounds to date and this is the first report of dual-acting inhibitors that are general to both anthrax toxins. A recent report of oligomerization inhibitors identified two compounds that were mildly protective of cells at 300 μM; however, the compounds themselves were toxic to the cells with viabilities of 40 and 70 % at the therapeutic concentrations 26. These compounds have a backbone similar to that of the lead inhibitors identified in this paper, with linked phenyl rings making up the majority of the structure. Furthermore, both sets of compounds target the same pocket. However, the compounds identified in this work are between 30- and 100-fold more effective than those previously identified.
Other small molecule and peptide inhibitors that target PA work by preventing furin cleavage or blocking LF and EF binding and translocation. Peptide inhibitors of furin cleavage are built on a poly-arginine motif that is often combined with other amino acids at either end of the motif 24, 49. These inhibitors have EC50 values against LT in vitro in the range of 5–100 μM, making the inhibitors described here more effective than the majority of those with a poly-arginine core. One of the poly-arginine inhibitors, In-2-LF, also displays a dual mode of action, blocking furin cleavage and LF catalytic activity, but this compound is subject to degradation by furin and does not act against EF’s catalytic activity. Modified cyclodextrans that target the binding and translocation of EF and LF have been shown to protect against the toxins at nanomolar concentrations 50–53. However, those compounds are heptavalent in nature and their efficacy against octameric PA63, which has been found to be more stable at physiological pH and temperature, would likely be diminished 2, 54.
It is generally considered challenging to inhibit protein-protein interactions with small molecules because the large interacting protein surfaces confer high affinity on the binding event. Thus, the ability of the compounds found here to block with EC50 values of nearly 1 μM may be viewed as surprising. With the goal of drug design in mind, care needs to be taken with these lead compounds because 35 contains a chromone group which is known for its promiscuous binding to protein targets and the other three are symmetrical, which can pose problems for lead optimization. In this study we screened a relatively modest sized database of 10,000 compounds and therefore screening a larger database to the furin pocket may identify new, more drug-like hits in the future.
In conclusion, this work identifies two lead compounds which are able to protect RAW264.7 and HT1080 cells from PA-delivered protein toxins in vitro by preventing cleavage of PA by furin and oligomerization of PA. Aside from the possible toxicity of long-term exposure to inhibitor 17, the only drawback of the two lead compounds reported in this work is their hydrophobicity. Further work is necessary to identify the mechanism of cleavage inhibition, as well as to optimize the identified backbones to decrease their hydrophobicity.
EXPERIMENTAL SECTION
Reagents
2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one dihydrate (Chembridge #5117235), 3,3'-methylenebis{6-[(2-hydroxybenzylidene)amino]phenol} (Chembridge #5180717), N,N'-bis[2-(aminocarbonyl)phenyl]isophthalamide (Chembridge #5181385) and N,N'-bis[2-(aminocarbonyl)phenyl]terephthalamide (Chembridge #5181401) were purchased from Hit2Lead (San Diego, CA). The manufacturer stated that the compounds were >95% pure, and we confirmed this by C-18 reverse-phase HPLC (see Supplementary Data). The compounds were dissolved in DMSO (Sigma, St. Louis, MO) at a concentration of 10 mM. An anti-PA rabbit serum #5308 produced in our lab was used for Western blots 33. Anti-rabbit infrared (IR) 800CW dye secondary antibody was purchased from Rockland (Gilbertsville, PA). PA, PA-L1 (a matrix metalloprotease-activated PA 55), PA-U7 (a PA mutant that is not cleaved by any protease 56), FP59 (a fusion protein consisting of the N-terminal PA-binding domain of LF and the catalytic domain of Pseudomonas exotoxin A 57), and LF were purified from avirulent B. anthracis as previously described 58. PA N645C was produced in Escherichia coli BL-21 using the pET22b expression system for PA described previously 48. Soluble human furin was a gift of Iris Lindberg (University of Maryland, Baltimore, MD). TGFα-PE38, a fusion protein consisting of TGFα linked by the furin-cleavable sequence RKKR to a 38-kDa fragment of Pseudomonas exotoxin A, was provided by David Fitzgerald (National Cancer Institute, Bethesda, MD) 59.
Cell Culture
RAW264.7 mouse macrophages and HT1080 human fibroblasts were purchased from American Type Culture Collection (Manassas, VA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) with Glutamax supplemented with fetal bovine serum (FBS) to 10% (both from Invitrogen, Carlsbad CA), 10 mM Hepes, pH 7.3 (Quality Biological, Gaithersburg, MD), and 50 μg/mL gentamicin (Invitrogen) (hereafter DMEM Complete). Chinese Hamster Ovary (CHO) C4 cells, which overexpress CMG2, were developed in our lab 60, and were cultured in αMEM supplemented with FBS to 10%, 10 mM Hepes, pH 7.3, 50 μg/mL gentamicin, and 500 μg/mL hygromycin B (Invitrogen) (hereafter αMEM Complete). Binding and cytotoxicity assays were performed with RAW264.7 and HT1080 in DMEM and with CHO C4 in αMEM supplemented with 10 mM Hepes, pH 7.3 and 50 μg/mL gentamicin (hereafter serum-free complete media).
Ligand Binding Pocket Prediction
The ICM Pocket Finder method 43, 44, 61 in the ICM-Pro software (MolSoft LLC, San Diego) uses only the protein structure for the prediction of cavities and clefts. No prior knowledge of the substrate is required. The position and size of the ligand-binding pocket are determined based on a transformation of the Lennard-Jones potential by convolution with a Gaussian kernel of a certain size, a grid map of binding potential, and construction of equipotential surfaces along the maps. The pockets are displayed graphically as a surface and the dimensions of each pocket are presented in an interactive table and plot.
Virtual Screening
All in silico molecular modeling and virtual screening experiments were undertaken using ICM-Pro version 3.5 (MolSoft LLC, San Diego, CA) 62–64. Full atom models of the monomeric (PDB code: 1T6B 65) and heptameric (PDB code: 1TZO 66) PA crystal structures were prepared in internal coordinates. In both crystal structures the furin loop region (residues 158–175) is disordered and no attempt was made to predict its conformation; however recent crystal structures have been solved with this loop ordered 67. Three potential ligand-binding pockets in each crystal form were identified on the heptamerization interface using ICMPocketFinder 43, 44. A diverse library of 10,000 compounds from ChemBridge (San Diego, CA) was screened to all three pockets in both structures using ICM-VLS (MolSoft LLC, San Diego, CA). Each pocket was represented by five potential grid maps: (1) electrostatics, (2) directional hydrogen bond, (3) hydrophobic interactions, (4) Van der Waals interactions for steric repulsion and dispersion attraction, and (5) soft Van der Waals potential to limit the effect of minor steric clashes. This method uses the ECEPP/3 68 force field parameters for proteins and MMFF 69 for chemicals. Each flexible ligand was docked to a pocket three times and its positional and internal torsions sampled using the ICM Biased Probability Monte Carlo methods which includes a local minimization after each random step 64. The docked ligand is then assigned a score which includes the internal force-field energy of the ligand, conformational entropy loss of the ligand, receptor-ligand hydrogen-bond interaction, solvation electrostatic energy change, hydrogen-bond donor/acceptor desolvation, and hydrophobic energy 70. Ranked hitlists of the top-scoring compounds were generated and the top-scoring compounds were selected based on their availability and predicted interactions with the receptor.
Preparation of fluorophore-labeled, nicked PA
PA-N645C was labeled with Alexa Fluor 488 C5 maleimide or Alexa Fluor 594 C5 maleimide (Invitrogen) using the manufacturer’s instructions. Unincorporated fluorophores were removed by gel filtration in a buffer containing 20 mM Tris-HCl, pH 8.5, 150 mM NaCl, 1 mM CaCl2. The labeled PAs were nicked using TPCK-treated trypsin (Sigma) by incubating PA (1–2 mg/mL) in 20 mM Tris-HCl, pH 8.5, 150 mM NaCl, and 1 mM CaCl2 for 35 min at room temperature at 1:1000 w/w trypsin:PA, followed by addition of a 100-fold excess of soybean trypsin inhibitor (Sigma) to prevent further proteolysis.
FRET assay of PA63 oligomerization
To monitor oligomerization of the mixture of the two fluorophore-conjugated, nicked PA proteins (nPA83), 96-well plates were prepared containing (in each well) 200 μL of 250 nM each of nPA83 N645C*488 and nPA83 N645C*594, in 20 mM Tris-HCl, pH 8.5, 150 mM NaCl, and 1 mM CaCl2. Compounds identified in the virtual ligand screen (VLS) were added at a concentration of 100 μM and LF was added to a final concentration of 50 nM to drive oligomerization 45. The process was monitored by measuring FRET emission intensity at 610 nm on a Spectromax Gemini EM spectrofluorometer. Data were calculated as differences between the results for the indicated inhibitor and for the no inhibitor control. Prism 5 (GraphPad Software, La Jolla, CA) was used to determine significance.
RAW264.7 cell cytotoxicity assays of LT inhibition
RAW264.7 macrophages were plated in DMEM Complete in a 96-well plate and allowed to grow to 50 % confluence overnight. The medium was changed to serum-free DMEM Complete containing LF, PA and serially-diluted inhibitors (from stocks in DMSO) to obtain final concentrations of 0.90 nM (75 ng/mL) PA, 5.6 nM (500 ng/mL) LF, and 0.1–100 μM inhibitor. The cells were incubated for 5 h at 37°C and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) was added at 500 μg/mL and the cells were incubated an additional 1 h. The medium was aspirated and MTT was solubilized in 91% isopropanol containing 0.5% SDS and 0.038 M hydrochloric acid and read using a SpectraMax 190 plate reader and SoftMax Pro 4.7.1 software (Molecular Devices, Sunnyvale, CA) at 570 and 490 nM. The difference of the absorbances was used to determine percent survival compared to a no treatment control.
RAW264.7 cell cytotoxicity assays of PA + FP59 toxin inhibition
RAW264.7 macrophages were treated as above except that the serially-diluted medium containing FP59, PA, and inhibitors dissolved in DMSO was added to obtain final concentrations of 0.060 nM (5 ng/mL) PA, 1.88 nM (100 ng/mL) FP59, and 0.1–100 μM inhibitor. The cells were incubated for 5 h at 37°C and the medium was changed to DMEM Complete with 10 mM ammonium chloride lacking toxin and inhibitor. The cells were allowed to grow another 23 h before MTT was added and the cells were cultured an additional 1 h before lysis.
HT1080 cell cytotoxicity assays of PA or PA-L1 + FP59 toxin inhibition
HT1080 fibroblasts were treated the same as the RAW264.7 cells above except the medium contained final concentrations of 0.60 nM (50 ng/mL) or 0.30 nM (25 ng/mL) PA-L1, 1.88 nM (100 ng/mL) FP59, and 0.1–100 μM inhibitor. The cells were incubated for 5 h at 37°C and the media were changed to DMEM Complete with 10 mM ammonium chloride and lacking toxin and inhibitors. The cells were allowed to grow another 47 h before MTT was added for another 1 h of growth before lysis.
CHO C4 assay for PA binding
CHO C4 cells were plated in 6-well plates in αMEM Complete and allowed to grow to confluence overnight. The medium was changed to serum-free αMEM Complete containing 12 nM (1 μg/mL) PA, 12 nM (1 μg/mL) PA and 11.1 nM (1 μg/mL) LF, or 12 nM (1 μg/mL) PA and 11.1 nM (1 μg/mL) each PA-U7 and LF, and 100 μM inhibitor. The cells were incubated for 1 h at 37°C. The cells were washed quickly three times with cold, phosphate-buffered saline (pH 7.4), and all liquid was thoroughly removed. The cells were lysed in modified RIPA lysis buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) containing Complete protease inhibitor (Roche, Basel, Switzerland) and boiled with 6x SDS protein gel loading dye (0.35 M Tris-HCl, pH 6.8, 10 % SDS, 36 % glycerol, 0.6 M dithiothreitol, 0.01% bromophenol blue). The proteins were separated on a Novex Tris-Glycine 1.0 mm 4–20% gradient gel (Invitrogen), transferred to nitrocellulose and probed with rabbit anti-PA serum #5308 (1:5000). The primary antibody was detected with Rockland anti-rabbit 800 (1:5000) and imaged on a Li-Cor Odyssey IR imager (Li-Cor, Lincoln, NE).
Cell-free cleavage inhibition assay
PA or TGFα-PE38 at 100 nM was mixed with 100, 10, or 1 μM inhibitor dissolved in DMSO, or 1 % DMSO as control, in 50 mM Hepes, pH 7.5, 2 mM CaCl2, 0.5 mM EDTA, and 0.2% octyl glucoside. Soluble human furin (1 nM final concentration) or PBS was added and the reaction was incubated at 37°C for 1 h. The reaction was quenched by boiling with SDS loading dye and the proteins were separated on a Novex 4–20 % gel. The proteins were transferred to nitrocellulose with an iBlot system (Invitrogen) and the membrane was blocked with Li-Cor Blocking Buffer. The blot was probed with anti-PA serum #5308 (1:5000) or Pseudomonas exotoxin rabbit antiserum P2318 (1: 2500, Sigma). The primary antibody was detected with Rockland anti-rabbit 800 (1:5000) and imaged on a Li-Cor Odyssey IR imager.
Supplementary Material
Acknowledgments
Portions of this research were supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases. MP and DP were supported, in part, by NIH grant MD001091-01. The authors thank Dr. Rodney Tweten for providing the pET vector encoding PA N645C, Rasem Fattah for protein purification, Dr. Iris Lindberg for the gift of furin, Dr. David Fitzgerald for TGFα-PE38, and Dr. Clinton E. Leysath for thoughtful discussions.
Abbreviations used
- PA
protective antigen
- PA83
83-kDa protective antigen
- PA63
63-kDa protective antigen
- nPA83
trypsin-nicked protective antigen
- LF
lethal factor
- EF
edema factor
- LT
lethal toxin
- ET
edema toxin
- CMG2
capillary morphogenesis protein 2
- TEM8
tumor endothelial marker 8
- TGFα
transforming growth factor α
- DMEM
Dulbecco’s modified Eagle’s media
- αMEM
α minimum essential media
- VLS
virtual ligand screen
- TPCK
tosyl phenylalanyl chloromethyl ketone
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
Reference List
- 1.Frankel AE, Kuo SR, Dostal D, Watson L, Duesbery NS, Cheng CP, Cheng HJ, Leppla SH. Pathophysiology of anthrax. Front Biosci. 2009;14:4516–4524. doi: 10.2741/3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kintzer AF, Thoren KL, Sterling HJ, Dong KC, Feld GK, Tang II, Zhang TT, Williams ER, Berger JM, Krantz BA. The protective antigen component of anthrax toxin forms functional octameric complexes. J Mol Biol. 2009;392:614–629. doi: 10.1016/j.jmb.2009.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moayeri M, Leppla SH. Cellular and systemic effects of anthrax lethal toxin and edema toxin. Mol Aspects Med. 2009;30:439–455. doi: 10.1016/j.mam.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koehler TM. Bacillus anthracis physiology and genetics. Mol Aspects Med. 2009;30:386–396. doi: 10.1016/j.mam.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu S, Miller-Randolph S, Crown D, Moayeri M, Sastalla I, Okugawa S, Leppla SH. Anthrax toxin targeting of myeloid cells through the CMG2 receptor Is essential for establishment of Bacillus anthracis infections in mice. Cell Host Microbe. 2010;8:455–462. doi: 10.1016/j.chom.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collier RJ. Membrane translocation by anthrax toxin. Mol Aspects Med. 2009;30:413–422. doi: 10.1016/j.mam.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier RJ, Young JAT. Anthrax toxin. Ann Rev Cell Dev Biol. 2003;19:45–70. doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 8.Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 9.Pellizzari R, Guidi-Rontani C, Vitale G, Mock M, Montecucco C. Lethal factor of Bacillus anthracis cleaves the N-terminus of MAPKKs: analysis of the intracellular consequences in macrophages. Int J Med Microbiol. 2000;290:421–427. doi: 10.1016/S1438-4221(00)80056-9. [DOI] [PubMed] [Google Scholar]
- 10.Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages. Biochem Biophys Res Commun. 1998;248:706–711. doi: 10.1006/bbrc.1998.9040. [DOI] [PubMed] [Google Scholar]
- 11.Levinsohn JL, Newman ZL, Hellmich KA, Fattah R, Getz MA, Liu S, Sastalla I, Leppla SH, Moayeri M. Anthrax lethal factor cleavage of Nlrp1 is required for activation of the inflammasome. PLoS Pathog. 2012;8:e1002638. doi: 10.1371/journal.ppat.1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang WJ, Guo Q. The adenylyl cyclase activity of anthrax edema factor. Mol Aspects Med. 2009;30:423–430. doi: 10.1016/j.mam.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 15.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Crown D, Miller-Randolph S, Moayeri M, Wang H, Hu H, Morley T, Leppla SH. Capillary morphogenesis protein-2 is the major receptor mediating lethality of anthrax toxin in vivo. Proc Natl Acad Sci U S A. 2009;106:12424–12429. doi: 10.1073/pnas.0905409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogridge J, Cunningham K, Lacy DB, Mourez M, Collier RJ. The lethal and edema factors of anthrax toxin bind only to oligomeric forms of the protective antigen. Proc Natl Acad Sci U S A. 2002;99:7045–7048. doi: 10.1073/pnas.052160199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacy DB, Mourez M, Fouassier A, Collier RJ. Mapping the anthrax protective antigen binding site on the lethal and edema factors. J Biol Chem. 2002;277:3006–3010. doi: 10.1074/jbc.M109997200. [DOI] [PubMed] [Google Scholar]
- 19.Thoren KL, Krantz BA. The unfolding story of anthrax toxin translocation. Mol Microbiol. 2011;80:588–595. doi: 10.1111/j.1365-2958.2011.07614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little SF, Leppla SH, Cora E. Production and characterization of monoclonal antibodies to the protective antigen component of Bacillus anthracis toxin. Infect Immun. 1988;56:1807–1813. doi: 10.1128/iai.56.7.1807-1813.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai C, Che J, Xu L, Guo Q, Kong Y, Fu L, Xu J, Cheng Y, Chen W. Tumor endothelium marker-8 based decoys exhibit superiority over capillary morphogenesis protein-2 based decoys as anthrax toxin inhibitors. PLoS ONE. 2011;6:e20646. doi: 10.1371/journal.pone.0020646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komiyama T, Swanson JA, Fuller RS. Protection from anthrax toxin-mediated killing of macrophages by the combined effects of furin inhibitors and chloroquine. Antimicrob Agents Chemother. 2005;49:3875–3882. doi: 10.1128/AAC.49.9.3875-3882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gujraty K, Sadacharan S, Frost M, Poon V, Kane RS, Mogridge J. Functional characterization of peptide-based anthrax toxin inhibitors. Mol Pharm. 2005;2:367–372. doi: 10.1021/mp050040f. [DOI] [PubMed] [Google Scholar]
- 24.Peinado JR, Kacprzak MM, Leppla SH, Lindberg I. Cross-inhibition between furin and lethal factor inhibitors. Biochem Biophys Res Commun. 2004;321:601–605. doi: 10.1016/j.bbrc.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Sarac MS, Peinado JR, Leppla SH, Lindberg I. Protection against anthrax toxemia by hexa- D-arginine In vitro and in vivo. Infect Immun. 2004;72:602–605. doi: 10.1128/IAI.72.1.602-605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez CR, Lopez-Perez D, Chmielewski J, Lipton M. Small molecule inhibitors of anthrax toxininduced cytotoxicity targeted against protective antigen. Chem Biol Drug Des. 2012;79:260–269. doi: 10.1111/j.1747-0285.2011.01285.x. [DOI] [PubMed] [Google Scholar]
- 27.Moayeri M, Wiggins JF, Lindeman RE, Leppla SH. Cisplatin inhibition of anthrax lethal toxin. Antimicrob Agents Chemother. 2006;50:2658–2665. doi: 10.1128/AAC.01412-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vance D, Shah M, Joshi A, Kane RS. Polyvalency: a promising strategy for drug design. Biotechnol Bioeng. 2008;101:429–434. doi: 10.1002/bit.22056. [DOI] [PubMed] [Google Scholar]
- 29.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Designing a polyvalent inhibitor of anthrax toxin. Nat Biotechnol. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 30.Joshi A, Kate S, Poon V, Mondal D, Boggara MB, Saraph A, Martin JT, McAlpine R, Day R, Garcia AE, Mogridge J, Kane RS. Structure-based design of a heptavalent anthrax toxin inhibitor. Biomacromolecules. 2011;12:791–796. doi: 10.1021/bm101396u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basha S, Rai P, Poon V, Saraph A, Gujraty K, Go MY, Sadacharan S, Frost M, Mogridge J, Kane RS. Polyvalent inhibitors of anthrax toxin that target host receptors. Proc Natl Acad Sci U S A. 2006;103:13509–13513. doi: 10.1073/pnas.0509870103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abrami L, Kunz B, van der Goot FG. Anthrax toxin triggers the activation of src-like kinases to mediate its own uptake. Proc Natl Acad Sci U S A. 2010;107:1420–1424. doi: 10.1073/pnas.0910782107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Leppla SH. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J Biol Chem. 2003;278:5227–5234. doi: 10.1074/jbc.M210321200. [DOI] [PubMed] [Google Scholar]
- 34.Johnson SL, Chen LH, Pellecchia M. A high-throughput screening approach to anthrax lethal factor inhibition. Bioorg Chem. 2007 doi: 10.1016/j.bioorg.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson SL, Jung D, Forino M, Chen Y, Satterthwait A, Rozanov DV, Strongin AY, Pellecchia M. Anthrax lethal factor protease inhibitors: synthesis, SAR, and structurebased 3D QSAR studies. J Med Chem. 2006;49:27–30. doi: 10.1021/jm050892j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaddis BD, Rubert Perez CM, Chmielewski J. Inhibitors of anthrax lethal factor based upon Noleoyldopamine. Bioorg Med Chem Lett. 2008;18:2467–2470. doi: 10.1016/j.bmcl.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 37.Gaddis BD, Avramova LV, Chmielewski J. Inhibitors of anthrax lethal factor. Bioorg Med Chem Lett. 2007;17:4575–4578. doi: 10.1016/j.bmcl.2007.05.089. [DOI] [PubMed] [Google Scholar]
- 38.Chiu TL, Solberg J, Patil S, Geders TW, Zhang X, Rangarajan S, Francis R, Finzel BC, Walters MA, Hook DJ, Amin EA. Identification of novel non-hydroxamate anthrax toxin lethal factor inhibitors by topomeric searching, docking and scoring, and in vitro screening. J Chem Inf Model. 2009;49:2726–2734. doi: 10.1021/ci900186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taha HM, Schmidt J, Gottle M, Suryanarayana S, Shen Y, Tang WJ, Gille A, Geduhn J, Konig B, Dove S, Seifert R. Molecular analysis of the interaction of anthrax adenylyl cyclase toxin, edema factor, with 2(3')--(-(methyl)anthraniloyl)-substituted purine and pyrimidine nucleotides. Mol Pharmacol. 2009;75:693–703. doi: 10.1124/mol.108.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen D, Ma L, Kanalas JJ, Gao J, Pawlik J, Jimenez ME, Walter MA, Peterson JW, Gilbertson SR, Schein CH. Structure-based redesign of an edema toxin inhibitor. Bioorg Med Chem. 2012;20:368–376. doi: 10.1016/j.bmc.2011.10.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Booth MG, Hood J, Brooks TJ, Hart A. Anthrax infection in drug users. Lancet. 2010;375:1345–1346. doi: 10.1016/S0140-6736(10)60573-9. [DOI] [PubMed] [Google Scholar]
- 42.Leysath CE, Chen KH, Moayeri M, Crown D, Fattah R, Chen Z, Das SR, Purcell RH, Leppla SH. Mouse monoclonal antibodies to anthrax edema factor protect against infection. Infect Immun. 2011;79:4609–4616. doi: 10.1128/IAI.05314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.An J, Totrov M, Abagyan R. Comprehensive identification of "druggable" protein ligand binding sites. Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- 45.Christensen KA, Krantz BA, Melnyk RA, Collier RJ. Interaction of the 20 kDa and 63 kDa fragments of anthrax protective antigen: kinetics and thermodynamics. Biochemistry. 2005;44:1047–1053. doi: 10.1021/bi047791s. [DOI] [PubMed] [Google Scholar]
- 46.Mourez M, Lacy DB, Cunningham K, Legmann R, Sellman BR, Mogridge J, Collier RJ. 2001: a year of major advances in anthrax toxin research. Trends Microbiol. 2002;10:287–293. doi: 10.1016/s0966-842x(02)02369-7. [DOI] [PubMed] [Google Scholar]
- 47.Christensen KA, Krantz BA, Collier RJ. Assembly and disassembly kinetics of anthrax toxin complexes. Biochemistry. 2006;45:2380–2386. doi: 10.1021/bi051830y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mourez M, Yan M, Lacy DB, Dillon L, Bentsen L, Marpoe A, Maurin C, Hotze E, Wigelsworth D, Pimental RA, Ballard JD, Collier RJ, Tweten RK. Mapping dominant-negative mutations of anthrax protective antigen by scanning mutagenesis. Proc Natl Acad Sci U S A. 2003;100:13803–13808. doi: 10.1073/pnas.2436299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remacle AG, Gawlik K, Golubkov VS, Cadwell GW, Liddington RC, Cieplak P, Millis SZ, Desjardins R, Routhier S, Yuan XW, Neugebauer WA, Day R, Strongin AY. Selective and potent furin inhibitors protect cells from anthrax without significant toxicity. Int J Biochem Cell Biol. 2010;42:987–995. doi: 10.1016/j.biocel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nestorovich EM, Karginov VA, Berezhkovskii AM, Bezrukov SM. Blockage of anthrax PA63 pore by a multicharged high-affinity toxin inhibitor. Biophys J. 2010;99:134–143. doi: 10.1016/j.bpj.2010.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nestorovich EM, Karginov VA, Popoff MR, Bezrukov SM, Barth H. Tailored ß,-cyclodextrin blocks the translocation pores of binary exotoxins from C. Botulinum and C. Perfringens and protects cells from intoxication. PLoS ONE. 2011;6:e23927. doi: 10.1371/journal.pone.0023927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karginov VA, Nestorovich EM, Yohannes A, Robinson TM, Fahmi NE, Schmidtmann F, Hecht SM, Bezrukov SM. Search for cyclodextrin-based inhibitors of anthrax toxins: synthesis, structural features, and relative activities. Antimicrob Agents Chemother. 2006;50:3740–3753. doi: 10.1128/AAC.00693-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karginov VA, Nestorovich EM, Schmidtmann F, Robinson TM, Yohannes A, Fahmi NE, Bezrukov SM, Hecht SM. Inhibition of S. aureus alpha-hemolysin and B. anthracis lethal toxin by beta-cyclodextrin derivatives. Bioorg Med Chem. 2007;15:5424–5431. doi: 10.1016/j.bmc.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kintzer AF, Sterling HJ, Tang II, Abdul-Gader A, Miles AJ, Wallace BA, Williams ER, Krantz BA. Role of the protective antigen octamer in the molecular mechanism of anthrax lethal toxin stabilization in plasma. J Mol Biol. 2010;399:741–758. doi: 10.1016/j.jmb.2010.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Netzel-Arnett S, Birkedal-Hansen H, Leppla SH. Tumor cell-selective cytotoxicity of matrix metalloproteinase-activated anthrax toxin. Cancer Res. 2000;60:6061–6067. [PubMed] [Google Scholar]
- 56.Liu S, Bugge TH, Leppla SH. Targeting of tumor cells by cell surface urokinase plasminogen activator-dependent anthrax toxin. J Biol Chem. 2001;276:17976–17984. doi: 10.1074/jbc.M011085200. [DOI] [PubMed] [Google Scholar]
- 57.Arora N, Klimpel KR, Singh Y, Leppla SH. Fusions of anthrax toxin lethal factor to the ADP-ribosylation domain of Pseudomonas exotoxin A are potent cytotoxins which are translocated to the cytosol of mammalian cells. J Biol Chem. 1992;267:15542–15548. [PubMed] [Google Scholar]
- 58.Pomerantsev AP, Pomerantseva OM, Moayeri M, Fattah R, Tallant C, Leppla SH. A Bacillus anthracis strain deleted for six proteases serves as an effective host for production of recombinant proteins. Protein Expr Purif. 2011;80:80–90. doi: 10.1016/j.pep.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Draoui M, Siegall CB, FitzGerald D, Pastan I, Moody TW. TGF alpha-PE40 inhibits non-small cell lung cancer growth. Life Sci. 1994;54:445–453. doi: 10.1016/0024-3205(94)00403-x. [DOI] [PubMed] [Google Scholar]
- 60.Liu S, Leung HJ, Leppla SH. Characterization of the interaction between anthrax toxin and its cellular receptors. Cell Microbiol. 2007;9:977–987. doi: 10.1111/j.1462-5822.2006.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kufareva I, Ilatovskiy AV, Abagyan R. Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res. 2012;40:D535–D540. doi: 10.1093/nar/gkr825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orry AJ, Totrov M, Raush E, Abagyan RA. ICM User's Guide. MolSoft, LLC; La Jolla, CA: 2012. [Google Scholar]
- 63.Abagyan R, Totrov M, Kuznetsov D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J Computational Chem. 1994;15:488–506. [Google Scholar]
- 64.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 65.Santelli E, Bankston LA, Leppla SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- 66.Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation. Proc Natl Acad Sci U S A. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feld GK, Kintzer AF, Tang II, Thoren KL, Krantz BA. Domain flexibility modulates the heterogeneous assembly mechanism of anthrax toxin protective antigen. J Mol Biol. 2011;415:159–174. doi: 10.1016/j.jmb.2011.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. Energy parameters in polypeptides. 10. Improved geometrical parameters and nonbonded interactions for use in the ECEPP/3 algorithm, with application to proline-containing peptides. J Phys Chem. 1992;96:6472–6484. [Google Scholar]
- 69.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Computational Chem. 1996;17:490–519. [Google Scholar]
- 70.Totrov M, Abagyan R. Derivation of sensitive discrimination potential for virtual ligand screening. RECOMB '99. Proceedings of the third annual international conference on Computational molecular biology; 1999; New York, NY, USA, ACM. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.