Abstract
Purpose.
Many rhodopsin mutations that cause retinitis pigmentosa produce misfolded rhodopsin proteins that are retained within the endoplasmic reticulum (ER) and cause photoreceptor cell death. Activating transcription factor 6 (ATF6) and protein kinase RNA-like endoplasmic reticulum kinase (PERK) control intracellular signaling pathways that maintain ER homeostasis. The aim of this study was to investigate how ATF6 and PERK signaling affected misfolded rhodopsin in cells, which could identify new molecular therapies to treat retinal diseases associated with ER protein misfolding.
Methods.
To examine the effect of ATF6 on rhodopsin, wild-type (WT) or mutant rhodopsins were expressed in cells expressing inducible human ATF6f, the transcriptional activator domain of ATF6. Induction of ATF6f synthesis rapidly activated downstream genes. To examine PERK's effect on rhodopsin, WT or mutant rhodopsins were expressed in cells expressing a genetically altered PERK protein, Fv2E-PERK. Addition of the dimerizing molecule (AP20187) rapidly activated Fv2E-PERK and downstream genes. By use of these strategies, it was examined how selective ATF6 or PERK signaling affected the fate of WT and mutant rhodopsins.
Results.
ATF6 significantly reduced T17M, P23H, Y178C, C185R, D190G, K296E, and S334ter rhodopsin protein levels in the cells with minimal effects on monomeric WT rhodopsin protein levels. By contrast, the PERK pathway reduced both levels of WT, mutant rhodopsins, and many other proteins in the cell.
Conclusions.
This study indicates that selectively activating ATF6 or PERK prevents mutant rhodopsin from accumulating in cells. ATF6 signaling may be especially useful in treating retinal degenerative diseases arising from rhodopsin misfolding by preferentially clearing mutant rhodopsin and abnormal rhodopsin aggregates.
Rhodopsin mutations cause many heritable types of retinitis pigmentosa. ATF6 and PERK can reduce mutant rhodopsin protein levels.
Introduction
Retinitis pigmentosa (RP) comprises a group of diseases arising from photoreceptor cell death that commonly present with loss of night vision followed by progressive loss of peripheral vision.1,2 The most common causes of autosomal dominant RP (ADRP) are mutations in rhodopsin, the visual pigment of the rod photoreceptor cells. More than 120 distinct rhodopsin mutations have been identified in ADRP (RetNet [Retinal Information Network]; may be accessed at www.sph.uth.tmc.edu/Retnet). Rhodopsin is a membrane glycoprotein of the G-protein–coupled receptor superfamily, with seven transmembrane helices surrounding a retinol-binding pocket buried within the transmembrane portion of the protein.3 Wild-type (WT) rhodopsin is synthesized and folded in the endoplasmic reticulum (ER) prior to delivery to the outer segment of the photoreceptor cells. Many mutant rhodopsin proteins (“class II mutants”) cannot fold properly and are retained within the ER, where they elicit ER stress and ultimately lead to the death of photoreceptor cells.4–9
Cells respond to ER stress by activating the unfolded protein response (UPR), which is controlled by three ER-resident transmembrane proteins, the activating transcription factor 6 (ATF6), protein kinase RNA-like endoplasmic reticulum kinase (PERK), and inositol-requiring enzyme 1 (IRE1). All three UPR mediators monitor ER stress via luminal domains that are coupled to cytosolic effector domains, enabling them to activate distinct signal transduction cascades.10 In response to ER stress, ATF6 undergoes regulated intramembrane proteolysis,11–13 to release its cytosolic basic leucine zipper (bZIP) transcriptional activator domain, ATF6f. ATF6f binds to the promoters bearing ER stress response elements to transcriptionally upregulate genes involved in ER protein folding and ER homeostasis, such as BiP/Grp78.14–16 By contrast to ATF6, PERK has a kinase in its cytosolic domain that phosphorylates eukaryotic translation initiating factor 2 subunit α (eIF2α) in response to ER stress, thereby impairing ribosomal assembly on mRNAs and attenuating protein translation.17 PERK-mediated phosphorylation of eIF2α also enhances translation of select mRNAs that have small open reading frames in their 5′ untranslated regions, leading to the generation of proteins such as the ATF4 transcription factor whose targets include genes required for oxidative protein folding in the ER.18 IRE1 has a cytosolic kinase/endoribonuclease that initiates the nonconventional splicing of Xbp-1 mRNA.19–21 Spliced Xbp-1 mRNA encodes a transcription factor that upregulates genes involved in ER protein folding, ER protein delivery, and ER-associated protein degradation (ERAD).22 Activation of the ATF6, PERK, and IRE1 signaling pathways thus enhances cell survival under ER stress by facilitating ER protein folding and reducing misfolded protein levels.
Given the powerful abilities of UPR signaling pathways to regulate many steps of protein folding in the ER, we hypothesized that selective activation of these signaling pathways might prevent rhodopsin misfolding or promote the removal of misfolded rhodopsin from cells, thereby offering new approaches to promote photoreceptor survival. Indeed, our previous studies demonstrated that selective chemical–genetic activation of the IRE1 signaling pathway specifically promoted the degradation of misfolded P23H rhodopsin with minimal effect on the WT rhodopsin.23 Here, we used chemical–genetic approaches to selectively activate ATF6 or PERK signaling to compare their effects on WT versus mutant P23H, T17M, Y178C, C185R, D190G, K296E, and S334ter rhodopsins.
Methods
Cell Culture and Transfection
HEK293 cells were maintained at 37°C, 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), and 1% penicillin/streptomycin (Invitrogen). To generate stable HEK293 cells expressing TetON-ATF6f, a cDNA encoding a fragment of human ATF6 containing the bZIP-containing transcriptional activator domain (1–373), ATF6f14 was cloned into the pcDNA5/FRT/TO vector (Invitrogen) following the manufacturer's instructions. Isogenic stable HEK293 clones expressing TetON-ATF6f were generated and analyzed after 3 weeks.
The generation and use of 1NM-PP1 sensitized HEK293 stable cell lines expressing IRE1[I642G] and AP20187 sensitized HEK293 stable cell lines expressing Fv2E-PERK have been previously described.8,24 To express WT or mutant rhodopsins, VCAM-1, and GFP in our cell systems, plasmids containing the cDNA of those genes were transiently transfected to the indicated cell line using a transfection reagent (Lipofectamine 2000; Invitrogen).
Chemicals
Doxycycline A was obtained from Sigma-Aldrich (St. Louis, MO). Tunicamycin and thapsigargin were obtained from Calbiochem EMD Bioscience Inc. (Darmstadt, Germany). 1NM-PP1 was generously provided by C. Zhang and K. Shokat (University of California at San Francisco, San Francisco, CA).25 AP20187 was obtained from ARIAD Pharmaceuticals (Watertown, MA).
Molecular Biology
Cells were lysed and total RNA was collected (RNeasy Mini Kit; Qiagen, Hilden, Germany). PolyA mRNA (1 μg) was reverse-transcribed using a commercial cDNA synthesis kit (iScript cDNA Synthesis Kit; Bio-Rad, Hercules, CA). cDNA was used as a template for PCR amplification (Taq PCR Master Mix; Qiagen) across the fragment of the Xbp-1 cDNA bearing the intron target of IRE1′s RNAse activity. Primers used include: human Xbp-1, 5′-TTA CGA GAG AAA ACT CAT GGC-3′ and 5′-GGG TCC AAG TTG TCC AGA ATGC-3′. PCR conditions were previously described.26 For quantitative PCR, cDNA were used as a template in SYBR green qPCR supermix (Bio-Rad). Primers used include: human Rpl19 mRNA, 5′-ATG TAT CAC AGC CTG TAC CTG-3′ and 5′-TTC TTG GTC TCT TCC TCC TTG-3′; human BiP mRNA, 5′-CGG GCA AAG ATG TCA GGA AAG-3′ and 5′-TTC TGG ACG GGC TTC ATA GTA GAC-3′. Quantitative PCR was performed using a real-time thermal cycler (Chromo4 DNA Engine; Bio-Rad). Rpl19 mRNA levels, a transcript whose levels are not altered by ER stress, served as an internal normalization standard. qPCR conditions were previously described.26
Protein Biochemistry and Quantification
Cells were lysed in SDS lysis buffer (2% SDS, 62.5 mM Tris-HCl [pH 6.8], containing protease inhibitors [Sigma-Aldrich]). Protein concentrations of the total cell lysates were determined by BCA protein assay (Pierce, Rockford, IL). Total protein (5 μg) was loaded onto 4–15% precast gels (Mini-PROTEAN TGX; Bio-Rad) and analyzed by Western blot. The following antibodies and dilutions were used: 1D4 anti-rhodopsin and anti-VCAM-1 at 1:1000 (Santa Cruz Biotechnologies, Santa Cruz, CA); B630N anti-rhodopsin at 1:1000 (gift from Dr. W. C. Smith, University of Florida, Gainesville, FL); anti-CREB2/ATF4, anti-GFP, and anti-GAPDH at 1:5000 (Santa Cruz Biotechnologies); anti-ATF6α antibody at 1:1000 (BioAcademia, Kyoto, Japan); anti-β-actin antibody at 1:20,000 (Millipore, Billerica, MA); and anti-BiP at 1:1000 (GeneTex Inc., Irvine, CA). After 2 hours of incubation with primary antibody, membranes were washed in TBS with 0.1% Tween-20 (TBST) followed by incubation of a horseradish peroxidase–coupled secondary antibody (Promega, Madison, WI). Immunoreactivity was detected using chemiluminescent precipitating substrates for Western blotting (SuperSignal West Dura Chemiluminescent Substrate; Pierce). Protein quantifications were performed using a commercial image acquisition and analysis software program (VisionWorks Life Science Software; UVP Inc., Upland, CA). Rhodopsin protein levels were determined by measuring the area density within the bracket indicated in the figures after normalizing with the equivalent area density from the control lane.
Endoglycosidase H (Endo H; New England Biolabs, Ipswich, MA) digestion was performed on total cell lysate (10 μg) for 1.5 hours at 37°C in the buffer supplied by the manufacturer.
Protein Biotinylation
HEK293 cells expressing TetON-ATF6f were grown in 10-cm dishes coated with poly-d-lysine (Millipore) and transfected with WT or mutant P23H rhodopsin protein. Four hours after transfection, cells were treated with or without doxycycline for 24 hours. The cells were then washed with ice-cold PBS containing 1 mM MgCl2 and 0.1 mM CaCl2. After washing, 5 mL of NaIO4 (Pierce) was added to the dishes and agitated for 30 minutes at 4°C in the dark. After three washes with PBS containing MgCl2 and CaCl2, cells were labeled with biotin hydrazide (Pierce) at 4°C in the dark. Biotinylated cells were collected and pelleted by brief centrifugation and solubilized in 0.5 mL of ice-cold lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 8], and 150 mM NaCl, containing protease inhibitors [Sigma-Aldrich]). A 50% slurry of resin beads (300 μL neutrAvidin-agarose beads; Thermo Fisher Scientific/Pierce Protein Biology Products, Rockford, IL) were incubated with biotinylated cell lysates. The neutrAvidin-agarose beads were washed, resuspended in SDS sample buffer, and boiled for 5 minutes. Aliquots of recovered biotinylated proteins were used for Western blot analysis.
Statistical Analysis
All results are presented as mean ± SD of at least three independent experiments. One-way ANOVA with Bonferroni post hoc test (for multiple groups) or Student's two-tailed t-tests (for paired samples) were performed using a commercial software program (Prism 5 software; GraphPad Software, Inc., La Jolla, CA) to determine P values. A value of P < 0.05 was considered significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Selective Activation of ATF6 Reduces the Protein Levels of Class II Mutant Rhodopsins and Oligomerized WT Rhodopsin Levels
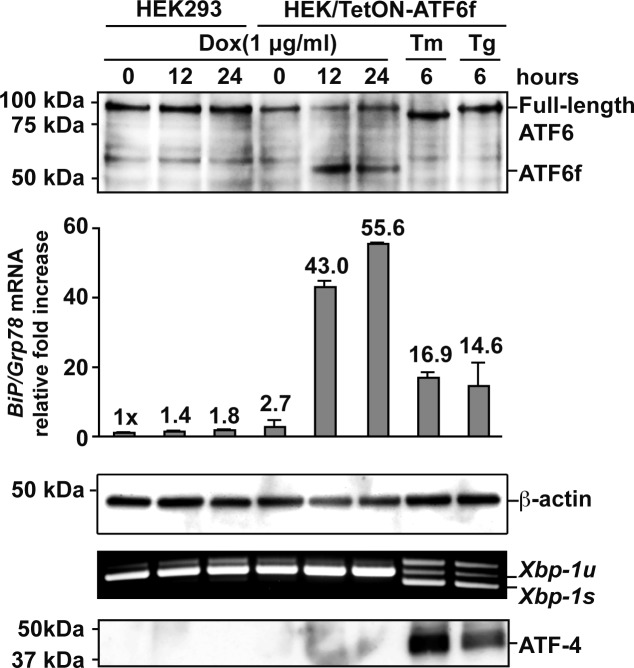
To determine how ATF6 signaling affects rhodopsin protein, we generated HEK293 cells stably expressing a tetracycline-inducible fragment of human ATF6, TetON-ATF6f. The addition of drug induced strong production of ATF6f and robustly upregulated the transcription of ATF6 target genes, such as the ER chaperone BiP/Grp78 (Fig. 1). Importantly, application of doxycycline did not activate other UPR signaling pathways, as indicated by the absence of ATF-4 protein and spliced Xbp-1 mRNA, molecular markers of the PERK and IRE1 signaling pathways, respectively (Fig. 1).8,24,27 By contrast, exposure of cells to ER stress–inducing agents, tunicamycin (Tm) or thapsigargin (Tg), activated all UPR signaling pathways (Fig. 1, last two lines). This system therefore enabled us to precisely evaluate the effects of ATF6 on rhodopsin protein.
Figure 1. .
Chemical–genetic activation of ATF6. WT and isogenic HEK293 cells stably expressing a tetracycline-inducible 373 amino acid cytosolic transcriptional activator domain of human ATF6 (ATF6f) were treated with doxycycline (Dox) (1 μg/mL), tunicamycin (Tm) (5 μg/mL), or thapsigargin (Tg) (1 μM) for the indicated durations. Endogenous full-length and induced ATF6 transcriptional activator domain, ATF6f, were detected by immunoblotting. β-Actin protein levels were assessed as a loading control. Levels of BiP/Grp78 mRNA, a gene robustly induced by ATF6, were assessed by real-time quantitative PCR. Xbp-1 mRNA splicing, a specific marker of IRE1 activation, was assessed by RT-PCR. Levels of ATF-4, a protein robustly induced by PERK signaling, were assessed by immunoblotting.
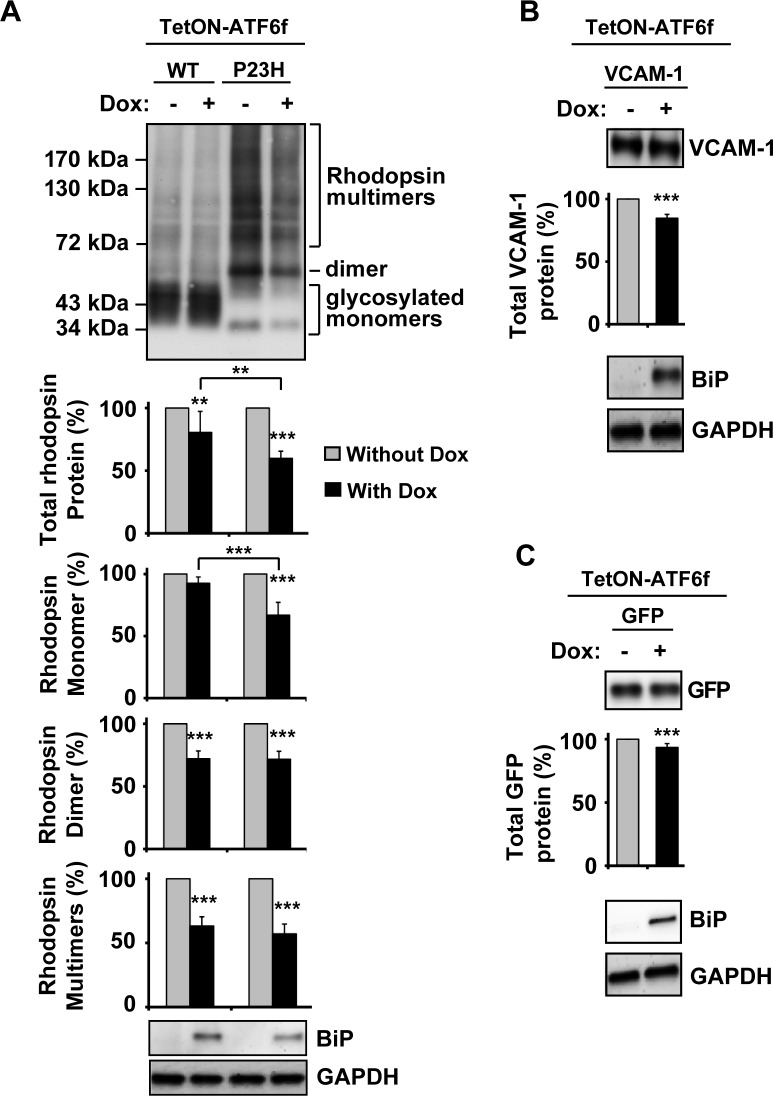
When we expressed P23H rhodopsin, the most common rhodopsin misfolding mutant in the North American population,2 and activated ATF6 signaling by adding drug to the cell culture media, we observed a pronounced reduction in total P23H rhodopsin protein levels (reduced to 59.8 ± 5.7% of levels seen in untreated cells after 24 hours of ATF6 induction; Fig. 2A). All species of P23H protein (monomer, dimer, and multimers) were affected by ATF6 activation (monomer reduced to 66.9 ± 10.3%, dimer reduced to 71.6 ± 6.4%, and multimers reduced to 57.1 ± 7.6%). By contrast, ATF6 activation had a small but significant effect on the total protein level of WT rhodopsin (Fig. 2A) and, interestingly, these effects were mostly limited to dimer and multimeric WT rhodopsin species (reduced to 72.1 ± 6.2% and 63.2 ± 7.2%, respectively), with milder effects on WT rhodopsin monomer protein levels (decreased to 92.6 ± 5.1%). We also examined how ATF6 activation affected levels of another WT membrane protein, vascular cell adhesion molecule 1 (VCAM-1), a single-pass membrane protein that has no propensity to dimerize or oligomerize. We saw small but significant alterations in VCAM-1 protein levels on ATF6 activation, similar to the effects we observed with WT rhodopsin (Fig. 2B). In addition, ATF6 activation also had a small but significant effect on the level of overexpressed cytosolic proteins such as GFP (Fig. 2C), but no effect on the level of endogenous cytosolic GAPDH. These findings indicated that ATF6 signaling preferentially reduced P23H rhodopsin levels affecting all conformations of the mutant P23H rhodopsin protein.
Figure 2. .
Chemical–genetic activation of ATF6 reduced misfolded P23H rhodopsin protein levels. (A) WT or P23H rhodopsin, (B) WT VCAM-1, or (C) GFP was transfected into cells expressing a tetracycline-inducible ATF6f (TetON-ATF6f), and Dox (1 μg/mL) was applied as indicated for 24 hours. Rhodopsin, VCAM-1, or GFP protein levels were detected by immunoblotting using 1D4 anti-rhodopsin, anti-VCAM-1, and anti-GFP antibody respectively and quantified by a commercial image acquisition and analysis software program (VisionWorks LS Software). Rhodopsin monomer, dimer, and multimers protein levels were determined by measuring the area density within the indicated line or bracket. Protein levels of BiP/Grp78, a downstream transcriptional target induced by ATF6 were assessed by immunoblotting. GAPDH levels were assessed as a loading control. Statistical significance was determined by one-way ANOVA with Bonferroni post hoc test (A, n = 7) or Student's t-test (B, C; n = 3), and is denoted by asterisks: **P < 0.01 and ***P < 0.001 compared with the cells expressing the transfected protein without the treatment of doxycycline or as indicated.
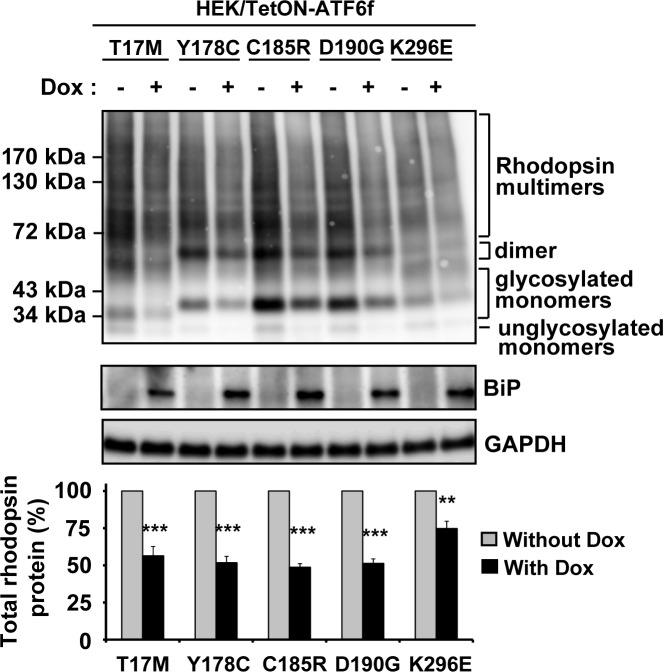
Next, we investigated the effects of selectively activating ATF6 on other class II mutant rhodopsin proteins also linked to ADRP. Similar to the effects we saw with P23H rhodopsin, we observed reductions in total protein levels of misfolded class II rhodopsins4,7,28–30 examined, including T17M, Y178C, C185R, D190G, and K296E (reduced to 56.4 ± 2.4%, 51.7 ± 3.0%, 48.7 ± 4.9%, 51.3 ± 8.6%, and 74.8 ± 9.2%, respectively; Fig. 3), after ATF6 activation compared with uninduced cells. In our system, expressing class II mutant rhodopsins mildly induced the UPR (unpublished data). To rule out the possibility that the reduction of class II mutant rhodopsin protein levels seen after ATF6 activation was due to inadvertent additional induction of PERK-mediated translational attenuation, we confirmed that ATF4, a marker of PERK signaling downstream of phosphor-eIF2α, was not upregulated by the doxycycline treatment in our studies with ATF6 (see Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/53/11/7159/suppl/DC1), indicating that the PERK branch of UPR was not further activated. Taken together, our results showed that ATF6 signaling preferentially reduced protein levels of class II mutant rhodopsins, with minimal effects on the protein levels of WT rhodopsin, or other WT membrane protein and cytosolic proteins.
Figure 3. .
Chemical–genetic activation of ATF6 reduced five additional class II mutant rhodopsin protein levels. T17M, Y178C, C185R, D190G, or K296E class II mutant rhodopsins were transfected into cells expressing TetON-ATF6f, and Dox (1 μg/mL) was applied as indicated for 24 hours. Rhodopsin protein levels were detected by immunoblotting using 1D4 anti-rhodopsin antibody and quantified by a commercial image acquisition and analysis software program (VisionWorks LS Software). Protein levels of BiP/Grp78, a downstream transcriptional target induced by ATF6, were assessed by immunoblotting. GAPDH levels were assessed as a loading control. Immunoblots are representative of three independent experiments. Statistical significance (mean ± SD; n = 3) was determined by Student's t-test, and is denoted by asterisks: **P < 0.01 and ***P < 0.001 compared with the cells expressing transfected mutant rhodopsin without the treatment of doxycycline.
Selective Activation of ATF6 or IRE1 Reduces the Protein Levels of S334ter Mutant Rhodopsin
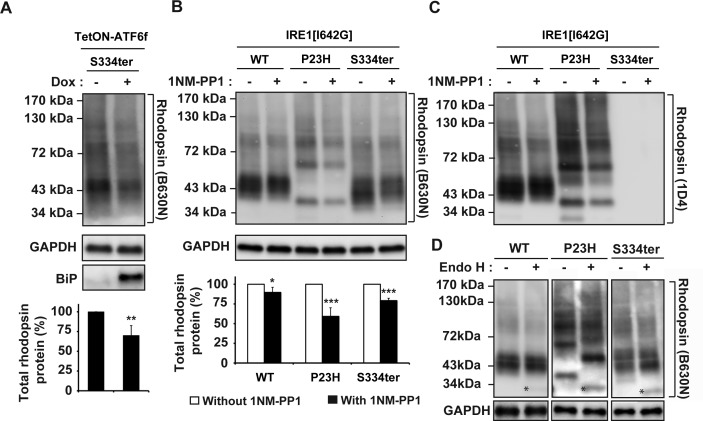
Next, we examined how ATF6 signaling affected a non–class II rhodopsin mutant bearing a nonsense mutation at serine residue 334 (S334ter). This mutation results in the synthesis of a truncated rhodopsin protein lacking the last 15 amino acid residues in the C-terminal tail. S334ter rhodopsin leads to severe retinal degeneration phenotypes in several animal models.31–35 By contrast to class II rhodopsins, which bear missense mutations that cause misfolding (e.g., enhanced oligomerization) and ER retention, S334ter rhodopsin lacks the 15 carboxyl-most residues required for proper intracellular trafficking as well as for precise termination of phototransduction signaling.36–38 For these studies, we used the B630N anti-rhodopsin antibody rather than the 1D4 anti-rhodopsin antibody used in our other experiments, because 1D4 recognized a carboxyl-terminal epitope of rhodopsin that was deleted in the S334ter truncation mutant, whereas B630N recognized an amino-terminal epitope still present in the S334ter mutant rhodopsin (Fig. 4C). To our surprise, activation of ATF6 also resulted in a significant reduction of S334ter rhodopsin protein levels (decreased to 70.0 ± 12.5%; Fig. 4A), when compared with uninduced cells. The decrease in S334ter protein levels was similar to the reduction of class II mutant rhodopsin protein levels that was detected after the induction of ATF6 (Figs. 2A, 3).
Figure 4. .
Chemical–genetic activation of ATF6 and IRE1 reduced S334ter rhodopsin protein levels. (A) S334ter rhodopsin was transfected into cells expressing TetON-ATF6f, and Dox (1 μg/mL) was applied as indicated for 24 hours. Rhodopsin protein levels were detected by immunoblotting using B630N anti-rhodopsin antibody and quantified by a commercial image acquisition and analysis software program (VisionWorks LS Software). Protein levels of BiP/Grp78, a downstream transcriptional target induced by ATF6, were assessed by immunoblotting. (B) WT or mutant rhodopsin was transfected into cells bearing IRE1[I642G], and 1NM-PP1 (5 μM) was applied as indicated for 24 hours. Rhodopsin protein levels were detected by immunoblotting using B630N anti-rhodopsin antibody and quantified by a commercial software program (VisionWorks LS Software). (A, B) GAPDH levels were assessed as a loading control. Immunoblots are representative of three independent experiments. Statistical significance (mean ± SD; n = 3) was determined by Student's t-test, and is denoted by asterisks: *P < 0.05, **P < 0.01, and ***P < 0.001 compared with the cells expressing WT or mutant rhodopsin without the treatment of doxycycline or 1NM-PP1. (C) For comparison, the same immunoblot from (B) was stripped and reprobed with 1D4 anti-rhodopsin antibody. (D) To measure the amount of Endo H–sensitive rhodopsin, 10 μg of total cell lysates expressing wild-type, P23H, or S334ter rhodopsin were treated with Endo H and the deglycosylated Endo H–sensitive species were resolved by SDS-PAGE and immunoblotting analysis. An asterisk (*) indicates the deglycosylated rhodopsin monomer isolated after Endo H treatment. GAPDH protein levels were assessed as a protein loading control.
We previously demonstrated that the chemical–genetic activation of IRE1 signaling pathway reduced levels of misfolded class II T17M, C185R, Y178C, D190G, and K296E rhodopsins.23 We therefore investigated how IRE1-XBP1 signaling affected this non–class II S334ter rhodopsin. The expression of S334ter rhodopsin in combination with IRE1 activation resulted in a reduction in S334ter protein levels (protein level of S334ter decreased to 79.2 ± 3.0%; Fig. 4B), when compared with IRE1-uninduced cells. Consistent with our prior studies, we also observed strong reduction in P23H rhodopsin protein levels and minimal effects on WT rhodopsin protein levels after selective IRE1 activation (protein level of WT rhodopsin decreased to 89.7 ± 6.5% and P23H rhodopsin decreased to 59.2 ± 10.9%; Fig. 4B). These findings showed that the selective activation of ATF6 or IRE1 could surprisingly reduce levels of non–class II rhodopsins in addition to their effects on misfolded class II rhodopsin such as P23H rhodopsin.
One possibility why S334ter rhodopsin protein was influenced by ER protein quality control pathways regulated by ATF6 and IRE1 could be that there was more S334ter rhodopsin present in the ER. Indeed, immunohistochemical studies had shown increased S334ter rhodopsin mislocalized to photoreceptor inner segment.36–38 To biochemically examine if there was increased S334ter rhodopsin in the ER, we analyzed S334ter rhodopsin susceptibility to Endo H, an enzyme that specifically cleaves high mannose N-linked glycans present only on proteins that have not matured beyond the ER.39,40 As a control and consistent with prior studies, we found that P23H rhodopsin showed pronounced Endo H sensitivity compared with wild-type rhodopsin, consistent with its retention within the ER (compare amounts of “*” labeled Endo H–sensitive protein between WT and P23H; Fig. 4D). Surprisingly, we found that S334ter rhodopsin showed increased Endo H sensitivity intermediate to those of WT and P23H rhodopsin (compare amounts of “*” labeled deglycosylated monomer bands between WT, P23H, and S334ter; Fig. 4D). These studies indicated that S334ter rhodopsin levels were increased in the ER in our experimental system and may therefore be more accessible to the effects of ATF6 and IRE1 ER protein quality control systems.
ATF6 Does Not Enhance P23H Rhodopsin Delivery to Surface Plasma Membrane
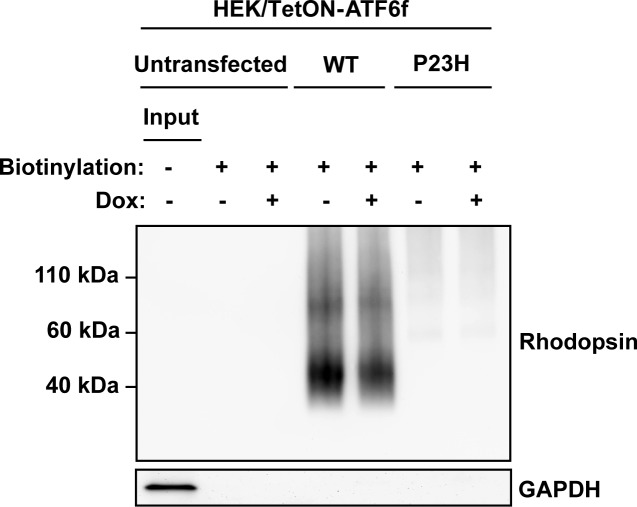
Pharmacologic and genetic chaperones partially correct P23H rhodopsin protein misfolding and promote delivery of P23H rhodopsin from ER to plasma membrane.41–44 ATF6 upregulates chaperones, such as BiP, and other genes that are involved in the transport of secreted and membrane proteins down the secretory pathway.15,16 Therefore, we examined whether selectively activating ATF6 signaling affected the delivery of P23H rhodopsin to the cell surface. We therefore used a cell surface biotinylation assay to measure changes in the amount of rhodopsin proteins at the cell surface plasma membrane after ATF6 activation. When we expressed WT rhodopsin, we detected large amounts of biotinylated rhodopsin protein irrespective of ATF6 activation (Fig. 5). By contrast, we saw minimal amounts of biotinylated P23H rhodopsin protein in cells consistent with its retention within the ER. Activation of ATF6 did not enhance the amount of biotinylated P23H rhodopsin (Fig. 5). Therefore, our findings demonstrated that ATF6 signaling did not enhance the delivery of P23H rhodopsin to the plasma membrane. Instead, the main effect of ATF6 signaling was to reduce intracellular P23H rhodopsin protein levels.
Figure 5. .
Chemical–genetic activation of ATF6 does not promote the delivery of rhodopsin to the cell surface. WT or P23H rhodopsin was transfected into cells expressing Tet-On-ATF6f, with or without application of Dox (1 μg/mL) for 24 hours. Surface membrane proteins were biotinylated, and rhodopsin protein levels in the biotinylated fraction were assessed by immunoblotting using 1D4 anti-rhodopsin antibody. GAPDH protein levels in the biotinylated protein fractions were assessed as a control for the specificity of surface protein biotinylation. Input: total cell lysate of the untransfected cells.
Selective Activation of PERK Reduces the WT and P23H Rhodopsin Protein Levels
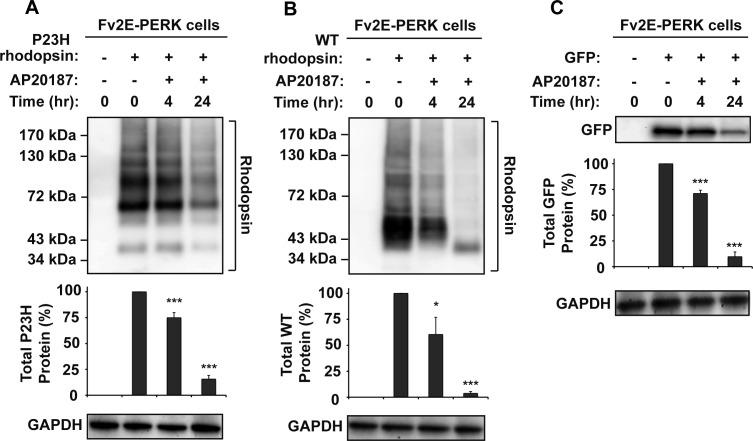
To determine how PERK signaling affects P23H rhodopsin protein, we used a chemical–genetic strategy that enabled artificial activation of a genetically altered PERK protein, consisting of PERK's eIF2α kinase domain fused to FK506 protein binding domains (Fv2E-PERK) by the FK506-like dimerizing molecule, AP20187. In this system, addition of AP20187 to the cell culture medium rapidly activated Fv2E-PERK's eIF2α kinase domain without activating the other endogenous UPR signaling pathway.24,45,46 When we expressed P23H rhodopsin in cells with Fv2E-PERK, we observed a pronounced decrease in P23H rhodopsin protein level after AP20187 treatment, compared with the untreated cells (decreased to 15.6 ± 3.6%; Fig. 6A). A similar reduction was observed in cells expressing WT rhodopsin after AP20187 treatment (down to 3.7 ± 1.7% of protein levels in untreated cells after 24 hours of drug exposure; Fig. 6B). A similar decrease was also observed in the protein level of cytosolic GFP to 9.7 ± 4.6% after 24 hours of AP20187 treatment (Fig. 6C). We also found that the transfected WT VCAM-1 protein level was also greatly reduced by selectively activating the PERK signaling pathway (unpublished data). However, endogenous GAPDH protein levels were not affected in these experiments, probably due to the long half-life of GAPDH. Interestingly, we observed a faster migrating WT rhodopsin band after 24 hours of PERK activation. This faster migrating WT rhodopsin band (∼37 kDa) corresponded to the core-glycosylated monomer and may be due to the PERK-induced translational attenuation of critical proteins required for protein glycosylation and maturation. PERK could also reduce levels of critical factors required for rhodopsin insertion into the ER or for rhodopsin folding in the ER, thereby also contributing to the sharp reduction seen in rhodopsin protein levels. Taken together, our findings demonstrated that PERK signaling strongly reduced levels of both mutant and WT rhodopsin proteins as well as the cytosolic GFP and WT VCAM-1, consistent with PERK's role in globally attenuating protein translation in the cell.
Figure 6. .
Chemical–genetic activation of PERK reduced WT and P23H rhodopsin protein levels. (A) P23H rhodopsin was transfected into cells expressing drug-sensitized Fv2E-PERK, and AP20187 (2 nM) was applied as indicated. (B) WT rhodopsin was transfected into cells expressing Fv2E-PERK, and AP20187 (2 nM) was applied as indicated. (C) GFP was transfected into HEK293 cells expressing Fv2E-PERK, and AP20187 (2 nM) was applied as indicated. (A–C) Rhodopsin or GFP protein levels were detected by immunoblotting and quantified by a commercial image acquisition and analysis software program (VisionWorks LS Software). GAPDH levels were assessed as a protein loading control. Immunoblots are representative of three independent experiments. Statistical significance (mean ± SD; n = 3) was determined by Student's t-test, and is denoted by asterisks: *P < 0.05 and ***P < 0.001 compared with the cells expressing WT or P23H rhodopsin without the treatment of AP20187.
Discussion
The unfolded protein response (UPR) signaling pathways are intracellular regulators of ER protein quality and ER homeostasis. UPR signaling could be useful in preventing or treating retinal diseases arising from protein misfolding and/or ER stress. To examine how UPR signaling affected mutant rhodopsin protein linked to ADRP, we applied chemical and genetic strategies to artificially activate the ATF6, PERK, and IRE1 signaling pathways. We found that the central effect of UPR signaling (either ATF6, PERK, or IRE1) on rhodopsin was to reduce its intracellular protein levels with varying degrees of selectivity for WT versus mutant rhodopsins. ATF6 signaling preferentially reduced the levels of misfolded class II rhodopsins and oligomerized WT rhodopsin, compared with WT monomeric rhodopsin, and other membrane or cytosolic proteins. Surprisingly, we found that ATF6 and IRE1 signaling also preferentially reduced non–class II S334ter mutant rhodopsin protein levels.23 By contrast, PERK signaling reduced levels of WT and mutant rhodopsins along with most cellular (cytosolic or ER) proteins. None of the UPR pathways “corrected” rhodopsin protein misfolding or enhanced delivery to surface plasma membrane.
Our prior studies showed that IRE1 signaling reduced mutant rhodopsin levels by promoting its degradation, in part through induction of genes that promote ER-associated degradation (ERAD).16,22,23,47 ATF6 signaling is also likely to promote the degradation of mutant rhodopsin protein because many ATF6 downstream target genes are also involved in ERAD and overlap with those induced by IRE1 signaling.15,48 Indeed, Atf6α−/− MEF cells show defects in degradation of other misfolded proteins such as null Hong Kong (NHK) variant of α1-antitrypsin.15 Furthermore, selective activation of the ATF6 pathway limited cell toxicity induced by another variant of α1-antitrypsin with an E342K (Z) mutation, by selectively promoting ERAD of the mutant protein in an HRD1 E3 ubiquitin-ligase–dependent manner.49 Similarly, selective activation of ATF6 induced the upregulation of HRD1-dependent ERAD of amyloid precursor protein (APP) and amyloid-β peptides (Aβs) associated with Alzheimer's disease.50 Taken together, these findings suggest that the selective activation of ATF6 may prevent the accumulation of misfolded rhodopsin by upregulating ERAD components such as HRD1 that facilitate proteasomal degradation of mutant rhodopsin.
By contrast to the selective reduction of mutant rhodopsin protein levels seen with ATF6 or IRE1 signaling, PERK signaling showed no preference between WT and mutant rhodopsins in reducing protein levels. PERK reduces protein levels using a mechanism distinct from that used by IRE1 or ATF6. Specifically, PERK phosphorylates translation initiation factor eIF2α, thereby inhibiting ribosomal assembly on 5′-UTR of mRNAs leading to translational attenuation.17 Some mRNAs escape PERK-mediated translational suppression by using unique “decoy” open reading frames in their 5′-UTRs.51,52 Rhodopsin mRNA is unlikely to bear these decoys because we observed reduction in protein levels for all WT and mutant rhodopsins tested in our studies.
Class II mutant rhodopsins misfold and accumulate in the ER, ultimately leading to photoreceptor cell death and causing the clinical symptoms of ADRP. No effective treatments exist to treat these patients. Our findings suggest that ATF6 signaling could be beneficial by preferentially removing mutant class II rhodopsins from cells. Furthermore, our findings suggest that ATF6 and IRE1 signaling may also be useful in removing non–class II mutant rhodopsins such as the S334ter rhodopsin. Genetic or chemical expression of a target of ATF6 signaling, the BiP/Grp78 ER chaperone, in P23H rat photoreceptors or RGC-5 cells, improved visual function and decreased cell death by unknown mechanisms.9,53 Coupled with our prior study showing beneficial effects for IRE1 signaling for cells expressing mutant rhodopsin or treated with ER stress-inducing agents,8,23 we propose that selective or combinatorial activation of the ATF6 or IRE1 signaling pathways may help enhance the survival of photoreceptors confronted with protein misfolding and/or ER stress.
Supplementary Material
Acknowledgments
The authors thank W. C. Smith and L. Wiseman for helpful comments and generous provision of reagents.
Footnotes
Supported in part by National Eye Institute/National Institutes of Health Grants EY018313 and EY020846; Fight-for-Sight Foundation (W-CC); and the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (NH).
Disclosure: W.-C. Chiang, None; N. Hiramatsu, None; C. Messah, None; H. Kroeger, None; J.H. Lin, None
References
- 1.Krauss HR, Heckenlively JR. Visual field changes in cone-rod degenerations. Arch Ophthalmol. 1982;100:1784–1790 [DOI] [PubMed] [Google Scholar]
- 2.Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34:1659–1676 [PubMed] [Google Scholar]
- 3.Park PS, Lodowski DT, Palczewski K. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu Rev Pharmacol Toxicol. 2008;48:107–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:8840–8844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaushal S, Khorana HG. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128 [DOI] [PubMed] [Google Scholar]
- 6.Illing ME, Rajan RS, Bence NF, Kopito RR. A rhodopsin mutant linked to autosomal dominant retinitis pigmentosa is prone to aggregate and interacts with the ubiquitin proteasome system. J Biol Chem. 2002;277:34150–34160 [DOI] [PubMed] [Google Scholar]
- 7.Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. J Cell Sci. 2002;115:2907–2918 [DOI] [PubMed] [Google Scholar]
- 8.Lin JH, Li H, Yasumura D, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbatyuk MS, Knox T, LaVail MM, et al. Restoration of visual function in P23H rhodopsin transgenic rats by gene delivery of BiP/Grp78. Proc Natl Acad Sci U S A. 2010;107:5961–5966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086 [DOI] [PubMed] [Google Scholar]
- 11.Ye J, Rawson RB, Komuro R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364 [DOI] [PubMed] [Google Scholar]
- 12.Okada T, Haze K, Nadanaka S, et al. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J Biol Chem. 2003;278:31024–31032 [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111 [DOI] [PubMed] [Google Scholar]
- 14.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Rutkowski DT, Dubois M, et al. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364 [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto K, Sato T, Matsui T, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376 [DOI] [PubMed] [Google Scholar]
- 17.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274 [DOI] [PubMed] [Google Scholar]
- 18.Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633 [DOI] [PubMed] [Google Scholar]
- 19.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96 [DOI] [PubMed] [Google Scholar]
- 20.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206 [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891 [DOI] [PubMed] [Google Scholar]
- 22.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang WC, Messah C, Lin JH. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell. 2012;23:758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bishop AC, Ubersax JA, Petsch DT, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401 [DOI] [PubMed] [Google Scholar]
- 26.Hiramatsu N, Joseph VT, Lin JH. Monitoring and manipulating mammalian unfolded protein response. Methods Enzymol. 2011;491:183–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529 [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Wang M, Xia CH, et al. Severe retinal degeneration caused by a novel rhodopsin mutation. Invest Ophthalmol Vis Sci. 2010;51:1059–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol Med. 2005;11:177–185 [DOI] [PubMed] [Google Scholar]
- 30.Rakoczy EP, Kiel C, McKeone R, Stricher F, Serrano L. Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J Mol Biol. 2011;405:584–606 [DOI] [PubMed] [Google Scholar]
- 31.Lee D, Geller S, Walsh N, et al. Photoreceptor degeneration in Pro23His and S334ter transgenic rats. Adv Exp Med Biol. 2003;533:297–302 [DOI] [PubMed] [Google Scholar]
- 32.Dykens JA, Carroll AK, Wiley S, et al. Photoreceptor preservation in the S334ter model of retinitis pigmentosa by a novel estradiol analog. Biochem Pharmacol. 2004;68:1971–1984 [DOI] [PubMed] [Google Scholar]
- 33.Anderson RE, Maude MB, McClellan M, Matthes MT, Yasumura D, LaVail MM. Low docosahexaenoic acid levels in rod outer segments of rats with P23H and S334ter rhodopsin mutations. Mol Vis. 2002;8:351–358 [PubMed] [Google Scholar]
- 34.Shinde VM, Sizova OS, Lin JH, LaVail MM, Gorbatyuk MS. ER stress in retinal degeneration in S334ter Rho rats. PLoS One. 2012;7:e33266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennesi ME, Nishikawa S, Matthes MT, Yasumura D, LaVail MM. The relationship of photoreceptor degeneration to retinal vascular development and loss in mutant rhodopsin transgenic and RCS rats. Exp Eye Res. 2008;87:561–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Concepcion F, Mendez A, Chen J. The carboxyl-terminal domain is essential for rhodopsin transport in rod photoreceptors. Vision Res. 2002;42:417–426 [DOI] [PubMed] [Google Scholar]
- 37.Green ES, Menz MD, LaVail MM, Flannery JG. Characterization of rhodopsin mis-sorting and constitutive activation in a transgenic rat model of retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:1546–1553 [PubMed] [Google Scholar]
- 38.Lee ES, Flannery JG. Transport of truncated rhodopsin and its effects on rod function and degeneration. Invest Ophthalmol Vis Sci. 2007;48:2868–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherblom AP, Smagula RM. High-mannose chains of mammalian glycoproteins. Methods Mol Biol. 1993;14:143–149 [DOI] [PubMed] [Google Scholar]
- 40.Trimble RB, Maley F. Optimizing hydrolysis of N-linked high-mannose oligosaccharides by endo-beta-N-acetylglucosaminidase H. Anal Biochem. 1984;141:515–522 [DOI] [PubMed] [Google Scholar]
- 41.Noorwez SM, Kuksa V, Imanishi Y, et al. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J Biol Chem. 2003;278:14442–14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noorwez SM, Malhotra R, McDowell JH, Smith KA, Krebs MP, Kaushal S. Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H. J Biol Chem. 2004;279:16278–16284 [DOI] [PubMed] [Google Scholar]
- 43.Kosmaoglou M, Kanuga N, Aguila M, Garriga P, Cheetham ME. A dual role for EDEM1 in the processing of rod opsin. J Cell Sci. 2009;122:4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendes HF, Cheetham ME. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Hum Mol Genet. 2008;17:3043–3054 [DOI] [PubMed] [Google Scholar]
- 45.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu PD, Jousse C, Marciniak SJ, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271 [DOI] [PubMed] [Google Scholar]
- 48.Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89 [DOI] [PubMed] [Google Scholar]
- 49.Smith SE, Granell S, Salcedo-Sicilia L, Baldini G, Egea G, Teckman JH. Activating transcription factor 6 limits intracellular accumulation of mutant alpha(1)-antitrypsin Z and mitochondrial damage in hepatoma cells. J Biol Chem. 2011;286:41563–41577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaneko M, Koike H, Saito R, Kitamura Y, Okuma Y, Nomura Y. Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-beta generation. J Neurosci. 2010;30:3924–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108 [DOI] [PubMed] [Google Scholar]
- 52.Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073 [DOI] [PubMed] [Google Scholar]
- 53.Inokuchi Y, Nakajima Y, Shimazawa M, et al. Effect of an inducer of BiP, a molecular chaperone, on endoplasmic reticulum (ER) stress-induced retinal cell death. Invest Ophthalmol Vis Sci. 2009;50:334–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.