SUMMARY
Spinal muscular atrophy (SMA) is a motor neuron disease caused by deficiency of the ubiquitous survival motor neuron (SMN) protein. To define the mechanisms of selective neuronal dysfunction in SMA, we investigated the role of SMN-dependent U12 splicing events in the regulation of motor circuit activity. We show that SMN deficiency perturbs splicing and decreases the expression of a subset of U12 intron-containing genes in mammalian cells and Drosophila larvae. Analysis of these SMN target genes identifies Stasimon as a novel protein required for motor circuit function. Restoration of Stasimon expression in the motor circuit corrects defects in neuromuscular junction transmission and muscle growth in Drosophila SMN mutants and aberrant motor neuron development in SMN-deficient zebrafish. These findings directly link defective splicing of critical neuronal genes induced by SMN deficiency to motor circuit dysfunction, establishing a molecular framework for the selective pathology of SMA.
INTRODUCTION
RNA splicing is a fundamental regulatory mechanism of eukaryotic gene expression that is crucial in the nervous system. Accordingly, mutations in proteins involved in RNA splicing have been associated with human neurodegenerative diseases (Cooper et al., 2009). However, an unsolved conundrum is how disruption of ubiquitously expressed splicing factors can cause selective dysfunction of specific subsets of neurons. The inherited neurodegenerative disease spinal muscular atrophy (SMA) is a prominent example of this enigma.
SMA is an autosomal recessive disorder characterized by degeneration of motor neurons and atrophy of skeletal muscle. SMA is caused by homozygous inactivation of the Survival Motor Neuron 1 (SMN1) gene (Lefebvre et al., 1995). The nearly identical SMN2 gene is unable to compensate for the loss of SMN1 as it produces low levels of functional SMN protein. Consistent with human pathology, in both invertebrate and vertebrate animal models low levels of SMN are sufficient for normal function of most cell types but not of motor neurons (Burghes and Beattie, 2009). However, the mechanisms that link ubiquitous SMN deficiency to selective neuronal dysfunction remain unclear.
The SMN protein forms a macromolecular complex whose only defined activity is in the biogenesis of small nuclear ribonucleoproteins (snRNPs) of the Sm-class (Neuenkirchen et al., 2008; Pellizzoni, 2007), essential components of the RNA splicing machinery composed of an snRNA molecule, seven common Sm proteins and additional snRNP-specific proteins. The SMN complex mediates the assembly of a heptameric ring of Sm proteins around a conserved sequence of each snRNA to form the Sm core required for snRNP stability and function (Meister et al., 2001; Pellizzoni et al., 2002).
Although SMN has been implicated in other cellular processes that could be relevant to SMA (Burghes and Beattie, 2009), increasing evidence support the hypothesis that SMN-dependent snRNP defects contribute to motor neuron dysfunction in the disease. First, cell lines from SMA patients show reduced snRNP assembly (Wan et al., 2005). Second, the degree of impairment of snRNP assembly correlates with disease severity in SMA mice (Gabanella et al., 2007). Third, SMN deficiency leads to a decrease in the levels of spliceosomal snRNPs (Gabanella et al., 2007; Zhang et al., 2008) and this reduction is more pronounced in motor neurons compared to other spinal cells in SMA mice (Ruggiu et al., 2012). Lastly, restoring normal snRNP levels provides phenotypic correction in both zebrafish and mouse models of SMA (Winkler et al., 2005; Workman et al., 2009). Consistent with snRNP dysfunction in SMA, widespread splicing changes have been found in tissues of SMA mice (Zhang et al., 2008). However, as this analysis was performed from late disease stages, it is difficult to discriminate direct effects of SMN deficiency from secondary consequences of degeneration (Baumer et al., 2009).
Insights into how perturbation of RNA splicing might lead to specific neuronal defects and possible ways to identify disease-relevant splicing events emerged from analysis of the effects of SMN deficiency on snRNP biology in vivo. Spliceosomal snRNPs comprise two distinct classes each dedicated to the removal of different intron types. Most eukaryotic introns are processed by the major (U2-dependent) spliceosome formed by U1, U2, U4/U6 and U5 snRNPs, while a small proportion of introns (~1%) are processed by the minor (U12-dependent) spliceosome comprised of U11, U12, U4atac/U6atac and U5 snRNPs (Patel and Steitz, 2003). Importantly, SMN deficiency changes the snRNP profile of tissues in a non-uniform manner and appears to preferentially reduce accumulation of minor snRNPs (Gabanella et al., 2007; Zhang et al., 2008). This has led to the hypothesis that genes containing U12 introns could be among the disease-relevant targets affected by SMN reduction in SMA (Gabanella et al., 2007). Consistent with this possibility, evidence for inefficient U12 splicing in cells from an SMA patient has recently been reported (Boulisfane et al., 2011). However, no SMN-dependent U12 splicing event has yet been directly linked to the SMA phenotype.
Here, we have used a combination of cellular and animal models to investigate the effects of SMN deficiency on U12 splicing and the potential link for disruption of this pathway to motor circuit dysfunction in SMA. We show that U12 splicing is disrupted in both SMN-deficient mammalian cells and Drosophila SMN mutant larvae. To link these splicing defects to motor circuit function in vivo, we exploited two advantages of the Drosophila model. First, Drosophila SMN loss-of-function mutants have selective defects in motor neuron electrophysiology and alterations in motor circuit function (Imlach et al., 2012). Second, whereas several hundred genes with U12 introns are present in the human and mouse genomes, Drosophila has only 23 genes with predicted U12 introns (Alioto, 2007; Lin et al., 2010), hence making their genome-wide functional analysis manageable. Capitalizing on these advantages, we have identified the gene stasimon as a U12 intron-containing SMN target that encodes a novel evolutionarily conserved transmembrane protein required for motor circuit function. We show that loss of Stasimon induces phenotypes that mirror aspects of SMN deficiency in Drosophila as well as zebrafish and that restoration of Stasimon can rescue SMN-dependent neuronal defects in both fly and zebrafish SMA models. We also show that SMN deficiency affects Stasimon U12 splicing and mRNA expression in the motor circuit of a mouse model of SMA that recapitulates many aspects of the human disease. Together, our data directly link defective splicing of a gene with essential functions in motor circuits to the phenotypic consequences of SMN deficiency, establishing a mechanistic basis for the neuronal selectivity of SMA.
RESULTS
SMN is required for U12 splicing in mammalian cells
We sought to investigate SMN requirement for U12 splicing based on the preferential reduction of minor snRNPs that occurs in SMA mice (Gabanella et al., 2007; Zhang et al., 2008). To study SMN-dependent U12 splicing events, we used a mouse NIH3T3 cell line that allows doxycycline (Dox)-inducible, RNAi-mediated depletion of SMN (see Supplemental Information). NIH3T3-SmnRNAi cells cultured in the presence of Dox for 5 days showed strong knockdown of SMN mRNA (Figure S1A–B) and reduction of SMN protein levels (Figure 1A) compared to untreated cells. SMN deficiency severely decreased snRNP assembly of snRNAs in vitro and caused a profound alteration of their expression in NIH3T3 cells (Figures 1B and S1C–E), including a reduction in the levels of all Sm-class snRNPs of the U12 spliceosome. Importantly, expression of RNAi-resistant human SMN in NIH3T3-SMN/SmnRNAi cells (Figure 1A) rescued these changes (Figures 1B and S1C–D), indicating that they are SMN-dependent.
Figure 1. SMN deficiency causes U12 splicing defects in mammalian cells.
(A) Western blot analysis of NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured without (−) or with (+) Dox for 5 days. (B) RT-qPCR analysis of snRNAs immunoprecipitated with anti-SmB antibodies from NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured as in (A). RNA levels in Dox-treated cells were expressed relative to untreated cells (dotted line). (C) RT-PCR analysis of U12 intron-containing genes in NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cells cultured as in (A). Genes and exons monitored by PCR are indicated on the left. Schematics of spliced and intron-containing mRNAs are shown on the right. Red lines highlight U12 introns. The asterisk indicates a Tmem41b mRNA spliced using donor and acceptor sites located at −30 and +25, respectively, relative to the U12 intron splice sites. (D) RT-qPCR analysis of U12 intron retention for a subset of genes in (C). (E) RT-qPCR analysis of mRNA expression for a subset of genes in (C). (F) RT-qPCR analysis of aberrantly spliced Tmem41b and exon-skipped Clcn7 mRNAs. For all RT-qPCR experiments, NIH3T3 cells were cultured as in (A) and RNA levels in Dox-treated cells were expressed relative to untreated cells (dotted line).
Data in all graphs are represented as mean and SEM. See also Figure S1 and Table S1.
Next, we analyzed 28 U12 introns from 25 genes (Table S1) representing a diverse spectrum of features such as i) splice site subtype, ii) intron length, iii) intron position, iv) number of U12 introns, v) total number of introns, vi) evolutionary conservation, and vii) gene function. RT-PCR experiments revealed a variety of U12 splicing defects in SMN-deficient NIH3T3-SmnRNAi cells (Figures 1C and S1F). These defects included: i) accumulation of unspliced U12 introns in pre-mRNAs for Parp1, Nol1, Vps16 (both U12 introns 9 and 13) and C19orf54; ii) skipping of the two exons flanking the U12 intron in Clcn7, Tmem41b and Tspan31 mRNAs; and iii) aberrant splicing of Harsl and Tmem41b mRNAs. In the latter events, splicing of the U12 intron is bypassed through the use of an upstream cryptic 5′ splice site that is spliced to a downstream U2-dependent 3′ splice site. All of these splicing defects were rescued by expression of human SMN in NIH3T3-SMN/SmnRNAi cells (Figure 1C) and Dox treatment had no effects in control cells (Figure S1F), demonstrating that the splicing defects are a direct consequence of reduced SMN levels. Increased U12 intron retention (Figure 1D) as well as decreased levels of mature mRNAs (Figure 1E) and accumulation of abnormally spliced mRNAs (Figure 1F) in SMN-deficient NIH3T3 cells were confirmed by RT-qPCR. However, not all U12 introns were affected by SMN deficiency as no splicing defects were observed in 19 of the 28 U12 introns we analyzed (Figure S1F). U2 splicing in genes that also had U12 introns appeared to be normal, whether or not the U12 introns in these genes were affected by SMN deficiency, as did the splicing of several mouse genes with only U2 introns (Figures S1F–G and S2A). These results established that SMN deficiency causes U12 splicing defects in mammalian cells, perturbing the expression of a subset of the genes with this type of intron.
Defects in U12 splicing events directly regulated by SMN would be expected to begin quickly after the onset of SMN reduction and increase in severity with progression of SMN depletion. To determine if this was the case, we carried out a temporal analysis in NIH3T3-SmnRNAi cells following Dox treatment over a period of 7 days. Analysis of representative target mRNAs showed that U12 intron retention began to accumulate in the first 3 days following RNAi induction and increased over time as SMN depletion progressed (Figure 2A–B). Inefficient U12 intron splicing was accompanied by accumulation of aberrantly spliced Tmem41b and exon skipped Clcn7 mRNAs (Figure 2B). The time of onset of these U12 splicing defects varied between genes indicative of a differential susceptibility of individual introns to SMN deficiency. We also found that SMN deficiency decreased the proliferation of NIH3T3-SmnRNAi cells (Figure 2C), while Dox alone had no effect in wild-type cells (Figure S2B). This reduced proliferation was SMN-dependent as it was corrected by expression of human SMN in NIH3T3-SMN/SmnRNAi cells (Figure S2C). The occurrence of SMN-dependent U12 splicing defects was detectable prior to onset of this decrease in cell proliferation, indicating that these defects were not a consequence of reduced cell proliferation. In support of this, decreasing the proliferation of NIH3T3 cells through the reduction of serum levels (Figure S2D) had no effect on SMN expression or on SMN-dependent U12 splicing events (Figure S2E–G). Thus, SMN deficiency results in both the early production and the accumulative increase of U12 intron splicing defects in mammalian cells, consistent with regulation of these events by SMN.
Figure 2. Early onset and time-dependent increase of U12 splicing defects in SMN-deficient mammalian cells.
(A) Western blot analysis of NIH3T3-SmnRNAi cells cultured with Dox for the indicated number of days. A two-fold serial dilution of the extract from uninduced cells is shown on the left. (B) RT-qPCR analysis of U12 intron retention in Clcn7, Parp1, Tspan31 and Tmem41b mRNAs as well as accumulation of abnormally spliced Tmem41b and Clcn7 mRNAs in NIH3T3-SmnRNAi cells cultured as in (A). RNA levels in Dox-treated cells were expressed relative to untreated cells (dotted line). (C) SMN deficiency decreases proliferation of NIH3T3 cells. Equal numbers of NIH3T3-SmnRNAi cells were cultured with or without Dox for the indicated number of days and cell number determined at each time point.
Data in all graphs are represented as mean and SEM. See also Figure S2.
SMN is required for expression of snRNAs and U12 intron-containing genes in Drosophila
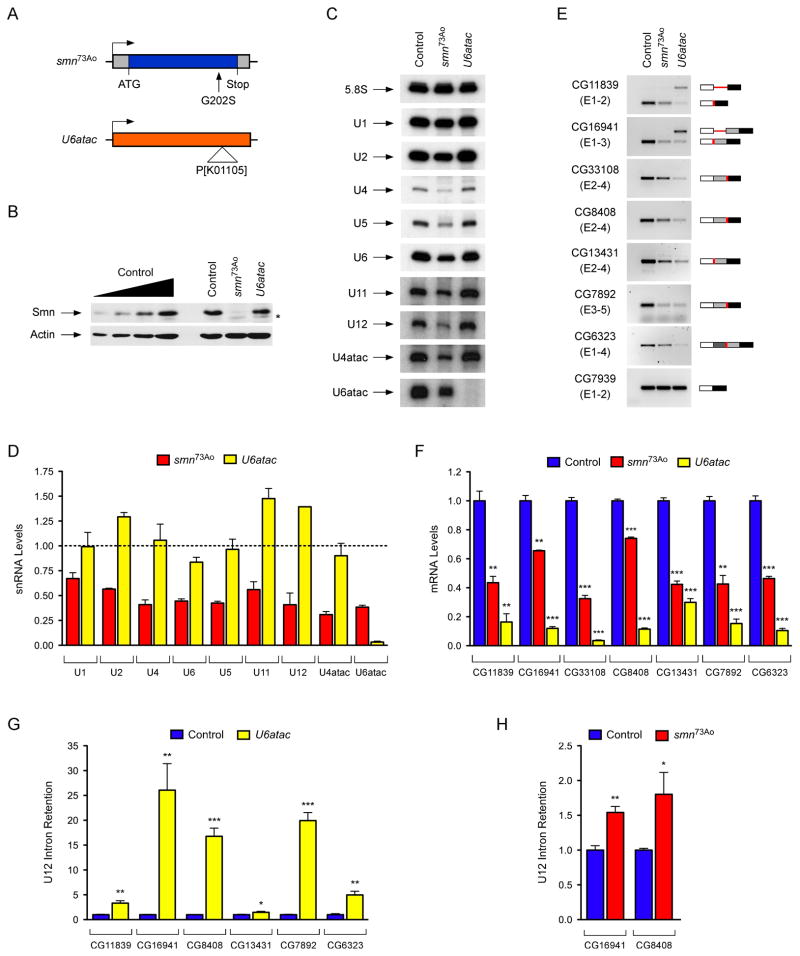
To determine the genome-wide effects of SMN deficiency on U12 splicing in vivo, we utilized Drosophila for both the availability of genetic mutants and the presence of only 23 putative U12 introns in the genome of this organism (Alioto, 2007; Lin et al., 2010) (Table S2). To deplete SMN, we utilized the previously characterized loss-of-function smn73Ao point mutant allele, which produces an unstable protein (Chan et al., 2003) (Figure 3A). As a control for intron excision by the U12 spliceosome, we used a mutant of the U6atac snRNA gene (U6atacK01105), which specifically disrupts U12 splicing (Otake et al., 2002) (Figure 3A). As expected, we found a large reduction of SMN levels in smn73Ao mutants but no change in U6atacK01105 mutants compared to wild-type third-instar larvae (Figure 3B).
Figure 3. SMN deficiency decreases snRNA levels and expression of U12 intron-containing genes in Drosophila.
(A) Schematic representation of Drosophila smn73Ao and U6atacK01105 mutants. (B) Western blot analysis of control, smn73Ao and U6atacK01105 larvae. A two-fold serial dilution of control extract is shown on the left. The asterisk indicates a non-specific protein. (C) Northern blot analysis of snRNA expression in control, smn73Ao and U6atacK01105 larvae. (D) snRNA levels in smn73Ao and U6atacK01105 relative to control (dotted line) larvae following normalization to 5.8S rRNA. (E) RT-PCR analysis of U12 intron-containing genes whose expression is affected in both smn73Ao and U6atacK01105 compared to control larvae. Genes and exons monitored by PCR are indicated on the left. Schematics of spliced and intron-containing mRNAs are shown on the right. Red lines highlight U12 introns. The –RT lanes correspond to RT-PCR reactions lacking reverse transcriptase. (F) RT-qPCR analysis of U12 intron-containing genes with decreased mRNA expression in both smn73Ao and U6atacK01105 compared to control larvae. (G) RT-qPCR analysis of genes with increased U12 intron retention in U6atacK01105 compared to control larvae. (H) RT-qPCR analysis of genes with increased U12 intron retention in smn73Ao compared to control larvae.
Data in all graphs are represented as mean and SEM. See also Figure S3 and Table S2.
We then investigated the consequences of SMN deficiency on snRNA expression in Drosophila. Northern blot analysis of U6atacK01105 mutant larvae showed that U6atac is depleted while other snRNAs appeared normal or slightly increased (Figure 3C). In contrast, the levels of all the snRNAs were decreased in smn73Ao mutants with differential effects of SMN deficiency on the accumulation of individual snRNAs ranging from a 70% reduction of U4atac to a 30% reduction of U1 (Figure 3C–D). Thus, SMN is required for normal expression of spliceosomal snRNAs in Drosophila.
To study the effects of SMN deficiency and decreased snRNA levels on U12 splicing in Drosophila, we next analyzed expression and splicing of all predicted U12 intron-containing genes by RT-PCR. This genome-wide analysis revealed a decrease in the levels of spliced mRNAs or an increase of pre-mRNAs containing unspliced U12 introns in 18 of the 23 predicted U12 intron-containing genes in U6atacK01105 larvae (Figures 3E and S3A), validating the presence of U12 introns in these 18 Drosophila genes. Remarkably, the mRNA levels of 7 of the 18 U12 intron-containing genes were also decreased in smn73Ao mutants (Figure 3E). RT-qPCR experiments using smn73Ao larvae as well as a second Drosophila smn mutant allele (smnX7)—which has a complete deletion of the smn gene (Chang et al., 2008)—confirmed that the expression of these 7 genes was SMN-dependent (Figures 3F and S3C). No changes in mRNA expression or U2 splicing were detected in the remaining U12 intron-containing genes or in additional genes that contained only U2 introns (Figure S3A–B). These experiments revealed that SMN is required for normal expression of ~40% of all the U12 intron-containing genes in the Drosophila genome.
To establish if excision of U12 introns was defective in the 7 SMN-target genes, we measured the levels of U12 intron retention in both U6atacK01105 and smn73Ao mutants by RT-qPCR. Consistent with inefficient U12 splicing, we found increased U12 intron retention for 6 of these genes in U6atacK01105 mutants and for 2 of these genes in smn73Ao mutants (Figure 3G–H). Notably, the mammalian homologs of 3 of the 4 Drosophila genes that were down regulated in smn mutants and have evolutionarily conserved U12 introns (Tmem41b/CG8408, Tspan31/CG6323 and C19orf54/CG33108) also had defective U12 intron splicing in SMN-deficient NIH3T3 cells (Figure 1). To further assess if the SMN-dependent regulation of target genes was associated with the presence of U12 introns, we examined the expression and splicing of 5 genes (CG8594/Clcn7, CG8545/Nol1, CG40411/Parp1, CG6335/Harsl and CG8454/Vps16) that have only U2 introns in Drosophila but both have a U12 intron in the homologous mouse genes and were perturbed by SMN deficiency in NIH3T3 cells (Figure 1C–E). The expression and splicing of all 5 of these genes appeared normal in Drosophila smn mutants (Figure S3B). Furthermore, 3 genes (CG13431/Mgat1, CG16941/Sf3a1, CG11839/Znf830) that have both U12 introns and reduced expression in Drosophila smn mutants but only have U2 introns in the mouse homolog had normal expression in SMN-deficient NIH3T3 cells (Figures 3E and S1G). Thus, SMN deficiency affects the expression of U12 intron-containing genes in Drosophila and some of these SMN-dependent U12 splicing events are conserved across evolution.
Stasimon is an SMN target gene required for normal synaptic transmission of Drosophila motor neurons
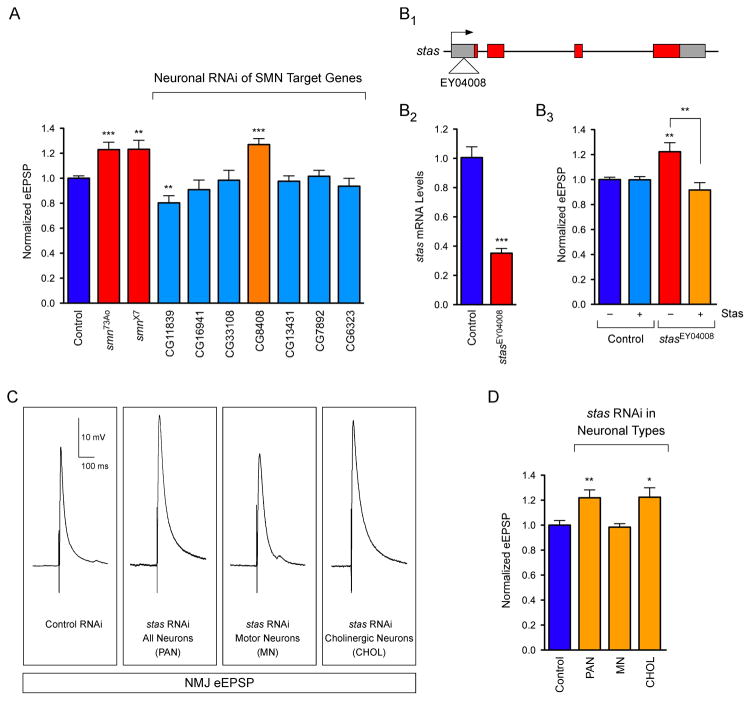
The identification of the subset of Drosophila genes with U12 introns that are regulated by SMN provided the foundation for functional analysis of their role in motor neurons. Drosophila smn73A0 and smnX7 mutants have an aberrant increase in evoked Excitatory Post-Synaptic Potential (eEPSP) amplitudes at the neuromuscular junction (NMJ) to ~125% of controls (Figures 4A and 5A–B). This aberrant NMJ neurotransmission is rapidly corrected by transgenic SMN expression in neurons (Imlach et al., 2012), consistent with this phenotype being an early consequence of SMN reduction. To determine the role of SMN-dependent U12 splicing events in motor neuron function, we analyzed if knockdown of any of the 7 U12 intron-containing genes that have reduced expression in Drosophila smn mutants would alter NMJ neurotransmission.
Figure 4. Functional analysis of SMN targets identifies CG8408/Stasimon as a novel gene required for synaptic transmission in Drosophila.
(A) Evoked Excitatory Post-Synaptic Potentials (eEPSPs) in smn73A0 and smnX7 mutant Drosophila larvae and following mRNA knockdown of the indicated genes by pan-neural expression of UAS-RNAi constructs with C155-Gal4 normalized to control. (B1–B3) Schematic representation of the stasEY04008 mutant showing the site of P-element insertion within the 5′ UTR region of the stasimon (CG8408) gene (B1). RT-qPCR analysis of Stasimon mRNA levels in control and stasEY04008 larvae (B2). Normalized eEPSP amplitude in control and stasEY04008 larvae with or without expression of UAS-Stasimon with the pan-neuronal nsyb-Gal4 driver relative to control (B3). (C) Representative eEPSP traces from larvae with Stasimon RNAi in all neurons (C155-Gal4; PAN), motor neurons (OK371-Gal4; MN) or cholinergic neurons (Cha-Gal4; CHOL) normalized to control. (D) Quantification of eEPSP amplitudes in larvae with Stasimon RNAi in specific neuronal types normalized to control.
Data in all graphs are represented as mean and SEM. See also Figure S4.
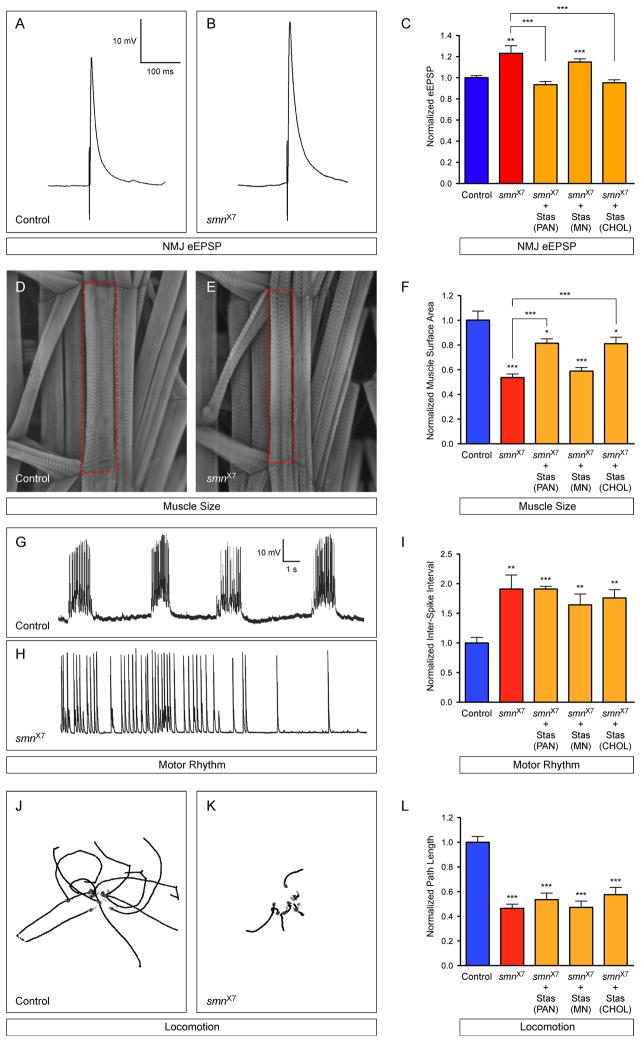
Figure 5. Expression of Stasimon rescues neurotransmitter release at the NMJ and ameliorates muscle size defects in Drosophila SMN mutants.
(A–B) Representative eEPSP traces recorded from muscle 6 of segment A3 in control and smnX7 larvae. (C) Normalized eEPSP amplitude of smnX7 mutants alone or with transgenic UAS-Stasimon expression in all neurons (nsyb-Gal4; PAN), motor neurons (OK371-Gal4; MN) or cholinergic neurons (Cha-Gal4; CHOL) relative to controls. (D–E) Representative images of muscles from segment A3 of control and smnX7 larvae labeled with TRITC-phalloidin. (F) Normalized muscle surface area of smnX7 mutants alone or expressing Stasimon with the same drivers described in (C) relative to controls. (G–H) Representative recordings of motor rhythms from control and smnX7 larvae. (I) Normalized inter-spike intervals of smnX7 mutants alone or expressing Stasimon with the same drivers described in (C) relative to controls. (J–K) Representative images of 10 superimposed locomotion path traces from control and smnX7 larvae. (L) Normalized path length of smnX7 mutants alone or expressing Stasimon with the same drivers described in (C) relative to controls.
Data in all graphs are represented as mean and SEM. See also Figure S5.
We first confirmed by RT-qPCR that transgenic UAS-RNAi constructs (Dietzl et al., 2007) were able to potently decrease the expression of their target mRNA in Drosophila larvae (Figure S4). We then measured the effect of pan-neuronal knockdown of each of these genes on NMJ neurotransmitter release. Strikingly, knockdown of CG8408 resulted in an increase in NMJ eEPSP amplitudes to 127% of controls, similar to the evoked neurotransmitter release defects observed in smn mutants (Figure 4A). In contrast, knockdown of CG11839 decreased eEPSP amplitudes at the NMJ (80.3% of control), unlike Drosophila smn mutants and therefore not pursued further, while knockdown of the 5 other SMN target genes had no effect on neurotransmitter release compared to controls (Figure 4A). As CG8408 and its mouse homologue Tmem41b (Figure 1) are both U12 intron-containing genes whose expression is regulated by SMN and knockdown of CG8408 in Drosophila neurons causes an electrophysiological phenotype similar to that of smn mutants, we renamed CG8408 stasimon (stymied in smn)—abbreviated as stas—and investigated its function further.
Homologs of Stasimon are found in humans, mice and zebrafish in addition to other species (Figure S5A), and display a remarkably high degree of amino acid conservation although their function is unknown. The Stasimon protein is predicted to contain six transmembrane domains and a SNARE-associated Golgi protein domain (Figure S5B). Furthermore, in situ hybridization revealed strong expression of Stasimon mRNA in the nervous system of Drosophila embryos as well as in the mouse spinal cord (Figure S5C–D) (Lein et al., 2007), consistent with a prominent role in neurons.
To confirm that Stasimon regulates neurotransmitter release at the Drosophila NMJ, we characterized a P-element mutant with an insertion in the 5′UTR of the stas gene (stasEY04008) (Bellen et al., 2004) (Figure 4B). RT-qPCR showed that Stasimon mRNA expression is reduced by ~65% in stasEY04008 mutants compared to control larvae (Figure 4B). Importantly, eEPSP amplitudes at the NMJ of stasEY04008 mutants were increased by 122% compared to controls (Figure 4B). To establish that reduced Stasimon expression was directly responsible for these effects, we carried out rescue experiments using a Gal4-driven Stasimon cDNA transgene. Neuronal expression of transgenic Stasimon had no effect on neurotransmitter release at the NMJ of control larvae (Figure 4B). Conversely, neuronal expression of transgenic Stasimon in stasEY04008 mutants fully restored NMJ eEPSP amplitudes to control levels (Figure 4B). These results established stasimon as an SMN target gene expressed in neurons that is required for normal synaptic transmission of motor neurons.
Stasimon is required in cholinergic neurons for normal synaptic transmission of Drosophila motor neurons
To investigate the requirement of Stasimon for the function of specific neurons in the motor circuit, we decreased Stasimon expression in subsets of Drosophila neurons by RNAi and analyzed the effects on synaptic transmission of motor neurons. Surprisingly, Stasimon knockdown in motor neurons did not change eEPSP amplitudes compared to controls (Figure 4C–D), suggesting that Stasimon is not cell autonomously required in glutamatergic motor neurons for this phenotype. In contrast, Stasimon knockdown in cholinergic neurons produced an increase in eEPSP amplitudes at motor neuron terminals (122% of control) remarkably similar to that of pan-neuronal Stasimon knockdown (Figure 4C–D). These results demonstrated that reduction of Stasimon perturbs the neurotransmitter release properties of motor neurons indirectly through disruption of the activity of cholinergic neurons in the motor circuit. Importantly, analogous NMJ neurotransmission defects in Drosophila smn mutants are caused by SMN deficiency in cholinergic neurons but not in motor neurons (Imlach et al., 2012). Thus, Stasimon has a role similar to that of SMN in the motor circuit function.
Stasimon expression rescues synaptic dysfunction in Drosophila smn mutants
To test the hypothesis that Stasimon deficiency contributes to the synaptic dysfunction in Drosophila smn mutants, we restored Stasimon expression by pan-neuronal transgenic expression of Stasimon cDNA in smnX7 mutants and then measured neurotransmitter release at the NMJ. Strikingly, restoring Stasimon expression in all neurons completely rescued the NMJ eEPSP amplitude of smn mutants to control levels (Figure 5A–C). This was consistent with the reduction of Stasimon function causing the neurotransmission defects at the NMJ of Drosophila smn mutants.
We next investigated the neuronal subtype in which Stasimon expression is required in order to rescue smn mutant NMJ electrophysiology. Similar to pan-neuronal Stasimon expression, transgenic Stasimon expression in the cholinergic neurons of Drosophila smnX7 mutants fully corrected NMJ eEPSP amplitudes to control levels (Figure 5C). In contrast, expression of transgenic Stasimon in the glutamatergic motor neurons of smnX7 mutants did not alter NMJ eEPSP amplitudes (Figure 5C). Thus, similar to SMN (Imlach et al., 2012), Stasimon expression must be restored in cholinergic neurons that provide excitatory input to motor neurons in order to rescue defects in NMJ neurotransmission in Drosophila smn mutants.
Reduced muscle size, decreased locomotion and altered rhythmic motor activity are additional phenotypes of Drosophila smn mutants (Figure 5D–L), all of which are rescued by SMN expression in cholinergic neurons (Imlach et al., 2012). To address if Stasimon contributed to these phenotypes, we analyzed the effects of Stasimon restoration in the neurons of Drosophila smn mutants on these defects. Muscle surface area in smnX7 mutants is reduced by 50% compared to controls (Figure 5D–F). Remarkably, pan-neuronal restoration of Stasimon in smnX7 mutants resulted in a robust increase in muscle area to ~80% of that in control larvae (Figure 5F). Expression of Stasimon in the cholinergic neurons but not in the motor neurons of smnX7 mutants produced an identical increase in muscle area to pan-neuronal expression (Figure 5F). These results indicated that Stasimon deficiency contributes to the muscle growth phenotype in Drosophila smn mutants. In contrast, locomotion and rhythmic motor activity were not significantly improved by pan-neuronal, motor neuron or cholinergic neuron expression of Stasimon in Drosophila smnX7 mutants (Figure 5G–L). Recently it was proposed that splicing disruption may not contribute to the locomotion phenotype of Drosophila smn mutants (Praveen et al., 2012). However, as this study did not directly evaluate the role of SMN-dependent genes, the contribution of defective splicing of genes other than Stasimon to additional Drosophila phenotypes remains an issue for future investigation. Our results establish that SMN-dependent, decreased expression of Stasimon in cholinergic neurons can account for the dysfunction of neurotransmitter release at the NMJ and contributes to defects of muscle growth in Drosophila smn mutants.
Stasimon expression rescues SMN-dependent motor neuron defects in zebrafish
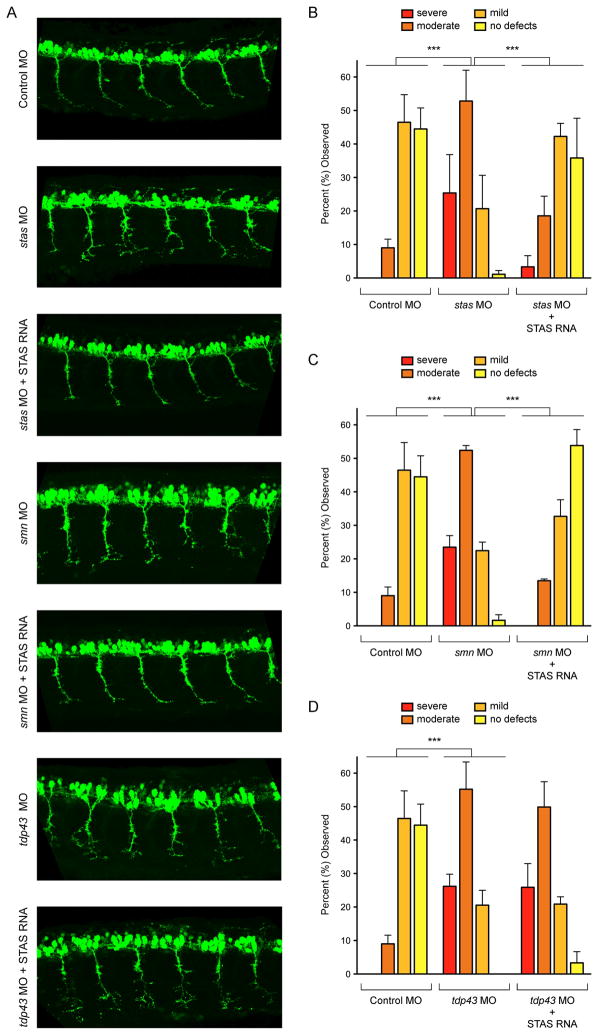
To determine if the contribution of Stasimon to SMN-dependent motor system dysfunction was conserved across species, we investigated its role in a vertebrate model of SMN deficiency (Beattie et al., 2007). Knockdown of SMN in zebrafish embryos causes developmental defects in motor neuron axonal outgrowth that include truncations and abnormal branching (McWhorter et al., 2003), which can be corrected by injection of mRNAs encoding wild-type human SMN but not SMN mutants associated with SMA (Carrel et al., 2006). We first examined the requirement of Stasimon for motor neuron development by decreasing Stasimon expression with an antisense morpholino oligonucleotide (MO) in a zebrafish transgenic line expressing GFP in motor neurons (Figure S6A). We found this severely disrupted the normal outgrowth of motor axons without other detectable alterations in the morphology of developing embryos (Figure 6A–B). Injection of a control MO caused no significant change of motor axons (Figure 6A–B). Furthermore, while injection of human Stasimon mRNA alone had no significant effects on normal motor axon development (Figure S6E), co-injection of this mRNA rescued the motor axon defects induced by stas MO injection (Figure 6A–B). These results established that Stasimon is required for normal motor neuron development in zebrafish.
Figure 6. Stasimon is required for motor neuron development and rescues SMN-dependent motor axon defects in zebrafish embryos.
(A) Representative lateral views of motor axons in Tg(mnx1:GFP) zebrafish embryos expressing GFP in motor neurons and injected with Control MO as well as stas MO, smn MO and tdp43 MO either with or without co-injected human STAS RNA. (B) Quantification of the effects of Stasimon deficiency on motor axon development in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with Control MO, stas MO or stas MO + STAS RNA. Embryos were classified as severe, moderate, mild or no defects based on the severity of motor axons defects and the percentage for each group is shown. (C) Quantification of Stasimon effects on SMN-dependent motor axons defects in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with Control MO, smn MO or smn MO + STAS RNA and embryos were classified as in (B). (D) Quantification of Stasimon effects on TDP43-dependent motor axons defects in zebrafish. Motor axons were scored in Tg(mnx1:GFP) embryos injected with Control MO, tdp43 MO or tdp43 MO + STAS RNA and embryos were classified as in (B).
Data in all graphs are represented as mean and SEM. See also Figure S6.
Next, we investigated the role of Stasimon in SMN-deficient zebrafish embryos. Consistent with previous studies (Carrel et al., 2006; McWhorter et al., 2003), injection of smn MO caused severe motor axon abnormalities compared to control embryos (Figure 6A and 6C) while knockdown of a control RNA binding protein did not (hnRNP Q, Figure S6B–D). Remarkably, co-injection of Stasimon mRNA with smn MO resulted in a robust correction of the SMN-dependent motor axon defects as measured by a strong reduction in the degree of abnormal motor axon branching (Figure 6A and 6C). This was not observed upon injection of an unrelated control mRNA (Bcl2, Figure S6F–G) and SMN levels were similarly reduced by the smn MO with and without co-injection of either Stasimon or control mRNAs (Figure S6H), indicating that mRNA co-injection did not interfere with the activity of the smn MO. To determine if the rescuing effect of Stasimon was specific to SMN-dependent phenotypes in zebrafish, we analyzed the effect of Stasimon on motor neuron phenotypes induced by depletion of the amyotrophic lateral sclerosis associated gene TDP43 (Kabashi et al., 2011). Injection of tdp43 MO in zebrafish embryos induced motor axonal defects (Figure 6A and 6D) as previously reported (Kabashi et al., 2011). However, co-injection of human Stasimon mRNA had no beneficial effects on the TDP43-dependent motor neuron phenotypes (Figures 6A and 6D). Collectively, these results established that increasing Stasimon expression can specifically rescue neuronal dysfunction in this vertebrate model of SMN deficiency.
SMN deficiency disrupts Stasimon U12 splicing and mRNA expression in the motor circuit of SMA mice
We next investigated if Stasimon expression was altered in a mouse SMA model (SMA-Δ7) that recapitulates many features of the human disease (Le et al., 2005). We first analyzed Stasimon mRNA splicing and U12 intron retention by RT-qPCR (Figure S7A) in tissues of SMA-Δ7 and control mice at postnatal day 1 (P1), P6 and P11, corresponding to pre-, early- and late-symptomatic stages of disease in this model. While there was no apparent difference in Stasimon U12 splicing at P1, aberrantly spliced Stasimon mRNA was detectable in the spinal cord and first lumbar Dorsal Root Ganglia (L1 DRG) of SMA-Δ7 mice compared to controls at the early-symptomatic P6 stage (Figures 7A–B and S7B–C). Stasimon aberrant splicing further accumulated in late-symptomatic P11 SMA-Δ7 mice and was accompanied by increased levels of U12 intron retention (Figure 7A–B). Significant accumulation of Stasimon U12 intron retention but not of the aberrantly spliced mRNA was also found in brain and non-neuronal kidney tissue (Figure S7B–C). These results established that SMN deficiency caused a progressive alteration in Stasimon U12 splicing in the spinal cord and DRG of SMA mice.
Figure 7. SMN deficiency disrupts Stasimon U12 splicing and mRNA expression in the sensory-motor circuit of SMA mice.
(A) RT-qPCR analysis of aberrantly spliced Stasimon mRNA in the spinal cord and L1 DRG from control and SMA mice at the indicated post-natal days. (B). RT-qPCR analysis of Stasimon U12 intron retention in the spinal cord and L1 DRG from control and SMA mice at the indicated post-natal days. (C) Strategy for labeling motor neurons and proprioceptive neurons of the motor circuit by CTb-488 injection in the iliopsoas muscle. (D) Confocal image of CTb-488-labelled iliopsoas motor neurons and DRG neurons from a control mouse. Scale bar, 100 μm. (E) Confocal image showing co-localization of CTb-488 (green) and ChAT (red) in motor neurons from the ventral horn of a CTb-injected mouse. Scale bar, 50 μm. (F) Confocal image showing co-localization of CTb-488 (green) and parvalbumin (blue) in proprioceptive neurons from the DRG of a CTb-injected mouse. Scale bar, 20 μm. (G) RT-qPCR analysis of Stasimon U12 intron retention and mRNA levels in motor neurons and proprioceptive neurons isolated by LCM from CTb-injected control and SMA mice at P6. (H) Model for the sequence of SMN-dependent molecular events necessary for normal motor circuit function (blue) which are disrupted in SMA (red).
Data in all graphs are represented as mean and SEM. See also Figure S7.
We examined Stasimon mRNA levels in brain, spinal cord, DRG and kidney of SMA-Δ7 mice but found no changes compared to normal littermates (Figure S7D), with the exception of a reduction of Stasimon mRNA in L1 DRG of SMA-Δ7 mice at P11 that did not reach statistical significance. As whole tissue analysis was not sufficient to determine mRNA expression changes in the subset of spinal and DRG neurons that constitute the motor circuit, we sought to label and isolate these neurons specifically. The iliopsoas muscle participates in the righting reflex, which is severely affected in SMA-Δ7 mice (Mentis et al., 2011). We injected the iliopsoas of SMA-Δ7 and control mice at P2 with a fluorescently conjugated Cholera Toxin b (CTb) to label the motor neurons and proprioceptive neurons that connect to this muscle (Figure 7C–D). At P6, immunohistochemistry against ChAT and parvalbumin demonstrated proper localization of CTb in the motor neurons and proprioceptive neurons associated with the iliopsoas muscle (Figure 7E–F). We then isolated the soma of these CTb-labeled neurons by laser capture microdissection (LCM) (Figure S7E–F) and carried out RT-qPCR analysis. We found strong and specific enrichment of ChAT mRNA in LCM motor neurons and parvalbumin mRNA in proprioceptive neurons (Figure S7G), confirming we had isolated the correct neuronal types. We then compared Stasimon U12 intron splicing and mRNA expression in motor circuit neurons isolated from control and SMA-Δ7 mice. Remarkably, Stasimon U12 intron retention was strongly increased in both the motor neurons and proprioceptive neurons of SMA-Δ7 mice relative to controls (Figure 7G), with the increase being most pronounced in proprioceptive neurons. Furthermore, we found that Stasimon mRNA levels were also reduced in both types of motor circuit neurons from SMA-Δ7 mice compared to controls, again with a larger reduction observed in proprioceptive neurons. These results demonstrated that SMN deficiency disrupts Stasimon U12 splicing and mRNA expression in the constituent neurons of the sensory-motor circuit in a mouse model of SMA.
DISCUSSION
Our findings establish that SMN regulation of splicing is essential for motor circuit function in vivo. Reduced SMN levels lead to altered U12 splicing and decreased expression of a discrete set of U12 intron-containing genes in both mammalian cells and Drosophila larvae. Stasimon, one of these SMN target genes, encodes an evolutionarily conserved transmembrane protein that is required for normal neurotransmitter release by motor neurons in Drosophila and motor axon outgrowth in zebrafish. We also observed defective U12 splicing and reduced mRNA levels of Stasimon in the motor circuit neurons of SMA mice. Rescue experiments demonstrate that in Drosophila Stasimon is not required cell autonomously in motor neurons but regulates the neurotransmitter release properties of motor neurons indirectly through activities in other motor circuit neurons that provide excitatory input to motor neurons, similar to the cellular requirement for SMN in this model (Imlach et al., 2012). Although multiple genes are affected by SMN deficiency, restoration of Stasimon levels alone rescues key motor neuron defects in both invertebrate and vertebrate models of SMA, establishing that altered expression of individual genes can account for specific aspects of motor circuit dysfunction in vivo. Thus, SMN-dependent splicing defects of select genes provide a cohesive explanation for the chain of events that produce motor circuit dysfunction in SMA and a molecular framework to understand the specific neuronal effects that result from the ubiquitous disruption of snRNP assembly by the disease (Figure 7H).
The requirement of SMN for U12 splicing
Using mammalian cells and Drosophila larvae, we demonstrate that the function of SMN in the assembly of spliceosomal snRNPs is required for efficient U12 splicing and that SMN deficiency decreases the expression of a subset of genes with this type of intron in vivo. While leaving open the possibility that SMN can also influence the activity of the U2-dependent spliceosome, as suggested by previous studies (Jodelka et al., 2010; Ruggiu et al., 2012; Zhang et al., 2008), our findings establish direct regulation of U12 splicing events by SMN. Under normal conditions, U12 splicing is thought to represent a rate-limiting step in the expression of genes that contain U12 introns, which are processed more slowly than U2 introns (Patel and Steitz, 2003). This, together with reduced availability of the snRNP components of the U12 spliceosome induced by low SMN levels, can explain the accumulation of SMN-dependent U12 splicing defects we observe. Decreased mRNA levels of U12 intron-containing genes are likely due to degradation by surveillance mechanisms of incorrectly processed mRNAs resulting from SMN-dependent disruption of U12 splicing. In addition to increased U12 intron retention and decreased mRNA expression, exon skipped and aberrantly spliced forms of some SMN target mRNAs also accumulate in SMN-deficient mammalian cells and, in the case of Stasimon, in the motor circuit neurons of SMA mice. Conceivably, these abnormalities are caused by poor exon definition consequent to inefficient binding of minor snRNPs to U12 introns.
Our results also highlight elements of selectivity in the effects of SMN depletion on U12 splicing. First, SMN deficiency causes selective rather than general defects in splicing, affecting some but not all of the U12 introns both in Drosophila larvae and mammalian cells. Second, the time of onset and degree of disruption in U12 intron splicing of SMN targets is variable and causes differential reduction of mRNA expression, which might have distinct functional consequences depending on the specific requirement of individual genes in vivo. Lastly, low SMN levels can affect evolutionarily conserved U12 introns of homologous genes in different species, pointing to a conservation of SMN splicing targets across evolution.
Stasimon is an SMN target gene required for motor circuit function
To gain insight into the biological relevance of SMN-dependent U12 splicing events in motor circuit function, we investigated the role of SMN target genes on NMJ neurotransmission in Drosophila larvae. Our results identify a novel U12 intron-containing gene, stasimon, which has reduced expression in Drosophila smn mutants and is required for the regulation of synaptic transmission of motor neurons. Decreased Stasimon activity elicits an increase in evoked neurotransmitter release at the NMJ. The effects of Stasimon deficiency on neurotransmission from glutamatergic motor neurons are caused by a dysfunction of cholinergic neurons in Drosophila. These findings indicate that the effects of loss of Stasimon on motor neurons are non-cell autonomous and suggest that Stasimon is required for proper regulation of motor circuit activity.
Our results reveal a striking similarity in the effects of both Stasimon and SMN deficiency on the electrophysiological properties of Drosophila motor neurons. Evoked neurotransmitter release from motor neurons is increased in Drosophila smn mutants and can be corrected by transgenic SMN expression in cholinergic neurons (Imlach et al., 2012). The non-cell autonomous increase in neurotransmitter release at the Drosophila NMJ is consistent with a hyperexcitable state of motor neurons resulting from reduced excitatory proprioceptive and interneuron input from the motor circuit. This is reminiscent of motor neurons in SMA mice where an imbalance between excitatory and inhibitory inputs is correlated with a homeostatic increase in motor neuron excitability presumably to compensate for the decreased presynaptic input (Mentis et al., 2011). It is conceivable that similar events take place in the presence of reduced Stasimon function in the motor circuit.
The precise mechanism by which reduced Stasimon perturbs motor circuit activity is presently unknown. However, Stasimon expression profile and protein structure suggest some possibilities. Stasimon is a ubiquitously expressed gene with a prominent expression in the Drosophila and mouse central nervous system and encodes a highly evolutionarily conserved protein containing six transmembrane domains and a region with homology to SNARE-associated Golgi proteins. These features are consistent with a neuronal function of Stasimon in transport or docking of vesicular cargo whose impairment in neurons could disrupt neuronal activity.
Stasimon contributes to motor circuit dysfunction in animal models of SMA
SMN deficiency disrupts motor circuit activity in Drosophila. Importantly, proper regulation of motor circuit activity in Drosophila requires SMN function in cholinergic neurons but not motor neurons (Imlach et al., 2012). Our results reveal that restoration of Stasimon expression in cholinergic neurons is necessary and sufficient to fully rescue aberrant neurotransmitter release at the NMJs and to robustly improve muscle growth defects in SMN loss-of-function mutants, mirroring the cellular requirement for SMN in the Drosophila motor circuit. Therefore, decreased Stasimon function in cholinergic neurons directly contributes to disruption of motor circuit activity triggered by SMN deficiency and has non-cell autonomous effects in motor neurons and muscle. These findings directly link selective neuronal effects of ubiquitous SMN deficiency to defective splicing of a gene with essential functions in motor circuits.
Our results further implicate Stasimon dysfunction in motor neuron phenotypes of a vertebrate model of SMA. SMN deficiency elicits motor axon defects in zebrafish embryos (Carrel et al., 2006; McWhorter et al., 2003). We show here that Stasimon is required for normal motor axon outgrowth during zebrafish development and that Stasimon overexpression corrects the axonal defects in motor neurons with low SMN levels. Importantly, Stasimon does not rescue TDP-43-dependent motor neuron defects in a zebrafish model of ALS (Kabashi et al., 2011), providing evidence that Stasimon is a specific downstream target of SMN. Previous studies showed that the combined injection of major and minor snRNPs rescued motor axon defects in SMN-deficient zebrafish embryos (Winkler et al., 2005), linking snRNP dysfunction to this phenotype. Based on these and our results, targeted ablation of minor snRNP components from the injected pool of snRNPs would be predicted to prevent correction of the motor neuron phenotype.
Splicing-dependent mechanisms of selective neuronal dysfunction in SMA
Our findings have important implications for understanding how mutations in ubiquitously expressed proteins cause the demise of selective neuronal types. Since the identification of SMN as the SMA-determining gene product and the discovery of its critical role in snRNP assembly, an unresolved problem has been how SMN-dependent snRNP biogenesis defects cause the selective dysfunction of the motor system. Particularly difficult to reconcile is the disruption of SMN ubiquitous activity in splicing with a selective pathology of motor function. Our findings address this issue and provide evidence mechanistically linking disruption of SMN activity in snRNP assembly to motor neuron dysfunction through a cascade of molecular events with cause-effect relationships (Figure 7H). First, SMN deficiency impairs Sm core formation leading to a decrease in snRNP levels with effects that are tissue-specific and particularly prominent on components of the U12 splicing machinery. Second, this reduction in snRNP levels causes selective splicing defects in a limited set of genes resulting in alterations in their normal profile of expression. Third, a subset of these SMN target genes, including but not necessarily limited to those with U12 introns, perform functions that are critical for specific neuronal classes. Lastly, disruption of the activity of these genes, such as Stasimon, results in selective defects of neuronal function that collectively generate the SMA phenotype.
Splicing defects have been thought to be too general to explain the highly discrete SMA phenotype, leading some to propose motor neuron-specific functions of SMN. However, our findings show that specificity can emerge through the combination of multiple mechanistic filters that act upon SMN’s ubiquitous role in splicing. The combination of these events accounts for the selectivity of the effects of SMN deficiency on the motor circuit in vivo. Our results link SMN-dependent impairment of snRNP assembly to alterations in the expression of selected genes that cause motor neuron dysfunction, consistent with SMA being a disease of RNA splicing.
EXPERIMENTAL PROCEDURES
NIH3T3 cell lines
The NIH3T3-SmnRNAi and NIH3T3-SMN/SmnRNAi cell lines were generated through transduction with lentiviral vectors (Figure S1A). Further details are provided in Supplemental Experimental Procedures.
Drosophila genetics and analysis
We utilized smn73Ao (Chan et al., 2003), smnX7 (Chang et al., 2008), U6atac (Otake et al., 2002) and stasEY04008 (Bellen et al., 2004) mutants. Analysis of NMJ electrophysiology, rhythmic motor activity, locomotion and muscle size was performed as described (Imlach et al., 2012). Further details on phenotypic analysis and additional UAS, RNAi and Gal4 stocks are described in Supplemental Experimental Procedures.
Zebrafish motor axon outgrowth
Transgenic Tg(mnx1:GFP) zebrafish that express GFP in ventrally projecting motor axons (Dalgin et al., 2011) were used in this study. One-two cell stage Tg(mnx1:GFP) embryos were injected with MO (Table S3) and RNA as previously described (McWhorter et al., 2003). Motor axons from each side of the embryo were scored at 28 hours post-fertilization and used to classify the embryo as severe, moderate, mild, or no defects based on number and type of motor axon abnormalities (Carrel et al., 2006). Further details are provided in Supplemental Experimental Procedures.
SMA mice and LCM
SMA-Δ7 (Smn−/−; SMN2+/+; SMNΔ7+/+) mice were obtained using JAX Stock No:005025 (Le et al., 2005). Tissues from control and SMA mice were rapidly dissected, immediately frozen in liquid nitrogen and stored at −80°C until use. To label motor neurons and proprioceptive neurons of the motor circuit, Alexa 488-conjugated CTb was injected in the iliopsoas muscle of control and SMA-Δ7 mice at P2. Spinal cord and DRG were rapidly dissected at P6 and processed by LCM or immunohistochemistry as described in Supplemental Experimental Procedures.
Supplementary Material
Research Highlights.
SMN is required for U12 splicing
Stasimon is a SMN-dependent U12 gene required for motor circuit function
Stasimon restoration corrects motor neuron dysfunction in animal models of SMA
Stasimon U12 splicing is disrupted in motor circuit neurons of SMA mice
Acknowledgments
We wish to thank Chris Henderson for invaluable comments. We are grateful to Spyros Artavanis-Tsakonas and Greg Matera for Drosophila stocks and reagents. This work was supported by grants from NIH-NINDS R01NS069601 and R21NS077038 (LP) and R01NS050414 (CEB), DoD W81XWH-08-1-0009 and W81XWH-11-1-0753 (BDM) and W81XWH-11-1-0689 (GZM), SMA Foundation (LP and CEB), MDA USA (LP), FSMA A. Lewis Young Investigator Award (GZM) and Columbia University Motor Neuron Center (LP and BDM).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35:D110–115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie CE, Carrel TL, McWhorter ML. Fishing for a mechanism: using zebrafish to understand spinal muscular atrophy. J Child Neurol. 2007;22:995–1003. doi: 10.1177/0883073807305671. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonne R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel TL, McWhorter ML, Workman E, Zhang H, Wolstencroft EC, Lorson C, Bassell GJ, Burghes AH, Beattie CE. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YB, Miguel-Aliaga I, Franks C, Thomas N, Trulzsch B, Sattelle DB, Davies KE, van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- Chang HC, Dimlich DN, Yokokura T, Mukherjee A, Kankel MW, Sen A, Sridhar V, Fulga TA, Hart AC, Van Vactor D, et al. Modeling spinal muscular atrophy in Drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgin G, Ward AB, Hao le T, Beattie CE, Nechiporuk A, Prince VE. Zebrafish mnx1 controls cell fate choice in the developing endocrine pancreas. Development. 2011;138:4597–4608. doi: 10.1242/dev.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- Imlach LW, Beck ES, Choi BJ, Lotti F, Pellizzoni L, McCabe BD. SMN is required for sensory-motor circuit function in Drosophila. Cell. 2012 doi: 10.1016/j.cell.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodelka FM, Ebert AD, Duelli DM, Hastings ML. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Bercier V, Lissouba A, Liao M, Brustein E, Rouleau GA, Drapeau P. FUS and TARDBP but not SOD1 interact in genetic models of amyotrophic lateral sclerosis. PLoS Genet. 2011;7:e1002214. doi: 10.1371/journal.pgen.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lin CF, Mount SM, Jarmolowski A, Makalowski W. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol Biol. 2010;10:47. doi: 10.1186/1471-2148-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L, Alvarez FJ, Sumner CJ, O’Donovan MJ. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Otake LR, Scamborova P, Hashimoto C, Steitz JA. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol Cell. 2002;9:439–446. doi: 10.1016/s1097-2765(02)00441-0. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Praveen K, Wen Y, Matera AG. A Drosophila Model of Spinal Muscular Atrophy Uncouples snRNP Biogenesis Functions of Survival Motor Neuron from Locomotion and Viability Defects. Cell reports. 2012;1:624–631. doi: 10.1016/j.celrep.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, McGovern VL, Lotti F, Saieva L, Li DK, Kariya S, Monani UR, Burghes AH, Pellizzoni L. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol Cell Biol. 2012;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes & Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E, Saieva L, Carrel TL, Crawford TO, Liu D, Lutz C, Beattie CE, Pellizzoni L, Burghes AH. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.