Abstract
A cDNA (Cel1) encoding an endo-1,4-β-glucanase (EGase) was isolated from ripe fruit of strawberry (Fragaria × ananassa). The deduced protein of 496 amino acids contains a presumptive signal sequence, a common feature of cell wall-localized EGases, and one potential N-glycosylation site. Southern- blot analysis of genomic DNA from F. × ananassa, an octoploid species, and that from the diploid species Fragaria vesca indicated that the Cel1 gene is a member of a divergent multigene family. In fruit, Cel1 mRNA was first detected at the white stage of development, and at the onset of ripening, coincident with anthocyanin accumulation, Cel1 mRNA abundance increased dramatically and remained high throughout ripening and subsequent fruit deterioration. In all other tissues examined, Cel1 expression was invariably absent. Antibodies raised to Cel1 protein detected a protein of 62 kD only in ripening fruit. Upon deachenation of young white fruit to remove the source of endogenous auxins, ripening, as visualized by anthocyanin accumulation, and Cel1 mRNA accumulation were both accelerated. Conversely, auxin treatment of white fruit repressed accumulation of both Cel1 mRNA and ripening. These results indicate that strawberry Cel1 is a ripening-specific and auxin-repressed EGase, which is regulated during ripening by a decline in auxin levels originating from the achenes.
Fruit ripening is a complex developmental program in which senescing tissues undergo programmed changes in firmness, texture, coloration, flavor, and susceptibility to microbial infection. Changes in firmness and texture are largely attributed to alterations in the composition and structure of cell wall polysaccharides. Because these modifications influence the postharvest properties (i.e. storage time and expense, handling damage, and desirability to the consumer) of important food crops and, consequently, are of great commercial importance, research in recent years has focused on identifying enzyme activities that are rate limiting in the promotion of fruit deterioration. In the climacteric species, which are characterized by the autocatalytic production of the ripening hormone ethylene and a ripening-related transient burst in CO2 evolution, the antisense suppression of ACC synthase (Oeller et al., 1991) and ACC oxidase (Picton et al., 1993) in tomato has provided fruit in which ripening and softening are retarded and can be controlled by the application of ethylene. Similar approaches have been taken in efforts to diminish the activities of cell wall-associated hydrolases (Sheehy et al., 1988; Smith et al., 1988), which may play a central role in fruit cell wall breakdown during ripening (Brady, 1987).
In nonclimacteric species such as strawberry (Fragaria × ananassa), much less is known about the ripening process. Because these plants lack a respiratory climacteric and because ethylene appears to play a minimal, if any, role in fruit ripening, there is growing interest in identifying the factor(s) that mediates ripening. Strawberry fruit (actually an enlarged receptacle rather than a true fruit) exhibit a low level of ethylene production, which is rather constant during ripening (Knee et al., 1977), and there is no observable stimulation of ripening upon application of exogenous ethylene (Iwata et al., 1969). Although there is no evidence for a ripening-related role for ethylene, strawberry fruit ripening has been shown to be negatively regulated by auxins that originate in the receptacle-borne achenes (Given et al., 1988b; Manning, 1994). As auxin levels decline, fruit exhibit a characteristic ripening profile, one of the major hallmarks of which is rapid deterioration once fruit achieve the red-ripe stage. In general, strawberry fruit ripening is typified by the induction of enzyme markers for anthocyanin pigment biosynthesis (e.g. Phe ammonia-lyase), a concomitant decrease in chlorophyll and increase in anthocyanin pigments, and a progressive decrease in tissue firmness (Woodward, 1972; Given et al., 1988a).
Efforts to reveal the molecular basis of changes in firmness, which are a major contributing factor to fruit quality, have focused on cell wall-associated enzymes, which are believed to mediate and/or contribute to cell wall breakdown. The most studied of these activities, endopolygalacturonase, is absent or below the limit of detection in ripening strawberry fruit (Neal, 1965; Barnes and Patchett, 1976; Huber, 1984). Although strawberry fruit is a rich source of pectin, this observation is consistent with cell wall studies that have shown that total extractable polyuronides remain constant as a proportion of cell wall material during ripening and do not show detectable depolymerization (Huber, 1984). In contrast to these findings, however, the hemicellulosic fraction of cell walls prepared from ripening fruit demonstrates a progressive shift from high- to low-Mr polymers (Huber, 1984). Whereas there is no discernible change in the neutral sugar composition of hemicelluloses isolated from the small-sized green to red-ripe stages, the average net Mr change is quite dramatic, suggestive of an active, developmentally regulated endohydrolyase. It is interesting that this observed hemicellulose depolymerization correlates well with a soluble CMCase activity measured in extracts prepared from ripening strawberry fruit (Barnes and Patchett, 1976). In ripening fruit of avocado (Hatfield and Nevins, 1986) and pepper (Harpster et al., 1997), CMCase activity is attributed to an EGase (EC 3.2.1.4).
Although largely correlative, there is considerable evidence for the importance of EGases in a wide variety of physiological processes involving changes in cell wall architecture, which range from cell wall expansion to disassembly (for review, see Brummell et al., 1994). For example, in abscission-zone formation the infusion of antiserum raised against an abscission-zone-related EGase into explants, which had been induced to abscise by ethylene, was observed to inhibit cell separation (Sexton et al., 1980). Furthermore, the induction of EGase gene expression in fruit of tomato (Lashbrook et al., 1994), avocado (Christoffersen et al., 1984), and pepper (Ferrarese et al., 1995; Harpster et al., 1997) correlates well with the onset and development of ripening. Recently, a partial cDNA showing homology to EGases was isolated from strawberry fruit and shown to be expressed in a ripening-related manner (Manning, 1998). As a first step toward determining whether the in vivo suppression of EGase gene expression is a viable strategy for enhancing firmness in harvested strawberry fruit, we describe the cloning of a full-length strawberry EGase cDNA (Cel1), an analysis of the hormonal and developmental regulation of Cel1 gene expression, and the identification and quantitation of Cel1 protein.
MATERIALS AND METHODS
Plant Material
Strawberry (Fragaria × ananassa Duch. cv Chandler) plants were grown in 3-gallon plastic bags and irrigated with fertilized water daily. Greenhouse temperatures ranged from 22°C during the day to 12°C at night. To promote synchronous flowering, potted runners were subjected to 1 week of preconditioning at 10°C during the day and 5°C at night. This regime was immediately followed by 3 weeks of vernalization at −2°C.
Unless indicated otherwise, all analyses were conducted using tissue that was harvested directly into liquid nitrogen and stored at −80°C. Fruit were staged by size and color. Color readings were conducted with a colorimeter (model CR-300, Minolta, Ramsey, NJ) and are described by the a* value on the Commission Internationale de l'Eclairnge L*a*b* scale, which is a measure of green (negative) to red (positive) reflectance of the visible spectrum. Although there was some variability between fruit, the average times taken for fruit to attain a specific stage of development in the ripening process and their color readings (means of at least four readings ± sd) were as follows: small green (14 dpa, a* = −14.4 ± 0.4), large green (20 dpa, a* = −12.8 ± 0.5), small white (28 dpa, a* = −11.3 ± 1.5), large white (35 dpa, a* = −9.4 ± 1.1), turning (40 dpa, a* = 1.7 ± 8.1), red ripe (45 dpa, a* = 24.8 ± 1.2), and overripe (>55 dpa, a* = 21.5 ± 2.9).
cDNA Cloning
The isolation of EGase cDNAs was conducted by screening a Uni-Zap XR cDNA library (Stratagene) constructed from red fruit poly(A+) mRNA. Hybridization conditions were empirically determined by probing replicate northern blots of red fruit total RNA over a range of temperatures with end-labeled ([γ-32P]ATP, >5000 Ci mmol−1) degenerate oligonucleotides corresponding to the conserved amino acid domain CWERPEDM (see Fig. 1B; for sequences, see Harpster et al., 1997). Hybridization at 55°C in 7% (w/v) SDS, 0.25 m sodium phosphate (pH 7.4), 1 mm EDTA, and 1% (w/v) BSA (type V) provided the recognition of a single mRNA species of the appropriate size for EGase (approximately 1.8 kb). Using these same conditions library lifts were then hybridized and washed in 2× SSC (1× SSC is 0.15 m NaCl and 15 mm sodium citrate, pH 7.0) and 0.1% (w/v) SDS at 55°C. After the purification of phage from hybridizing plaques and the in vivo excision of cloned inserts into phagemids, double-stranded DNA minipreps were partially characterized by dideoxy sequencing (Sanger et al., 1977) and restriction endonuclease mapping. The two longest cDNA clones were completely sequenced on both strands.
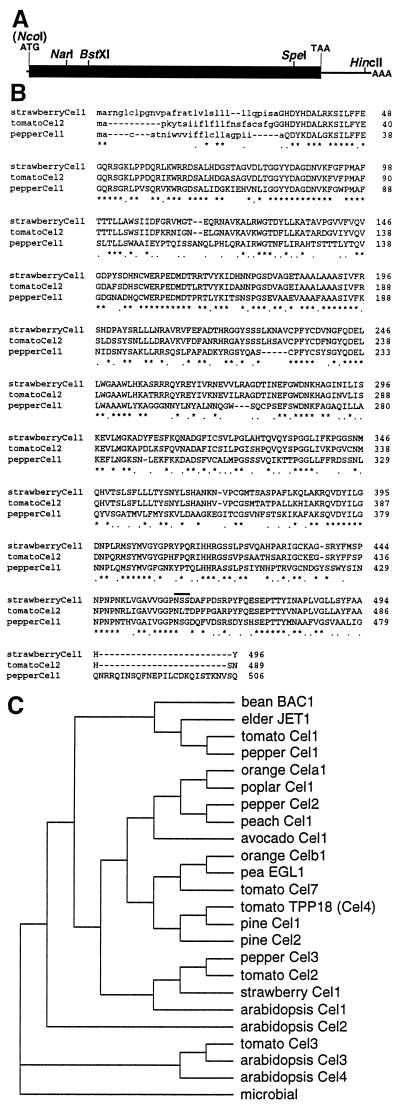
Figure 1.
A, Schematic map of strawberry Cel1 cDNA clone (drawn to scale) showing relevant restriction endonuclease sites. The NcoI site at the ATG initiation codon was introduced by in vitro mutagenesis (M. Harpster and P. Dunsmuir, unpublished data). B, Alignment of the deduced amino acid sequence of strawberry Cel1 with the ripening-related EGases tomato Cel2 (Lashbrook et al., 1994) and pepper PCEL1 (Harpster et al., 1997). Amino acid identity is indicated by asterisks and conservative changes by dots. Amino acids constituting putative signal sequences are shown in lowercase. A potential N-glycosylation sequence in strawberry Cel1 is overlined. C, Phylogenetic comparison of plant EGase mature proteins. The sequences used, with accession numbers in parentheses, are: Arabidopsis Cel1 (X74290), Cel2 (AC003033), Cel3 (U37702), and Cel4 (AC001229); avocado Cel1 (M17634); bean BAC1 (N57400); elder JET1 (X74290); orange Cela1 (AF000135) and Celb1 (AF000136); pea EGL1 (L41046); peach Cel1 (X96853); pepper PCEL1 (X87323), Cel2 (X97190), and Cel3 (X97189); pine Cel1 (U76725) and Cel2 (U76756); poplar Cel1 (D32166); and tomato Cel1 (U13054), Cel2 (U13055), Cel3 (U78526), TPP18/Cel4 (U20590), and Cel7 (Y11268).
Phylogenetic Analysis
Mature EGase protein sequences (i.e. after removal of signal sequences) were aligned using Clustal V (Higgins et al., 1992), followed by analysis using PAUP (Swofford, 1993) with a bacterial cellulase (accession no. X60545) as the outgroup. A heuristic search was employed using the random stepwise addition of taxa with 100 replicates and global (tree bisection and reconnection) branch swapping.
Southern-Blot Analysis
Strawberry genomic DNA was isolated using a small-scale extraction procedure that uses urea as a denaturant (Greene et al., 1994). Contaminating pectins were removed by selective precipitation with 2-butoxyethanol (Sigma). DNA digestion, gel and electrophoresis conditions, and probe hybridization conditions have been described elsewhere (Harpster et al., 1997). High-stringency blot washes were done at 65°C in 0.1× SSC and 0.1% (w/v) SDS.
RNA Isolation and Expression Analysis
Total RNA was isolated from different tissues of strawberry plants using a hot borate/phenol method (Wan and Wilkins, 1994) with several modifications. Strawberry tissue (1–2 g) was first ground to a fine powder in liquid nitrogen and then transferred to a 15-mL polypropylene tube containing 5 mL of borate extraction buffer (0.2 m borax, 30 mm EGTA, 1% [w/v] sodium deoxycholate, 1% [w/v] SDS, and 10 mm DTT) adjusted to 85°C. The sample was vortexed for 10 s, after which an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v) was added and the sample vortexed for an additional 30 s. The sample was then transferred to a 15-mL Corex tube and centrifuged at 10,000g for 10 min at 4°C. After removal of the aqueous layer and reextraction with an equal volume of chloroform:isoamyl alcohol (24:1, v/v), the sample was centrifuged at 1,500g for 5 min and the aqueous layer transferred to a Corex tube containing an equal volume of 4 m lithium acetate. The sample was incubated at 4°C overnight, centrifuged at 10,000g for 20 min, and the supernatant discarded. To remove pigments, the pellet was resuspended in 0.5 mL of cold 2 m lithium acetate, transferred to a microcentrifuge tube, and then repeatedly pelleted (16,000g for 5 min at 4°C) and resuspended in 2 m lithium acetate until the supernatant was clear of pigmentation (twice for fruit RNA and three to four times for leaf RNA). After the last precipitation, the pellet was resuspended in 0.3 mL of water with vigorous vortexing, adjusted to 0.2 m potassium acetate (pH 5.5), and vortexed for 20 to 30 s with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v). Finally, the sample was centrifuged (5 min at 4°C), the aqueous layer was transferred to a microcentrifuge tube containing 2.5 volumes of ethanol, and the RNA was precipitated and resuspended in RNase-free water. Based on A260 readings, yields of total RNA were 600 to 1200 μg g−1 fresh weight from leaf tissue and 40 to 100 μg g−1 fresh weight from fruit tissue. Northern-blot analysis was conducted as described elsewhere (Harpster et al., 1997), and high-stringency washes of blots were performed as described for Southern blots. To ensure that equal amounts of RNA per lane were loaded for each northern blot, parallel blots were stained with ethidium bromide and individual lanes evaluated for comparable levels of fluorescence upon exposure to a short-wave UV light source (data not shown).
Hormone Treatment
Deachenation was conducted by removing the achenes from longitudinal halves of green/small-white fruit (on the vine) with needle-nose forceps. For hormone treatment, NAA was applied to longitudinal halves of similarly staged but otherwise intact fruit (possessing achenes) at a concentration of 0.2 mm in a lanolin paste containing 1% (v/v) DMSO. Seven days after deachenation or auxin treatment, tissue encompassing the zone separating treated from untreated halves (i.e. deachenated/achenated and +NAA/−NAA) was discarded (slice of approximately 5 mm in thickness) and the remaining halves were wiped clean of the lanolin, frozen separately, and stored at −80°C. Color readings of the individual fruit halves on the L*a*b* scale were recorded as described above.
Protein Extraction and CMCase Activity Measurements
Soluble-protein extracts used for both CMCase activity measurements and analysis by SDS-PAGE were prepared from strawberry tissues by first grinding frozen samples to a fine powder in liquid nitrogen. The powders were then ground with a mortar and pestle to a thick slurry by the addition of cold 100% acetone, which was gravity filtered using Whatman no. 1 filter paper, and extensively washed with acetone (50 mL g−1 fresh weight of tissue) until the filtrates were clear of pigment. After air drying the retentates to a powder, samples were frozen in liquid nitrogen and stored at −80°C until further use, or processed immediately. To extract soluble proteins, acetone powders were first ground in liquid nitrogen and then ground further in extraction buffer (0.1 m sodium phosphate [pH 6.0], 9.2 mm borax, 13 mm boric acid, 5 mm EDTA, 5 mm DTT, 0.15 m NaCl, and 0.5% Triton-X) containing a cocktail of proteinase inhibitors (see Harpster et al., 1997). The resultant slurry was centrifuged at 10,000g (4°C) for 15 min to remove insoluble material and the supernatant assayed directly for relative CMCase activity by viscometry (Harpster et al., 1997). For preparation of samples for electrophoresis, the protein concentration of supernatants was determined using the dye-binding method of Bradford (1976), after which the desired amount of protein was adjusted to 80% (v/v) acetone and incubated on ice for a minimum of 30 min. Samples were then centrifuged (16,000g for 10 min at 4°C) and the protein pellets dried and resuspended in a sample loading buffer (20 mm Tris-Cl [pH 6.8], 3% [w/v] SDS, 10% [v/v] glycerol, and 0.01% [w/v] bromophenol blue) by boiling for 10 min. To prevent band smearing during electrophoresis, residual insoluble material was removed from samples by centrifugation at 16,000g for 10 min before loading. Molecular-mass markers were purchased from BRL.
Antibody Preparation and Western-Blot Analysis
Polyclonal antiserum was raised against strawberry EGase by popliteal lymph node injections of New Zealand White rabbits with a protein A/Cel1 fusion protein. The fusion consisted of a 1.53-kb NarI-HincII fragment of the Cel1 cDNA containing the entire ORF (except for a 189-bp deletion at the 5′ end deleting 63 amino acids at the amino terminus of the Cel1 protein) and 224 bp of the 3′ untranslated sequence, which was translationally fused to a protein A fusion-protein vector (Harpster et al., 1997). The resultant construct produces a fusion protein that is localized in inclusion bodies and inefficiently binds IgG-Sepharose (Pharmacia). Therefore, partial purification of the fusion protein for antibody production was performed by preparative SDS-PAGE, as described previously (Harpster et al., 1997).
Western-blot analysis of ripe-fruit protein extracts was carried out as described by Taylor et al. (1987) and provided a complex of several cross-reacting polypeptide species, despite the observation that parallel blots treated with preimmune serum demonstrated an absence of background hybridization (data not shown). Previous work using antiserum raised against a ripening-related pepper EGase suggests that the majority of these cross-reacting polypeptides are cell wall-associated proteins with shared antigenic epitopes (Harpster et al., 1997). To enrich for antibodies specific to strawberry Cel1, contaminating antibodies were selectively removed by passing antiserum over an affinity column containing covalently coupled protein isolated from small, green fruit tissue, which shows an absence of Cel1 expression. This was accomplished by first desalting a soluble-protein extract from small, green fruit on a column (PD-10, Pharmacia) and then covalently coupling the protein to a HiTrap column according to the manufacturer's instructions (Pharmacia). Finally, antiserum was loaded onto the column, which was then sealed and incubated overnight at 10°C. Unbound antiserum was collected from the column according to the manufacturer's instructions. When used in subsequent western-blot analysis of fruit protein extracts, the antiserum showed the selective enrichment of antibodies recognizing a single 62-kD polypeptide species.
RESULTS
Isolation of EGase cDNAs from Strawberry and Characterization of Encoded Protein
Using degenerate oligonucleotides corresponding to a conserved amino acid domain shared by plant EGases (see Methods), 55 putative EGase cDNAs were identified in the screening of a phage cDNA library constructed from red fruit poly(A+) mRNA. Because this library was not amplified and 80,000 plaques were screened, we estimate that strawberry EGase expression accounts for 0.07% of poly(A+) mRNA isolated from red fruit. Plasmid cross-hybridization experiments using all cDNA clones and dideoxy sequencing analysis at the 5′ and 3′ ends of a subset of these clones demonstrated that they were all derived from the expression of a single gene, designated strawberry Cel1. Analysis at the 5′ end of the longest cDNA (see Fig. 1A for a schematic map) showed that it contained a single ORF encoding 496 amino acids and 14 nucleotides of sequence upstream of the presumptive ATG initiation codon. Analysis of the 3′ ends of all of the cDNAs identified an untranslated region of approximately 260 nucleotides and a minimum of three alternative, closely spaced polyadenylation sites.
Translation of the ORF provided a predicted polypeptide of 496 amino acids with a Mr of 54,600, possessing a putative signal sequence of 32 amino acids and a single potential site for N-glycosylation (Fig. 1B). Alignment of the strawberry Cel1-deduced amino acid sequence with tomato Cel2 and pepper Cel1, two other fruit-ripening-related EGases, showed the presence of several highly conserved amino acid domains (Fig. 1B). To compare the amino acid sequence of strawberry Cel1 with all plant EGase sequences determined to date, signal-sequence cleavage sites were predicted using the rules of von Heijne (1986), and the subsequent mature protein sequences were used to construct a phylogenetic tree (Fig. 1C). Based on their phylogenetic relatedness, there are three major branches of plant EGases, with one branch containing EGases possessing a membrane-spanning domain such as Arabidopsis Cel3 and Cel4, and tomato Cel3 (Brummell et al., 1997b), another branch containing Arabidopsis Cel2 alone, and the third branch containing all of the remaining sequences. Strawberry Cel1 is within this last major branch and, with pepper Cel3, tomato Cel2, and Arabidopsis Cel1 (all show 80%–82% amino acid identity with strawberry Cel1), forms a subgroup.
Genomic Southern-Blot Analysis of Cel1
F. × ananassa Duch. cv Chandler is a highly cultivated octoploid variety, which is an interspecific hybrid of two diploid species, Fragaria chiloensis and Fragaria virginiana (Hancock et al., 1996). Consequently, because of the polyploid nature of the F. × ananassa genome, genomic Southern-blot analysis of Cel1 yields a complex banding pattern that does not provide an accurate assessment of the gene copy number (Fig. 2). To estimate the number of genes closely related to Cel1, genomic DNA was analyzed from Fragaria vesca, a diploid line. Based on the hybridization profile seen after washing the blot at high stringency, and the observation that all of the bands revealed in the digestion of F. vesca appeared to be a subset of the bands provided by the digestion of F. × ananassa, we estimate that strawberry Cel1 is probably encoded by a single gene per diploid genome. Although the hybridization patterns provided by digests with BclI, EcoRI, HindIII, and NcoI showed two strongly hybridizing bands, their sizes are more consistent with a single gene and can be accounted for by invoking the presence of these sites within introns. The presence of faint bands in both blots after washing at high stringency indicates the presence of related sequences that collectively constitute a multigene family for EGase in strawberry.
Figure 2.
Southern blot of genomic DNA isolated from leaves of F. × ananassa (octoploid) and F. vesca (diploid). Each lane contained 15 μg of DNA digested with the indicated restriction endonuclease, none of which cut within the hybridization probe. The random-primer-labeled ([α-32P]dCTP, 800 Ci mmol−1) probe is a 1.0-kb BstXI-SpeI fragment from the ORF of the Cel1 cDNA (see Fig. 1A). Autoradiograph exposure time was 1 week.
Expression of Cel1 in Strawberry Tissues
Northern-blot analysis of total RNA isolated from developing fruit revealed Cel1 expression as a 1.8-kb transcript, which was first detectable in small-sized white fruit (Fig. 3A). Thereafter, Cel1 transcript levels increased as fruit enlarged, and attained maximum size at the red-ripe stage, with a gradual increase in Cel1 mRNA in white fruit being followed by a larger increase coincident with the development of red color. In red-ripe fruit steady-state Cel1 mRNA levels were at their highest, and we observed no significant change in expression levels during ripening, even in fruit that had evident signs of deterioration (i.e. tissue rotting and liquefaction; see Fig. 3A, lane over ripe 3). Although the pattern of Cel1 mRNA accumulation generally showed an abrupt increase in steady-state levels between large-sized white and red-ripe fruit, we occasionally observed higher levels in large-sized white fruit and a more gradual increase in transcript accumulation (data not shown). This suggests that the increase in Cel1 mRNA abundance and the development of red coloration are not completely coupled. Autoradiograph exposure of the blot shown here in excess of 2 weeks revealed an apparent absence of Cel1 mRNA expression in developing green fruit (data not shown).
Figure 3.
Northern-blot analysis of RNA isolated from developing fruit (A) and different tissues (B) of flowering strawberry plants. In A, results are shown for two fruit from each stage of fruit development, except for overripe fruit, for which results are shown for three successive stages of fruit deterioration. Each blot contained 10 μg of total RNA per lane and was probed with a random-primer-labeled, 1.7-kb NcoI-HincII fragment comprising the ORF and the 3′ untranslated sequence of the Cel1 cDNA (see Fig. 1A). Autoradiograph exposure time was 24 h.
The analysis of Cel1 transcript levels in other tissues of mature, flowering strawberry plants showed that Cel1 mRNA expression is restricted to developing fruit. We detected no expression in leaves regardless of size and age, nor in floral stem tissue, vegetative stem tissue, roots, the primary tissues of dissected flowers, or callus (Fig. 3B). The high-level mRNA expression observed in red fruit of cv Chandler did not appear to show any cultivar specificity, insofar as expression levels were comparably high in ripe fruit of cv Camarosa (Fig. 3B, lane 1).
Auxin Effects on Cel1 Expression
As with other nonclimacteric fruit, ethylene production levels in ripening strawberry were exceptionally low and have been reported to decrease on a per unit fresh weight basis when harvested fruit turn from white to red (Knee et al., 1977). To investigate the role of auxin in Cel1 expression, three auxins were tested for their capacity to inhibit ripening: IAA and the two auxin analogs 2,4-D and NAA. Based on a demonstrable reduction in visible anthocyanin accumulation relative to control fruit, NAA was almost completely effective in retarding ripening when applied to small-sized white fruit on the vine, 2,4-D was moderately effective, and IAA performed poorly (data not shown). Presumably, the higher percentage of fruit showing a substantial decrease in anthocyanin accumulation after NAA treatment is a consequence of enhanced hormone uptake and/or stability relative to 2,4-D and IAA.
To test whether Cel1 mRNA expression is affected by the removal of endogenous auxin, achenes were manually removed from the longitudinal halves of individual, small-sized white fruit (on the vine), which were then harvested 7 d later. As shown in the northern blot in Figure 4A, fruit treated in this manner showed a dramatic increase in Cel1 transcript levels in the deachenated halves. Furthermore, this was accompanied by an enrichment in anthocyanin levels in the deachenated halves, as measured by colorimetry (Fig. 4A, upper panel). In conducting this experiment, we discarded any fruit that showed signs of an induced wound response, which was observed to develop within hours after deachenation and was evident as extensive browning within the outer epidermal layers of the fruit tissue.
Figure 4.
Flesh (receptacle) color readings and northern-blot analysis of Cel1 expression in longitudinal halves of whole fruit after deachenation (A) or treatment with auxin (B). Whole fruit on the vine were either deachenated on one half and the other half left untreated, or one half of otherwise intact fruit was left untreated and the other half coated with lanolin alone or lanolin containing 0.2 mm NAA. After 7 d, two fruit per treatment were harvested, dissected longitudinally, and untreated and treated halves analyzed separately. Color readings (a* on the Commission Internationale de l'Eclairnge L*a*b* scale) are given on a green (negative) to red (positive) scale, and each value is the mean of two readings ± sd. For northern-blot analysis, each lane contained 10 μg of total RNA, and the probe used was the same as described in the legend of Figure 3. Autoradiograph exposure time was 22 h.
To examine further the effect of auxin on Cel1 expression, transcript levels were measured by northern-blot analysis in NAA-treated and control small-sized white fruit. Figure 4B shows the results obtained at 7 d after application, in which total RNA was isolated from the separately treated longitudinal halves of control (with or without lanolin) and hormone-treated (with or without lanolin and NAA) fruit. At this time, Cel1 mRNA levels were low to undetectable in fruit sectors treated with NAA and high in tissue lacking exogenous hormone treatment. This reduction in the Cel1 steady-state transcript levels was correlated with an absence of anthocyanin pigment accumulation, whereas untreated halves showed a reddening of tissue (Fig. 4B, upper panel).
Because it is well established that achenes are a rich source of auxins, the concentration of which declines during strawberry fruit ripening (Dreher and Pooviah, 1982; Given et al., 1988b), these results provide corollary evidence that Cel1 expression is inhibited by auxins. Clearly, the removal of endogenous auxin through deachenation facilitates the premature initiation of the ripening program, which is accompanied by the accumulation of Cel1 mRNA. More work is required, however, to determine whether auxin control of Cel1 expression is at the transcriptional or the posttranscriptional level.
Cel1 Protein and EGase Activity in Mature Strawberry Plants
Using polyclonal antiserum raised against a protein A/Cel1 fusion protein, western-blot analysis of soluble-protein extracts prepared from a variety of strawberry tissues shows a major cross-reacting protein species of 62 kD, which is restricted to those tissues demonstrating expression of Cel1 mRNA (Fig. 5). Whereas all fruit samples, except for small-sized green fruit, showed similar accumulation of this protein, there was a clear absence in all of the other tissues we examined. The detection of a second cross-reacting polypeptide species of approximately 60 kD in fruit stems, leaf stems, and leaves may indicate the presence of a diverged isoform that shares common antigenic epitopes with the Cel1 protein. Although these data suggest that the 62-kD polypeptide is encoded by the Cel1 locus, the molecular mass is significantly larger than what was predicted for the mature, processed protein (approximately 51 kD). This can be reconciled, however, by aberrant migration on SDS gels or extensive posttranslational modification, which for avocado EGase has been shown to be attributed in part to glycosylation (Bennett and Christoffersen, 1986; Kanellis and Kalaitzis, 1992). In an extreme case, tomato Cel3 encodes a predicted 68.5-kD polypeptide, which is detected as 88- and 93-kD polypeptide species upon SDS-PAGE (Brummell et al., 1997b).
Figure 5.
Western blot of soluble-protein extracts prepared from different tissues of flowering strawberry plants probed with a 1:5000 dilution of antiserum raised to a Cel1 fusion protein. Each lane contained 100 μg of acetone-precipitated protein electrophoresed on a 10% SDS-PAGE gel.
Although Cel1 protein accumulation is limited to ripening fruit beyond the green stage of development, CMCase activity measurements in protein extracts demonstrate the likelihood that EGases are present in all of the major tissues of strawberry plants (Fig. 6). In leaves we detected the highest levels of CMCase activity, but Cel1 protein, if present, was below the limit of detection by western-blot analysis. Relatively high levels of enzyme activity were also recorded for green fruit, leaf stems, flower stems, flowers, and roots, none of which has detectable Cel1 protein. These data support the existence of a highly divergent and potentially large gene family for strawberry EGase, with Cel1 as the ripening-related member.
Figure 6.
Histogram of relative CMCase activities measured in soluble-protein extracts prepared from various strawberry tissues. Relative specific activities are an average of three independent measurements (for a definition of activity units as provided here, see Harpster et al., 1997). sd values were less than 10% of the values shown.
DISCUSSION
We have isolated and characterized full-length cDNAs encoding a ripening-related EGase (Cel1) from strawberry, which correspond to a partial-length EGase cDNA recently reported by Manning (1998). As observed for cell wall-localized EGases from a wide variety of plant species, the amino-terminal sequence of the predicted protein (32 amino acids) is predominantly hydrophobic and shows homology with eukaryotic signal sequences (von Heijne, 1986). Although the sequence alignment of all EGases to date reveals several conserved amino acid domains, it is becoming clear that amino acid sequence homology and phylogenetic similarity do not necessarily predict mRNA expression pattern. This is illustrated by the observation that the closest related EGases to strawberry Cel1 are tomato Cel2, Arabidopsis Cel1, and pepper Cel3, three genes that exhibit quite different patterns of expression. Whereas tomato Cel2 mRNA is most similar to strawberry Cel1 in that it accumulates predominantly during fruit ripening (Lashbrook et al., 1994), Arabidopsis Cel1 mRNA is expressed mainly in expanding vegetative tissues (Shani et al., 1997) and pepper Cel3 mRNA is expressed in abscissing leaf-abscission zones (Ferrarese et al., 1995). Two other EGases with mRNA-accumulation patterns that are predominantly ripening related, avocado Cel1 (Christoffersen et al., 1984) and pepper PCEL1 (Harpster et al., 1997), encode proteins that are less related and that are found in other branches or subbranches of the phylogenetic tree (Fig. 1C).
Strawberry Cel1 mRNA appears early in fruit development at the small-sized white stage and accumulates to maximal levels in full-sized red fruit (Fig. 3A), an observation that is consistent with the data of Manning (1998). As shown in previous studies (Given et al., 1988a; Abeles and Takeda, 1990), the onset of fruit softening in strawberry begins during the white stage and slightly precedes the appearance of anthocyanin pigments. Thereafter, fruit progressively exhibit a loss of firmness, which nears a maximum at the red stage. Although correlative, these findings raise the possibility that Cel1 activity may contribute to fruit softening. This association between strawberry Cel1 expression and fruit softening is further supported by the absence of Cel1 mRNA (Fig. 3B) and Cel1 protein (Fig. 5) in other tissues. In other species in which a ripening-related EGase has been described, either significant mRNA levels were also found in abscission zones (e.g. tomato Cel2 [Lashbrook et al., 1994] and avocado Cel1 [Tonutti et al., 1995]) or low levels were also detected in stems and petioles (e.g. pepper Cel1 [Harpster et al., 1997]). Because levels of strawberry Cel1 mRNA are either absent or below the limit of detection in tissues other than fruit, strawberry Cel1 appears to show the most ripening-specific pattern of mRNA accumulation for an EGase described to date.
Whereas strawberry Cel1 may be the predominant EGase gene expressed in softening red fruit (it was the only sequence identified among 55 independent cDNAs isolated from a red fruit library), CMCase activity measurements of protein extracts prepared from developing fruit and a variety of other tissues (Fig. 6) indicate the expression of other EGase genes. In green fruit Cel1 expression is undetectable, yet total CMCase activity is approximately 80% of levels measured in red fruit. Furthermore, high levels of activity are also detected in the leaf and in the leaf stem, with lower levels measured in flowers, flower stems, roots, and fruit stems. Genomic Southern-blot analysis indicates that in strawberry EGases are encoded by a divergent gene family (Fig. 2), and it is likely that CMCase activities measured in different tissues reflect the expression of one or more differentially expressed members of this family. In tomato, in which EGases are encoded by a divergent gene family, there are at least seven members, each with its own characteristic pattern of tissue-specific expression and regulation (Lashbrook et al., 1994; Milligan and Gasser, 1995; del Campillo and Bennett, 1996; Brummell et al., 1997a, 1997b; Catala et al., 1997).
Based on the analysis of EGase gene expression in different plant species, it appears that EGases involved in organ abscission and fruit ripening exhibit a pattern of hormonal regulation distinct from that of EGases involved in cell expansion in vegetative tissues. Whereas auxin can suppress ethylene-mediated increases in EGase expression and cell wall breakdown and/or separation in fruit and abscission zones, auxin exhibits a stimulatory effect on EGase expression in cells undergoing growth and expansion. This inverse effect is illustrated by the auxin-mediated increases in steady-state levels of mRNAs encoded by the poplar Cel1 gene in suspension-cultured cells (Nakamura et al., 1995), the pea EGL1 gene in epicotyls (Wu et al., 1996), and the tomato Cel7 gene in hypocotyls (Catala et al., 1997), and decreases in the expression of the ethylene-regulated, abscission-related EGase genes of bean (BAC1; Tucker et al., 1988) and tomato (Cel1 and Cel5; del Campillo and Bennett, 1996). In ripening avocado fruit discs, suppression of ethylene-induced EGase gene expression by auxin is observed at the posttranscriptional level (Buse and Laties, 1993). Whereas the antagonistic control of EGase expression by auxin and ethylene is observed in many plant species, this relationship may not necessarily hold true in all cases. In the nonclimacteric plant species, for instance, attempts to demonstrate ethylene-mediated effects on gene expression and physiological processes have been largely inconclusive.
Although the high-level autocatalytic production of ethylene and its promotion of ripening is a hallmark of climacteric plants such as tomato and avocado, the control of ripening in nonclimacteric species such as grape and strawberry is less well understood. In strawberry, fruit produce extremely low and relatively constant levels of ethylene during ripening (Knee et al., 1977; Abeles and Takeda, 1990), and treatment with ethylene antagonists such as 2,5-norbornadiene, aminoethoxyvinylglycine, or silver did not affect ripening, as measured by anthocyanin accumulation (Given et al., 1988a). Furthermore, exposing strawberry fruit to high concentrations of exogenous ethylene had no obvious effect on ripening (Iwata et al., 1969). Whereas these findings question the importance of ethylene to nonclimacteric fruit ripening, the well-documented yet minor promotion by ethylene of EGase expression and fruit reddening in nonclimacteric pepper (Ferrarese et al., 1995; Harpster et al., 1997) suggests that ethylene may play some role in the ripening of nonclimacteric fruit. In most nonclimacteric ripening fruit, however, the potential role of ethylene is generally obscured by technical difficulties in monitoring the physiological consequences of exceptionally low-level endogenous ethylene production, and by the possibility that the low levels of endogenous ethylene production are saturating, in which case the application of exogenous ethylene would be without observable effect. Whatever the precise role ethylene may have, the main regulation of the ripening process in strawberry and grape appears to occur through declining levels of auxin in the fruit (Given et al., 1988b; Manning, 1994; Davies et al., 1997).
In immature strawberry fruit, receptacle expansion is controlled by auxins originating in and released by the achenes (Nitsche, 1950). Achenes produce large amounts of free and conjugated IAA, with peak levels achieved at 10 to 14 dpa, depending on the cultivar (Dreher and Pooviah, 1982; Archbold and Dennis, 1984). Removal of achenes from white fruit mimics the in vivo decline of endogenous auxin levels, albeit at an accelerated rate, in that there is a rapid accumulation of anthocyanins and a concomitant loss in chlorophyll content and tissue firmness, all of which are typical of ripening in strawberry fruit (Given et al., 1988a, 1988b; Manning, 1994). Because the application of the synthetic auxin NAA prevents these changes, it is concluded that strawberry ripening is regulated in part by a decline in the amount of auxin produced by the achenes (Given et al., 1988b; Manning, 1994).
In the experiments presented here we have manipulated endogenous auxin levels and found that, although the application of NAA to halves of small-sized white fruit evoked a suppression of Cel1 mRNA expression levels and anthocyanin accumulation relative to untreated control halves, the removal of achenes from similarly aged fruit caused an enhancement in expression and ripening. Thus, there is a negative correlation between high auxin levels and steady-state abundance of Cel1 mRNA. These observations suggest that Cel1 mRNA accumulation is repressed by auxin and is induced by a decline in endogenous auxin content, which, directly or indirectly, initiates the onset and development of fruit ripening.
The control of Cel1 expression by auxin contributes to a growing body of information that suggests that a decrease in fruit auxin levels triggers strawberry ripening and the concomitant derepression of a battery of ripening-related genes. In a study by Manning (1994), the removal of achenes from immature-green strawberry fruit and the subsequent analysis of in vitro translation products by two-dimensional PAGE showed the induction of several mRNAs that increase in abundance in normally ripening fruit. Additional studies with deachenated fruit have demonstrated the auxin-mediated repression of Phe ammonia-lyase and total anthocyanin accumulation in ripening fruit (Given et al., 1988b) and the isolation of an auxin-repressed cDNA of unknown function (Reddy and Pooviah, 1990). Medina-Escobar et al. (1997) have characterized a strawberry pectate-lyase gene that shares identical spatial, temporal, and hormonal patterns of regulation with those shown here for Cel1. In future experiments transgenic Cel1-suppressed plants will be monitored for phenotypic effects, and a reduction in the activity of a single enzyme in a complex developmental program such as ripening will have to be evaluated with the understanding that the activity of several genes may contribute to overall fruit firmness.
ACKNOWLEDGMENTS
We thank Dr. Rita Teutonico for her efforts in the construction of the cDNA library, Malini Nag for assistance with the isolation and characterization of cDNA clones, Dr. Paul Oeller for careful review of the manuscript, and Jay Maddox and Vincenzo Pirozzi of the greenhouse staff for maintenance of the plants.
Abbreviations:
- CMCase
carboxymethylcellulase
- dpa
days postanthesis
- EGase
endo-1,4-β-glucanase
- NAA
naphthalene acetic acid
- ORF
open reading frame
Footnotes
The accession number for the sequence reported in this article is AF074923.
LITERATURE CITED
- Abeles FB, Takeda F. Cellulase activity and ethylene in ripening strawberry and apple fruits. Sci Hortic. 1990;42:269–275. [Google Scholar]
- Archbold DD, Dennis FG. Quantification of free ABA and free and conjugated IAA in strawberry achene and receptacle tissue during fruit development. J Am Soc Hortic Sci. 1984;109:330–335. [Google Scholar]
- Barnes MF, Patchett BJ. Cell wall degrading enzymes and the softening of senescent strawberry fruit. J Food Sci. 1976;41:1392–1395. [Google Scholar]
- Bennett AB, Christoffersen RE. Synthesis and processing of cellulase from ripening avocado fruit. Plant Physiol. 1986;81:830–835. doi: 10.1104/pp.81.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brady CJ. Fruit ripening. Annu Rev Plant Physiol. 1987;38:155–178. [Google Scholar]
- Brummell DA, Bird CR, Schuch W, Bennett AB. An endo-1,4-β-glucanase expressed at high levels in rapidly expanding tissues. Plant Mol Biol. 1997a;33:87–95. doi: 10.1023/a:1005733213856. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Catala C, Lashbrook CC, Bennett AB. A membrane-anchored E-type endo-1,4-β-glucanase is localized on Golgi and plasma membranes of higher plants. Proc Natl Acad Sci USA. 1997b;94:4794–4799. doi: 10.1073/pnas.94.9.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Lashbrook CC, Bennett AB. Plant endo-1,4-β-glucanases. In: Himmel ME, Baker JO, Overend RP, editors. Enzymatic Conversion of Biomass for Fuels Production. ACS Symposium Series 566. Washington, DC: American Chemical Society; 1994. pp. 100–129. [Google Scholar]
- Buse EL, Laties G. Ethylene-mediated posttranscriptional regulation in ripening avocado (Persea americana) mesocarp discs. Plant Physiol. 1993;102:417–423. doi: 10.1104/pp.102.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala C, Rose JKC, Bennett AB. Auxin regulation and spatial localization of an endo-1,4-β-D-glucanase and a xyloglucan endotransglycosylase in expanding tomato hypocotyls. Plant J. 1997;12:417–426. doi: 10.1046/j.1365-313x.1997.12020417.x. [DOI] [PubMed] [Google Scholar]
- Christoffersen RE, Tucker ML, Laties GG. Cellulase gene expression in ripening avocado fruit: the accumulation of cellulase mRNA and protein as determined by cDNA hybridization and immunodetection. Plant Mol Biol. 1984;3:385–391. doi: 10.1007/BF00033386. [DOI] [PubMed] [Google Scholar]
- Davies C, Boss PK, Robinson SP. Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 1997;115:1155–1161. doi: 10.1104/pp.115.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E, Bennett AB. Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiol. 1996;111:813–820. doi: 10.1104/pp.111.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW, Pooviah BW. Changes in auxin content during development in strawberry fruits. J Plant Growth Regul. 1982;1:267–276. [Google Scholar]
- Ferrarese L, Trainotti L, Moretto P, de Laureto PP, Rascio N, Casadoro G. Differential ethylene-inducible expression of cellulase in pepper plants. Plant Mol Biol. 1995;29:735–747. doi: 10.1007/BF00041164. [DOI] [PubMed] [Google Scholar]
- Given NK, Venis MA, Grierson D. Phenylalanine ammonia-lyase activity and anthocyanin synthesis in ripening strawberry fruit. J Plant Physiol. 1988a;133:25–30. [Google Scholar]
- Given NK, Venis MA, Grierson D. Hormonal regulation of ripening in the strawberry, a non-climacteric fruit. Planta. 1988b;174:402–406. doi: 10.1007/BF00959527. [DOI] [PubMed] [Google Scholar]
- Greene B, Walko R, Hake S. Mutator insertions in an intron of the maize knotted1 gene result in dominant suppressible mutations. Genetics. 1994;138:1275–1285. doi: 10.1093/genetics/138.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Scott DH, Lawrence FJ. Strawberries. In: Janick J, Moore JN, editors. Fruit Breeding, Vol II: Vine and Small Fruits. New York: John Wiley & Sons; 1996. pp. 419–470. [Google Scholar]
- Harpster MH, Lee KY, Dunsmuir P. Isolation and characterization of a gene encoding endo-1,4-β-glucanase from pepper (Capsicum annuum L.) Plant Mol Biol. 1997;33:47–59. doi: 10.1023/a:1005795028489. [DOI] [PubMed] [Google Scholar]
- Hatfield R, Nevins D. Characterization of the hydrolytic activity of avocado cellulase. Plant Cell Physiol. 1986;27:541–552. [Google Scholar]
- Higgins DG, Bleasby AJ, Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Huber DJ. Strawberry fruit softening: the potential roles of polyuronides and hemicelluloses. J Food Sci. 1984;49:1310–1315. [Google Scholar]
- Iwata T, Omato I, Ogata K. Relationship between the ripening of harvested fruits and the respiratory pattern. III. Changes of ethylene concentration in fruits and responses to applied ethylene with relation to the respiratory pattern. J Jpn Soc Hortic Sci. 1969;38:64–72. [Google Scholar]
- Kanellis AK, Kalaitzis P. Cellulase occurs in multiple active forms in ripe avocado fruit mesocarp. Plant Physiol. 1992;98:530–534. doi: 10.1104/pp.98.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee M, Sargent JA, Osborne DJ. Cell wall metabolism in developing strawberry fruits. J Exp Bot. 1977;28:377–396. [Google Scholar]
- Lashbrook CG, Gonzalez-Bosch C, Bennett AB. Two divergent endo-1,4-β-glucanase genes exhibit overlapping expression in ripening fruit and abscissing flowers. Plant Cell. 1994;6:1485–1493. doi: 10.1105/tpc.6.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K. Changes in gene expression during strawberry fruit ripening and their regulation by auxin. Planta. 1994;194:62–68. [Google Scholar]
- Manning K. Isolation of a set of ripening-related genes from strawberry: their identification and possible relationship to fruit quality traits. Planta. 1998;205:622–631. doi: 10.1007/s004250050365. [DOI] [PubMed] [Google Scholar]
- Medina-Escobar N, Cardenas J, Moyano E, Caballero JL, Munoz-Blanco J. Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol Biol. 1997;34:867–877. doi: 10.1023/a:1005847326319. [DOI] [PubMed] [Google Scholar]
- Milligan SB, Gasser CS. Nature and regulation of pistil-expressed genes in tomato. Plant Mol Biol. 1995;28:691–711. doi: 10.1007/BF00021194. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Mori H, Sakai F, Hayashi T. Cloning and sequencing of a cDNA for poplar endo-1,4-β-glucanase. Plant Cell Physiol. 1995;36:1229–1235. [PubMed] [Google Scholar]
- Neal GE. Changes occurring in the cell walls of strawberries during ripening. J Sci Food Agric. 1965;16:604–608. [Google Scholar]
- Nitsche JP. Growth and morphogenesis of the strawberry as related to auxin. Am J Bot. 1950;37:211–215. [Google Scholar]
- Oeller PW, Wong L-M, Taylor LP, Pike DA, Theologis A. Reversible inhibition of tomato fruit senescence by antisense RNA. Science. 1991;254:437–439. doi: 10.1126/science.1925603. [DOI] [PubMed] [Google Scholar]
- Picton S, Barton SL, Bouzayen M, Hamilton AJ, Grierson D. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 1993;3:469–481. [Google Scholar]
- Reddy ASN, Pooviah BW. Molecular cloning and sequencing of a cDNA for an auxin-repressed mRNA: correlation between fruit growth and repression of the auxin-regulated gene. Plant Mol Biol. 1990;14:127–136. doi: 10.1007/BF00018554. [DOI] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulsen A. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton R, Durbin M, Lewis L, Thomson W. Use of cellulase antibodies to study leaf abscission. Nature. 1980;283:8743–8744. [Google Scholar]
- Shani Z, Dekel M, Tsabary G, Shoseyov O. Cloning and characterization of elongation specific endo-1,4-β-glucanase (cel1) from Arabidopsis thaliana. Plant Mol Biol. 1997;34:837–842. doi: 10.1023/a:1005849627301. [DOI] [PubMed] [Google Scholar]
- Sheehy RE, Kramer M, Hiatt WR. Reduction of polygalacturonase activity in tomato fruit by antisense RNA. Proc Natl Acad Sci USA. 1988;85:8805–8809. doi: 10.1073/pnas.85.23.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Ray J, Bird CR, Morris PC, Schuch W, Grierson D. Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature. 1988;334:724–726. [Google Scholar]
- Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony, Version 3.1.1. Champaign: Illinois Natural History Survey; 1993. [Google Scholar]
- Taylor JL, Jones JDG, Sandler S, Mueller GM, Bedbrook JR, Dunsmuir P. Optimizing the expression of chimeric genes in plant cells. Mol Gen Genet. 1987;210:572–577. [Google Scholar]
- Tonutti P, Cass LG, Christoffersen RE. The expression of cellulase gene family members during avocado fruit abscission and ripening. Plant Cell Environ. 1995;18:709–713. [Google Scholar]
- Tucker ML, Sexton R, del Campillo E, Lewis LN. Bean abscission cellulase. Characterization of a cDNA clone and regulation of gene expression by ethylene and auxin. Plant Physiol. 1988;88:1257–1262. doi: 10.1104/pp.88.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C-Y, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Woodward JR. Physical and chemical changes in developing strawberry fruits. J Sci Food Agric. 1972;23:465–473. doi: 10.1002/jsfa.2740230406. [DOI] [PubMed] [Google Scholar]
- Wu S-C, Blumer JM, Darvill AG, Albersheim P. Characterization of an endo-1,4-β-glucanase gene induced by auxin in elongating pea epicotyls. Plant Physiol. 1996;110:163–170. doi: 10.1104/pp.110.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]