Abstract
A large segment of the population suffers from addiction to alcohol, smoking, or illicit drugs. Not only do substance abuse and addiction pose a threat to health, but the consequences of addiction also impose a social and economic burden on families, communities, and nations. Genome-wide linkage and association studies have been used for addiction research with varying degrees of success. The most well-established genetic factors associated with alcohol dependence are in the genes encoding alcohol dehydrogenase (ADH), which oxidizes alcohol to acetaldehyde, and aldehyde dehydrogenase (ALDH2), which oxidizes acetaldehyde to acetate. Recently emerging genetic studies have linked variants in the genes encoding the α3, α5, and β4 nicotinic acetylcholine receptor subunits to smoking risk. However, the influence of these well-established genetic variants accounts for only a small portion of the heritability of alcohol and nicotine addiction, and it is likely that there are both common and rare risk variants yet to be identified. Newly developed DNA sequencing technologies could potentially advance the detection of rare variants with a larger impact on addiction risk.
Keywords: addiction, alcoholism, smoking, nicotinic acetylcholine receptors, alcohol metabolizing genes
INTRODUCTION
Substance abuse and addiction pose a worldwide threat to public health and have a devastating social and economic impact on individuals and their families. The World Health Organization (144) has estimated that there are 2 billion alcohol users, 1.3 billion tobacco users, and 185 million illicit drug users worldwide. In the 2010 National Survey on Drug Use and Health conducted by the Substance Abuse and Mental Health Administration (SAMHSA) (119a), 51.8% of Americans aged 12 or older (131.3 million people) reported being current drinkers of alcohol, and 23.1% reported participating in binge drinking (defined as having five or more drinks on the same occasion on at least 1 day in the 30 days prior to the survey). The World Health Organization (143) has also estimated that approximately 20%–30% of esophageal cancer, liver cancer, cirrhosis of the liver, homicide, epilepsy, and motor vehicle accidents worldwide result from the harmful use of alcohol.
In the 2010 SAMHSA survey (119a), approximately 58.3 million Americans aged 12 or older reported being current cigarette smokers, and a recent Surgeon General’s report (129) indicated that one-third of people who have tried smoking became daily smokers (defined as those who reported that they have smoked 100 or more cigarettes during their lifetime and currently smoke every day or some days). The SAMHSA survey (119a) also showed that 59.6% of current smokers aged 12 or older smoked daily and that this proportion increased with age---going from 16.5% among those aged 12–17, to 27.8% among those aged 18–25, to 48.9% among those aged 26 or older. The detrimental effects of tobacco use or exposure to secondhand smoke include an increased risk of cancer, chronic lung disease, heart disease, and stroke. In the United States, cigarette smoking accounts for 30% of deaths from cancer and nearly 80% of deaths from chronic obstructive pulmonary disease (COPD) (22, 95), and it is also the primary causal factor for early cardiovascular disease and deaths (22). Globally, cigarette smoking kills 5.4 million people every year and accounts for 10% of adult deaths (144).
Other psychoactive substances, such as cannabis, cocaine, and opioids, also cause significant health and social problems for both the people who use them and their families. According to data from the United Nations Office on Drugs and Crime, 149–271 million people worldwide aged 15–64 used an illicit drug in 2009; of these, 15–39 million were classified as problem users (31). In the SAMHSA survey (119a), an estimated 22.6 million Americans aged 12 or older reported having used an illicit drug in 2010 during the month prior to the survey interview; of these, 2.9 million abused or were dependent on both alcohol and illicit drugs, and 4.2 million abused or were dependent on illicit drugs but not alcohol. Major health consequences of illicit drug use include accidental and intended injury, drug-induced psychotic symptoms, and increased risk for heart, liver, and lung diseases (31). A World Health Organization report (144) indicated that in 2004, an estimated 0.7% of the global burden of disease resulted from cocaine and opioid use.
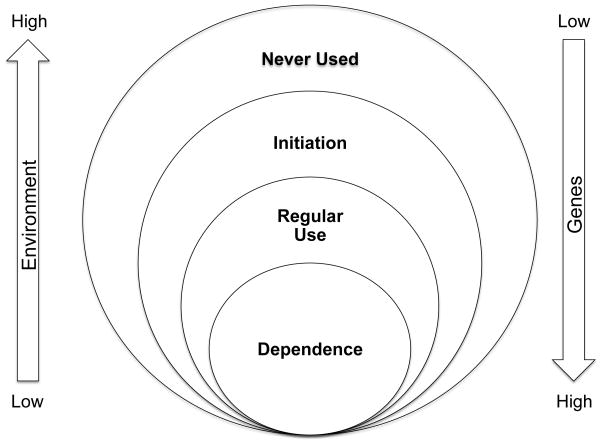
Drug addiction is a chronic psychiatric disorder characterized by the persistent, compulsive, and uncontrolled use of a drug despite harmful consequences. Scientific studies on addictive behaviors began in the 1930s and revealed that people with an addiction are not simply lacking in willpower; instead, they are unable to control their use of the drug (135). With advances in our understanding of the effects of alcohol, nicotine, and illicit drugs on brain physiology and behavior, it has become evident that addiction is a psychiatric disease attributable to biological and environmental factors (135). The development of addiction involves several steps: the initiation of substance use, the transition from experimental use to regular use, and the actual development of addiction. Environmental factors such as peer pressure, parental monitoring, and the accessibility of a substance play a major role in the initial decision to drink, smoke, or take illicit drugs. Beyond the initiation step, the transition from regular substance use to dependence differs from person to person and is largely under genetic control (Figure 1) (74, 133, 135).
Figure 1.
Interaction of genetic and environmental factors in the development of substance dependence. The initiation of substance use is influenced largely by environmental factors; the use of the addictive substance is affected largely by genetic factors.
GENETIC INFLUENCES ON THE RISK OF SUBSTANCE DEPENDENCE
Evidence for a genetic influence on substance dependence has been provided by many family, twin, and adoption studies. Family members of alcohol-dependent individuals have a higher probability of suffering from alcohol dependence (54). In a study of families severely affected by alcohol-abuse disorders, approximately 50% of brothers and 22%–25% of sisters of an alcohol-dependent proband were alcohol dependent (15). Similarly, siblings of marijuana-dependent, cocaine-dependent, or habitual-smoking probands were at increased risk (approximately 1.7-fold higher) of developing marijuana dependence, cocaine dependence, or habitual smoking compared with siblings of nondependent individuals (15). Studies with large twin cohorts have shown that the risk of alcohol dependence in the co-twin of an affected monozygotic twin is significantly higher than the risk in the co-twin of an affected dizygotic twin pair, which is similar to that of full siblings of the affected individual (117). In adoption studies, children of alcoholics adopted by nonalcoholics who grow up in a nondrinking environment have a higher risk of becoming alcoholic than do children of nonalcoholics adopted by the same parents; children of alcoholics raised by their alcoholic father have a similar risk of developing alcohol dependence as their full brothers who were adopted by nonalcoholics (117). Overall, studies have shown that the heritability of alcohol-use disorders ranges from 40% to 60% (117, 131).
Cigarette smoking commonly co-occurs with alcohol abuse. A meta-analysis of twin studies showed that both genetic and environmental factors affect smoking and smoking-related behaviors (83). In women, the initiation of smoking is influenced largely by genetic factors; in men, the genetic impact is more significant on the persistence of smoking than on the initiation of smoking (83). The heritability of smoking initiation and nicotine dependence is estimated to be 50% and 59%, respectively (83).
Studies have also indicated the familial transmission of illicit-substance-use disorders, with heritability estimates ranging from 30% to 80% (2, 83, 127). A recent meta-analysis of twin studies on marijuana use clearly indicated that vulnerability to both initiation and persistent use was significantly affected by both genetic and environmental factors (132). Genetic factors accounted for 48% and 40% of the total variance in initiation of marijuana use in men and women, respectively, and for 51% and 59% of the total variance in marijuana abuse in men and women, respectively.
Epidemiological and clinical studies have shown that many people subsequently use multiple drugs after their initiation of one drug (105, 109). Twin studies have demonstrated the presence of shared environmental factors that contribute to substance use. The shared environmental influence has a significant effect on tobacco initiation, alcohol use, and any drug use; however, genetic factors have a higher impact than shared environmental influences on tobacco use, tobacco problem use, and marijuana initiation (109). Studies on the etiology of the comorbidity of multiple substances in adolescents have suggested that genetic and environmental influences are common across substance classes (74, 109). In family studies involving adults, findings regarding general versus substance-specific familial risks have not been conclusive (15, 91, 128).
IDENTIFICATION OF GENETIC RISK FACTORS FOR ALCOHOL DEPENDENCE
Genome-Wide Linkage Studies
One of the earliest genome-wide approaches to identifying genetic risk factors for alcoholism employed linkage mapping in large extended families or in many sibling pairs affected by alcohol dependence. Studies using this method identified several chromosomal regions with LOD (logarithm of the odds, to the base 10) scores suggesting that they contain loci influencing risk for alcohol dependence. The Collaborative Study on the Genetics of Alcoholism (COGA) investigators performed linkage studies on a large sample from the general US population using multigenerational pedigrees densely affected by alcoholism (45, 108). In contrast, investigators at the National Institute on Alcohol Abuse and Alcoholism conducted a linkage study on a more homogeneous population from a southwestern Native American tribe (88). Both of these studies provided evidence that loci on human chromosome 4 increase the risk for alcohol dependence. However, the linkage peak detected in the COGA study is near the alcohol dehydrogenase (ADH) gene cluster, whereas the linkage signal observed in the Native American sample is near the GABRB1 gene.
The separate locations of these linkage signals may reflect differences in the underlying etiology across distinct populations, a suggestion supported by the observation of genome-wide significant linkage of alcohol dependence with markers on human chromosome 10 in an African American sample (47). Alternatively, this difference could reflect the inability of this kind of linkage study to accurately pinpoint the location of the gene(s) underlying a signal, or could mean that one or more of these linkage regions is associated with a false-positive signal.
Recently, linkage analysis of a community-based sample of Australian adults detected a suggestive linkage peak on human chromosome 5p with a LOD score of 2.2 (58). A genome-wide scan performed with community samples recruited through the University of California, San Francisco (UCSF) Family Alcoholism Study identified several suggestive regions linked with DSM-IV alcohol dependence (on human chromosomes 1, 2, 8, 9, 18, and 22) (50). Unfortunately, there is little consensus among studies regarding the location of linkage signals for alcohol dependence. This may be due to underlying genetic heterogeneity in the risk for alcohol dependence, with many genetic loci contributing to risk. This is probably compounded by the fact that all of these studies were underpowered to detect genes of small effect size (110).
Genome-Wide Association Studies
Several genome-wide association studies (GWAS) examining the risk for alcohol dependence have been completed using a variety of designs, including case- control series of male alcoholics recruited from inpatient treatment facilities (125), individuals selected from densely affected families with alcohol dependence (35), a mixed case-control series drawn from treatment- and community-based samples (14), subjects ascertained from community-based sibships, and individuals selected for heavier alcohol use (61). GWAS using quantitative traits derived from alcohol-consumption and alcohol-dependence symptomatology have also been examined in controls from a population-based sample recruited for schizophrenia (73) and an Australian population of related individuals (61). One study identified two correlated intergenic single-nucleotide polymorphisms (SNPs) on human chromosome 2q35 that met genome-wide significance in the combined analysis of the GWAS and follow-up data sets (125). The other studies did not observe any association that met conventional genome-wide significance, and the overlap of the top genetic signals across studies has been limited.
Although the results to date have been somewhat disappointing, they underscore the prior observations from linkage studies and support the hypothesis that alcohol dependence is a genetically heterogeneous disorder influenced by many genes of small effect. The power to detect statistically significant association is also an important consideration. The sample sizes (n < 5,000) in GWAS of alcohol dependence to date are much smaller than those of successful GWAS of other diseases such as type 2 diabetes and breast cancer, which used >30,000 subjects (140a).
Candidate Gene Studies
Genetically influenced metabolic factors have been implicated in the etiology of alcoholism in a number of ethnic groups. The conversion of alcohols to the corresponding aldehydes is catalyzed by ADHs. This is the rate-limiting step in the elimination of ethanol in humans and experimental animals (18). Seven ADH-encoding genes (ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, and ADH7) are located as a cluster on human chromosome 4q22–23 (33). The class 1 enzymes---encoded by ADH1A, ADH1B, and ADH1C (previously termed ADH3) in humans---have high affinity for ethanol and contribute the most to its conversion to acetaldehyde, particularly during the elimination phase. This class of ADH enzymes includes the most important ADH isoforms for oxidizing ethanol in humans (33). ADH7 acts early in the time course of alcohol metabolism in stomach mucosa that is exposed to high concentrations of alcohol (42).
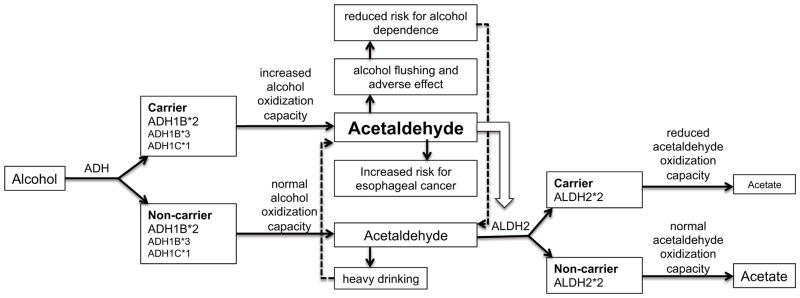
The majority of association studies investigating the role of alcohol-metabolizing genes in risk for alcohol-use disorders have focused on the well-characterized coding variants within ADH1B, ADH1C, and ALDH2 and on the phenotype of alcohol dependence. There are three different ADH1B alleles (33). The reference allele is ADH1B*1, which encodes the β1 subunit with an arginine at amino-acid positions 48 (Arg48) and 370 (Arg370). ADH1B*2, a common allele in Asians, encodes the β2 subunit with a histidine at position 48 (His48). The ADH1B*3 allele, which encodes the β3 subunit with a cysteine at position 370 (Cys370), is found primarily in people of African descent. Amino-acid substitutions at positions 48 (ADH1B*2) and 370 (ADH1B*3) result in 70–80-fold higher enzyme activity compared with that produced by the ADH1B*1 allele (33). The rapid conversion of ethanol to acetaldehyde causes facial flushing and aversive effects after alcohol consumption and is protective against alcohol dependence (103) (Figure 2). A meta-analysis of the ADH1B*2 allele in Han Chinese and Japanese showed that individuals who are homozygous for this variant (His48/His48) have a fivefold decrease in risk for alcohol dependence compared with individuals who are heterozygous for this variant (Arg48/His48) (142). In Europeans, the risk for developing alcohol dependence is twofold lower in His48/His48 carriers compared with Arg48/His48 carriers (142). Recently, a case-control study in populations of European and African ancestry demonstrated that the ADH1B*2 (His48) allele in these populations is associated with a lower maximum number of drinks in a 24-h period (p = 3 × 10−13) and has a strong protective effect on DSM-IV alcohol dependence in both populations (odds ratio = 0.34, p = 6.6 × 10−10) (16). The protective effect of ADH1B*2 was not detectable by a GWAS approach in studies involving populations of European or African descent because none of the variants on these genotyping chips showed high linkage disequilibrium with this rare variant. These studies demonstrate that the ADH1B*2 allele correlates with reduced alcohol consumption and risk for alcohol dependence in all populations, though the allele frequencies vary in people of different ethnicity. The ADH1B*3 allele also has a protective effect on risk for alcoholism in African American families and southwest California Native Americans (36, 136).
Figure 2.
The role of ADH and ALDH2 variants in the alcohol metabolic pathway.
The reference allele of the ADH1C gene is ADH1C*1, with an arginine at position 272 (Arg272) and an isoleucine at position 350 (Ile350). The ADH1C*2 allele, with a glutamine at position 272 (Gln272) and a valine at position 350 (Val350), is common in Europeans and African Americans. The ADH1C*3 allele, with a threonine at position 352 (Thr352), is found in Native Americans (33). Studies have shown that ADH1C*1 also has protective effects on the risk for alcohol dependence in people of Asian and African descent (33, 96). However, some studies showed that the protective effect of the ADH1C*1 allele is not an independent effect owing to the linkage disequilibrium of this allele with the ADH1B*2 allele (24, 103).
Another well-known polymorphism is in the ALDH2 gene encoding aldehyde dehydrogenase 2 family (mitochondrial). The ALDH2*2 allele, which substitutes lysine for glutamate at position 504 (Lys504), results in a nearly inactive protein subunit that is unable to metabolize acetaldehyde (149). This allele is relatively common in Asians but nearly absent in people of European or African descent (69, 102) and is strongly associated with a reduced risk for alcohol dependence (33). Polymorphisms in other ADH genes have also been associated with alcohol dependence (36, 77). Studies have shown that variation in the ADH genes contributes substantially to variation in alcohol metabolism and consequently affects the risk for alcohol dependence. Although the variants ADH1B Arg48His and ADH1C Arg272Gln/Ile350Val are known to have a major effect on enzyme activity in vitro, these variants account for only a very small amount of the genetic variance in in vivo metabolism (18, 94). In vivo studies in Europeans demonstrated that variants in ADH7 are associated with the early stages of alcohol metabolism, with additional effects in ADH1A, ADH1B, and ADH4 (18). Postabsorptive alcohol metabolism is affected by variants in the ADH7-ADH1C-ADH1B gene cluster. Approximately 20% of the total genetic variance for alcohol metabolism was attributed to the combined effects of variants in the ADH gene region (18). Because patterns of linkage disequilibrium across this genomic region vary among different ethnic populations (36, 104) and the frequencies of functional variants differ from one population to another, the effects of functional variants may be population specific.
Gamma amino butyric acid (GABA) is the main inhibitory neurotransmitter in the central nervous system, and its transmission is considered to mediate the pharmacological effects of alcohol in the brain. The modulatory actions of GABA are mediated through two types of receptors: the ionotropic GABAA receptor and the metabotropic GABAB receptor (57, 137). A family study from the COGA group identified multiple SNPs in GABRA2 (which encodes the GABAA receptor α2 subunit) associated with increased risk for alcohol dependence (34). Several subsequent studies using case-control samples replicated the association of GABRA2 and alcohol dependence, though the nature of the association and the specific variants associated with alcohol dependence differ in some samples (27, 40, 43, 80, 119). Furthermore, one study showed that GABRA2 alleles affect the SRE (self-rating of the effects of alcohol), suggesting that genetic variations in GABRA2 might play a role in the risk for alcohol-use disorders by moderating the SRE (111). Evidence from a functional MRI study suggested that a SNP in GABRA2 (rs279871) associated with alcohol dependence is also associated with the medial frontal response to alcohol cues (72).
Adjacent to GABRA2 is the GABRG1 gene, which encodes the GABAA receptor γ1 subunit. Several studies have reported association of GABRG1 variants with the risk for alcohol dependence and drinking behaviors (39, 107). Haplotype analyses have suggested that markers in the GABRA2 gene associated with alcohol dependence are in linkage disequilibrium with markers in the GABRG1 gene in many populations, indicating that the association with GABRA2 may be driven by variants in GABRG1 (28, 71). Despite multiple studies implicating SNPs in GABRA2 and GABRG1 in the risk for alcohol-related behaviors, the specific functional alleles underlying these associations have yet to be identified.
In summary, candidate gene studies have successfully detected functional variants in alcohol metabolism genes such as ADH1B, ADH1C, and ALDH2 associated with a risk of developing alcohol dependence in populations of Asian descent. Although alleles associated with reduced risk in Asians are rare in populations of African and European descent, they also reduce risk for alcohol dependence in these populations. Several GWAS of alcoholism have produced no conclusive evidence for specific genetic risk factors. The heterogeneous nature of the ascertainment strategies and the phenotypic measures used across studies could potentially explain the lack of a replicated association. Furthermore, the sample sizes in current alcohol studies are small compared with those in GWAS of other psychiatric disorders, limiting the power to detect genetic risk factors.
IDENTIFICATION OF GENETIC RISK FACTORS FOR NICOTINE DEPENDENCE
Genome-Wide Linkage Studies
To identify susceptibility loci for nicotine dependence, more than 20 linkage analyses across the entire genome have been conducted using a family-based and/or sib-pair design (for a review, see 83). Although a number of genomic regions were identified as significant or suggestive for harboring susceptibility loci for nicotine dependence or smoking-related phenotypes, only four linkage regions have been replicated in four or more independent samples---these reside on human chromosomes 9q, 10q, 11p, and 17p (82). Recently a genome-wide linkage scan suggested that a region on human chromosome 2q31.1 confers risk for the development of nicotine dependence with a broad range of dependence symptoms rather than a specific aspect of the disorder (51).
Genome-Wide Association Studies
In contrast to the studies of alcohol dependence, GWAS of smoking behavior have reported consistent and compelling genetic evidence for association. The first GWAS using a case-control sample reported evidence that variants within the nicotinic acetylcholine receptor (nAChR) subunit genes on the long arm of human chromosomes 15 (CHRNA5-CHRNA3-CHRNB4) and 8 (CHRNA6-CHRNB3) influence risk for nicotine dependence, as defined by scores on the Fagerström test for nicotine dependence (17). The chromosome 15 association has been replicated in subsequent GWAS either directly or indirectly using highly correlated SNPs (r2 > 0.8), with cigarettes per day (CPD) as a quantitative variable to define heavy- and light-smoking individuals (13, 17, 123, 140). Genome-wide association meta-analyses for CPD further confirmed that variants in CHRNA5, CHRNA3, and CHRNB4 are associated with the risk of developing heavy smoking (87, 124, 124a). In addition, the GWAS reported by Thorgeirsson et al. (124) showed that variation in the CHRNA6-CHRNB3 gene cluster on human chromosome 8 is associated with CPD at a genome-wide significance level.
Genetic variation in nicotine metabolism also plays an important role in cigarette consumption (93, 116) and nicotine dependence (8). Conversion of nicotine to cotinine typically accounts for 70%–80% of nicotine metabolism, the majority of which is catalyzed by the cytochrome P450 2A6 (CYP2A6) enzyme (67). Recent GWAS meta-analyses using subjects of European descent identified SNPs in the region of CYP2A6 associated with CPD (124, 124a).
Candidate Gene Studies
In parallel with these GWAS, several studies using a candidate gene approach have also reported the association of SNPs in the CHRNA5-CHRNA3-CHRNB4 gene cluster with nicotine dependence and smoking quantity (17, 113, 140). Furthermore, a fine mapping study (113) observed that the nonsynonymous SNP rs16969968 in exon 5 of CHRNA5 has consistent effects on the risk for nicotine dependence in both European (odds ratio of 1.40; 95% confidence interval, 1.23–1.59) and African (odds ratio of 2.04; 95% confidence interval, 1.15–3.62) populations, despite a large difference in allele frequency for the SNP. A second locus tagged by rs578776 in the 3′ untranslated region of CHRNA3 that has low linkage disequilibrium with rs16969968 is associated with nicotine dependence in European Americans but not in African Americans. Another linkage disequilibrium bin tagged by an intronic SNP in CHRNA5, rs588765, confers a protective effect for nicotine dependence in populations of European descent (Figure 3) (113, 139). A comprehensive meta-analysis involving more than 32,000 subjects confirmed the three unique loci in this gene cluster that affect smoking quantity (112). In Asians, a locus tagged by rs578776 overlapped with a locus tagged by rs588765, and variants in this distinctive linkage disequilibrium pattern were reported to influence smoking initiation, smoking cessation (84), and smoking quantity (84, 146).
Figure 3.
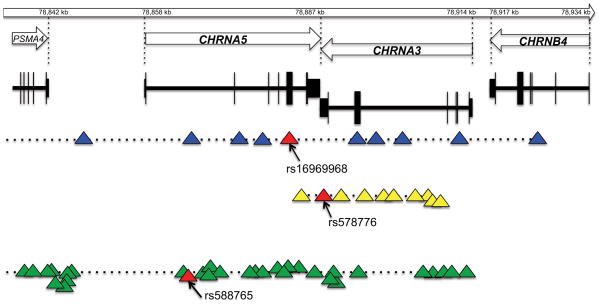
Three distinct loci across the CHRNA5-CHRNA3-CHRNB4 gene cluster on human chromosome 15. Triangles represent single nucleotide polymorphisms (SNPs). SNP position is not drawn to scale.
There are at least two distinct biological mechanisms in the nAChR gene cluster on chromosome 15 that alter the risk for developing nicotine dependence. One mechanism involves the variant rs16969968 (D398N), which likely alters protein structure and receptor function. An in vitro functional analysis demonstrated that the maximal response to agonist per receptor was twofold higher for the α4β2α5D398 nAChR variant relative to the α4β2α5N398 nAChR variant (17). The second potential mechanism is altered mRNA expression of CHRNA5 (139a)(139). Several variants located upstream of the coding region and intronic regions of CHRNA5 (i.e., rs588765) are strongly associated with the variability in CHRNA5 mRNA expression observed in the human frontal cortex. Subjects homozygous for the minor allele of rs588765 showed a 2.9-fold increase in CHRNA5 mRNA expression compared with subjects homozygous for the major allele (139a)(139). The rs588765 polymorphism and highly correlated variants are only weakly correlated with the D398N variant. The N398 variant, which greatly increases risk for nicotine dependence, occurs primarily on the background of low mRNA expression of CHRNA5. The nonrisk variant D398 occurs on both high- and low-expression alleles. The risk for nicotine dependence is significantly lower when D398 occurs on a background of low CHRNA5 mRNA expression than when it occurs on a background of high CHRNA5 mRNA expression (139).
Studies examining genetic and environmental risks for nicotine dependence have shown that there is an interaction between environmental factors and the rs16969968 variant that has an effect on smoking. The genetic risk associated with rs16969968 was reduced in subjects with high levels of parent monitoring and increased in subjects with low levels of parent monitoring (23). Interaction between childhood adversity and rs16969968 is also associated with the risk for nicotine dependence in men (148): Among men who experienced childhood adversity, individuals who carry the AA risk genotype have the highest risk of developing nicotine dependence compared with individuals who carry the GA or GG genotype.
A study that sequenced all genes encoding nicotinic receptor subunits has demonstrated that the low-frequency coding variants R37H in CHRNA3 and T375I and T91I in CHRNB4 decrease the risk for nicotine dependence among regular smokers (55). It further showed that the minor allele of each polymorphism increases the cellular response to nicotine (β4T375I p = 0.01, β4T91I p = 0.02, α3R37H p = 0.003), but the largest effect on in vitro receptor activity was seen in the presence of both CHRNB4 T91I and CHRNA3 R37H (p = 2 × 10−6), two SNPs in strong linkage disequilibrium in human populations (r2 = 0.89, n = 2,035 European Americans; r2 = 0.59, n = 710 African Americans).
Nicotine is the major substance in tobacco responsible for addiction among cigarette smokers (62). An in vivo study has shown that approximately 80% of nicotine consumed is metabolically inactivated to cotinine (12); approximately 90% of this conversion is mediated by CYP2A6 (92, 99). The next step of nicotine metabolism, which oxidizes cotinine to form trans-3′-hydroxycotinine, is entirely catalyzed by CYP2A6 (98). CYP2A6 is a highly polymorphic enzyme. Different CYP2A6 alleles have different functional consequences, and the frequency of CYP2A6 alleles varies among ethnic populations (97). A number of studies have reported association between reduced or absent CYP2A6 enzyme activity and lower risk of smoking, including decreased cigarette consumption, smoking intensity, and withdrawal symptoms; shorter smoking duration; and increased cessation. However, some studies have failed to detect any association between CYP2A6 variation and smoking status (64). A recent study using quantified measures of deuterated (D2)-cotinine/(D2-cotinine + D2-nicotine) following oral administration in 189 European Americans demonstrated that CYP2A6*12 is a loss-of-function allele indistinguishable from CYP2A6*4 and CYP2A6*2 alleles, and that the CYP2A6*1B 5′ untranslated region conversion has a negligible impact on metabolism (19). After controlling for the CYP2A6 genotype, the authors found modest associations between increased metabolism and both gender and current smoking (19).
In summary, genetic studies of nicotine dependence have successfully identified risk factors using both GWAS and candidate gene approaches. The consistent phenotypic measure---CPD---is easily obtained in large cohort studies and has been successfully used in meta-analyses of the genetics of smoking. These studies have greatly increased the power to detect genetic risk factors for nicotine consumption. However, these associated genetic factors explain only a small percentage of the variance in nicotine consumption, indicating that further research to detect other genetic factors influencing smoking is warranted.
IDENTIFICATION OF GENETIC RISK FACTORS FOR ILLICIT DRUG DEPENDENCE
Genome-Wide Linkage Studies
Cannabis is the most widely use illicit drug. A genome-wide linkage analysis of cannabis dependence and related phenotypes in individuals from the UCSF Family Alcoholism Study identified genome-wide significant linkage (LOD score of 3 or higher) for cannabis craving and withdrawal symptoms for regions on human chromosomes 1, 3, 6, 7, and 9; no evidence for linkage with cannabis dependence reached genome-wide significant (38). Loci on human chromosomes 3 (3q21) and 9 (9q34), which are close to the regions linked to cannabis withdrawal in the UCSF study, were also suggested to influence cannabis-dependence symptoms in adolescents who participated in a Colorado Center on Antisocial Drug Dependence study (65). A Native American community study detected genome-wide significant linkage with the severe cannabis use/antisocial subtype on human chromosomes 16 (LOD score of 4.4) and 19 (LOD score of 6.4) (37).
For other illicit drugs, significant linkage peaks have been identified on human chromosomes 9 (a region approximately 40 cM upstream of the region linked with cannabis use) and 12 for cocaine dependence (48), on chromosome 17 at 103.5 cM for a heavy-opioid-use cluster-defined trait (49), and on 14q for DSM-IV opioid dependence (79). Genome-wide linkage analysis of heroin dependence in Han Chinese reported several linkage regions, but none reached genome-wide significance (52). An analysis in families severely affected by alcohol-use disorders reported significant linkage on human chromosome 2 (LOD score of 3.2) with illicit drug dependence (1).
Genome-Wide Association Studies
There have not been many GWAS of illicit-drug-use disorders. A study using 708 DSM-IV cannabis-dependent cases and 2,346 cannabis-exposed nondependent controls from the Study of Addiction: Genetics and Environment data set showed a suggestive association between cannabis dependence and variants in the ANKFN1 gene on human chromosome 17 (4). In a sample of 325 methadone-stabilized, formerly severe heroin addicts and 250 control individuals, Nielsen et al. (100) used a pooled GWAS approach to find variants associated with vulnerability to heroin addiction.
Candidate Gene Studies
Genes involved in dopamine neurotransmission are biologically plausible candidates for association with cocaine dependence because dopamine pathways play a major role in drug reward (70, 78). Genetic association analysis of dopamine receptors and transporter genes found both positive and negative associations (76). These discrepancies may be due to small sample size as well as the complex nature of the phenotype.
OPRM1, which codes for the G protein--coupled mu opioid receptor, is the primary site of action of most opioids. A nonsynonymous SNP in exon 1 of OPRM1, A118G, is the most commonly studied variant for opioid dependence, but its association is controversial. Several studies have reported a positive association between variants in OPRM1 and opiate (including heroin) dependence (11, 20, 81, 120), whereas other studies did not detect an association (101, 138). A study using sensory neurons isolated from a humanized mouse model showed that the A118G missense variant of OPRM1 modulates the morphine and fentanyl pharmacological profile (89). Morphine is approximately fivefold less potent and 26% less efficacious in neurons with the 118GG genotype than it is in neurons with the 118AA genotype. However, there is no difference in the potency and efficacy of the agonist fentanyl in neurons with different genotypes.
Two well-characterized cannabinoid receptors associated with the endocannabinoid signaling system, CB1 (CNR1) and CB2 (CNR2), have been reported to be associated with vulnerability to psychiatric disorders, including substance abuse (130). Studies using CNR1-knockout mice have reported that the mice display alterations in reward- and drug-seeking behaviors in response to psychostimulants, including alcohol (25, 106), nicotine (32, 44), cocaine, and amphetamine (90). The most-studied genetic variant in CNR1 is the (AAT)n tri-nucleotide short-tandem repeat, which was reported to be associated with intravenous administration of drugs of abuse (26). However, other studies have not confirmed this finding (10, 29, 85). Several other variants in CNR1 have been reported to be associated with cannabis dependence (3), cannabis-dependence symptoms (66), cocaine dependence (151), and other substance dependences (63, 115, 150).
Interestingly, the rs16969968 nonsynonymous variant in the α5 nicotinic acetylcholine receptor is also associated with cocaine dependence, but the minor allele reduces the risk for cocaine dependence, which is the opposite of the effect reported for nicotine dependence (53).
SEQUENCING APPROACHES TO IDENTIFY VARIANTS THAT COULD EXPLAIN THE MISSING HERITABILITY FOR SUBSTANCE DEPENDENCE
One drawback of the GWAS method is its reliance on linkage disequilibrium. This means that this approach is good for identifying variants that are common in the general population (>1%) but misses rare variants with larger effects on risk that have low linkage disequilibrium with common variants detected with standard genotyping chips. As a way to uncover the missing heritability factors that influence the risk for psychiatric diseases, including addiction, next-generation DNA sequencing combined with the results of association and perhaps linkage studies holds the promise of identifying a larger set of susceptibility loci (9, 46). In contrast to GWAS, sequencing of targeted genomic regions identified from GWAS or linkage analysis, or whole-exome or whole-genome sequencing, improves the ability to discover novel causative or highly penetrant mutations for human diseases.
Rare variants have been shown to be risk factors for some complex disorders, but their role in psychiatric disorders and especially addiction-related phenotypes is largely unexplored (75). A few studies have shown associations between rare variation in nicotinic receptor genes and nicotine dependence (55, 141, 147).
Several variants in the alcohol metabolism genes (i.e., ADH1B) and nicotine metabolism genes (i.e., CYP2A6) have low frequencies (1%–5% minor allele frequency) and generally reduce risk for dependence, suggesting that human populations might be genetically predisposed to develop addiction, with rare variant alleles leading to reduced risk. This is supported in the nicotine literature, in which most people who smoke develop some symptoms of dependence, whereas only 20% smoke without developing any symptoms of dependence (14). It could be that as a result of some unknown evolutionary selection pressure, most people are predisposed to addiction when exposed to substances.
IMPLICATIONS OF RECENT FINDINGS ON HEALTH BEYOND ADDICTION
Nicotinic Receptors and Lung Cancer and Chronic Obstructive Pulmonary Disease
GWAS approaches have revealed that several SNPs within the nAChR gene cluster are significantly associated with the risk of lung cancer and COPD (5, 68, 118, 123). The most strongly associated SNPs are the same as those that show association with nicotine dependence and CPD in other studies (13, 112, 123, 124a). It is unclear whether the association of this locus with lung cancer is a direct biological effect on lung cancer susceptibility or is mediated through effects on increased risk of smoking. Although SNPs at this locus are only weakly associated with lung cancer risk in those who have never smoked, they are associated with risk for other smoking-associated cancers and diseases (86, 126). This implies that this locus predisposes individuals to increased tobacco consumption, leading to increased risk for cancer.
However, some studies have suggested a direct link between CHRNA5-CHRNA3-CHRNB4 variants and lung cancer: The risk of lung cancer that can be attributed to the CHRNA5-CHRNA3-CHRNB4 variants is higher than can be explained by the variant’s effect on smoking quantity (123), and the genetic risk for lung cancer and COPD remains after the risk associated with smoking has been statistically accounted for using CPD and the duration of smoking (86). Other studies, however, have shown that the amount of nicotine absorbed by smokers is not fully accounted for by CPD owing to differences in how individuals smoke (56, 60).
The α5 nAChR subunit is expressed in lung tissue, and a 30-fold upregulation of expression of CHRNA5 mRNA is seen in lung cancer tissue compared with normal lung tissue (41). In addition, tobacco smoke and nicotine can both mediate the stepwise overexpression of nAChR subtypes, which leads to increased Ca2+ permeability in exposed cells (6). Thus, a switch in the nAChR composition (involving the α3 and α5 subunits, among others) could change receptor function, leading to pathologic effects in nicotine-exposed cells. SNPs in the CHRNA5-CHRNA3-CHRNB4 gene cluster could therefore contribute to increased risk of nicotine dependence and to lung cancer independently and on two levels: (a) by increasing the number of cigarettes smoked and the likelihood of nicotine dependence, and (b) by inserting themselves into the pathophysiological cascade that leads to lung cancer (134).
ADH and ALDH2 Genes and Esophageal Cancer
Variants in ADH1B and ALDH2 that influence alcohol consumption and alcohol dependence also play a role in the risk for upper aero-digestive tract (UADT) cancer. The protein encoded by the ADH1B*2 allele, which is associated with reduced risk for alcohol dependence, has increased enzyme activity. Protein encoded by the ALDH2*2 allele, a protective allele for alcohol dependence, has almost zero enzyme activity (33). Individuals who carry these two alleles have much higher levels of acetaldehyde (a carcinogen) compared with noncarriers if they consume alcohol (Figure 2). Studies have shown that, owing to the accumulation of acetaldehyde in the blood, individuals who have the alcohol flushing response are at higher risk for esophageal cancer (21).
GWAS have also identified a significant association between esophageal squamous cell cancer and SNPs on human chromosomes 4q21–23 and 12q24, which include the functional variants rs1229984 in ADH1B and rs671 in ALDH2, respectively (30). Tanaka et al. (121) demonstrated that the interaction of ADH1B and/or ALDH2 risk alleles with smoking and alcohol consumption significantly increases the risk for the development of esophageal squamous cell carcinoma. Studies have also shown that the combination of alcohol consumption with the inactive heterozygous ALDH2 genotype (ALDH2*1/*2) and less-active homozygous ADH1B genotype (ADH1B*1/*1) increases the risk of UADT squamous cell carcinoma in central European (59) and Japanese (7) populations. The effect of ALDH2*1/*2 results from the high level of acetaldehyde; the effect of ADH1B*1/*1 is due to heavy drinking that leads to longer exposure of the UADT to salivary ethanol and acetaldehyde. These studies point to significant gene-environment interactions that potentially lead toward complex pathophysiological pathways for the development of such diseases.
CONCLUSION
Genomic approaches are beginning to provide clues to the underlying genetic etiology of addiction, and have demonstrated that exposure to these substances in combination with genetic vulnerability to addiction plays an important role in the risk for common cancers previously associated with substance use.
SUMMARY POINTS.
Genetic studies have identified functional alleles in alcohol metabolism genes (ADH, which encodes alcohol dehydrogenase, and ALDHL2, which encodes aldehyde dehydrogenase) that influence the risk for alcohol dependence. The interaction of genetic (polymorphisms of ADH and ALDH2 genes) and environmental (heavy drinking) factors is associated with risk for UADT cancers.
Recent GWAS have successfully identified variants in the α3, α5, and β4 subunits of nAChR associated with risk of nicotine dependence. However, GWAS on alcoholism have not provided conclusive evidence for specific genetic factors for this type of addiction. It is likely that CPD, a common phenotype used in nicotine-dependence GWAS, is a more consistent measurement than other quantitative measures of other substance-use disorders. Harmonized phenotypic measures provide a convenient method for combining small cohorts into a large sample with more than 10,000 subjects, increasing the power to detect statistically significant association.
Although the GWAS approach has been successfully used to investigate the genetic influence on smoking, the association of CHRNA5-CHRNA3-CHRNB4 with nicotine dependence explains only a small part of this addiction’s heritability. A significant fraction of the genetic variance remains unexplained despite the use of very large sample sizes. This missing heritability may be explained by rare variants with large effect. More robust DNA sequencing approaches can potentially identify this missing heritability.
The roles of alcohol metabolism genes in esophageal cancer and nicotinic receptors in lung cancer indicate significant gene-environment interaction in cancer vulnerability.
Acknowledgments
This work is supported by the following projects: the Collaborative Study on the Genetics of Alcoholism, funded by NIH grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism and the National Institute on Drug Abuse (NIDA); the Collaborative Genetic Study of Nicotine Dependence, funded by NIH grant P01 CA89392 from NIDA and the National Cancer Institute; and the Genetics of Vulnerability to Nicotine Addiction, funded by NIH grant 5R01 DA12854 from NIDA.
Footnotes
DISCLOSURE STATEMENT
A.M.G. and J.C.W. are listed as inventors on issued US patent 8,080,371, “Markers for Addiction,” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction.
Contributor Information
Jen-Chyong Wang, Email: jcwang@psychiatry.wustl.edu.
Manav Kapoor, Email: kapoorm@psychiatry.wustl.edu.
Alison M. Goate, Email: goatea@psychiatry.wustl.edu.
LITERATURE CITED
- 1.Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, et al. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008;93:12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103:1069–81. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A, Lynskey MT. Candidate genes for cannabis use disorders: findings, challenges and directions. Addiction. 2009;104:518–32. doi: 10.1111/j.1360-0443.2009.02504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, et al. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol. 2011;16:514–18. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arredondo J, Chernyavsky AI, Grando SA. SLURP-1 and -2 in normal, immortalized and malignant oral keratinocytes. Life Sci. 2007;80:2243–47. doi: 10.1016/j.lfs.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asakage T, Yokoyama A, Haneda T, Yamazaki M, Muto M, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis. 2007;28:865–74. doi: 10.1093/carcin/bgl206. [DOI] [PubMed] [Google Scholar]
- 8.Audrain-McGovern J, Al Koudsi N, Rodriguez D, Wileyto EP, Shields PG, Tyndale RF. The role of CYP2A6 in the emergence of nicotine dependence in adolescents. Pediatrics. 2007;119:e264–74. doi: 10.1542/peds.2006-1583. [DOI] [PubMed] [Google Scholar]
- 9.Avramopoulos D. Genetics of psychiatric disorders methods: molecular approaches. Psychiatr Clin N Am. 2010;33:1–13. doi: 10.1016/j.psc.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballon N, Leroy S, Roy C, Bourdel MC, Charles-Nicolas A, et al. (AAT)n repeat in the cannabinoid receptor gene (CNR1): association with cocaine addiction in an African-Caribbean population. Pharmacogenomics J. 2006;6:126–30. doi: 10.1038/sj.tpj.6500352. [DOI] [PubMed] [Google Scholar]
- 11.Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, et al. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–49. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benowitz NL, Jacob P., III Metabolism of nicotine to cotinine studied by a dual stable isotope method. Clin Pharmacol Ther. 1994;56:483–93. doi: 10.1038/clpt.1994.169. [DOI] [PubMed] [Google Scholar]
- 13.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, et al. α-5/α-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–87. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, et al. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55:982–88. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 16.Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2011 Oct 4; doi: 10.1038/mp.2011.124. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165:1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, et al. ADH single nucleotide polymorphism associations with alcohol metabolism in vivo. Hum Mol Genet. 2009;18:1533–42. doi: 10.1093/hmg/ddp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloom J, Hinrichs AL, Wang JC, von Weymarn LB, Kharasch ED, et al. The contribution of common CYP2A6 alleles to variation in nicotine metabolism among European-Americans. Pharmacogenet Genomics. 2011;21:403–16. doi: 10.1097/FPC.0b013e328346e8c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bond C, LaForge KS, Tian M, Melia D, Zhang S, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters β-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–13. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks PJ, Goldman D, Li TK. Alleles of alcohol and acetaldehyde metabolism genes modulate susceptibility to oesophageal cancer from alcohol consumption. Hum Genomics. 2009;3:103–5. doi: 10.1186/1479-7364-3-2-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cent. Dis. Control Prev. Smoking-attributable mortality, years of potential life lost, and productivity losses---United States, 2000–2004. Morb Mortal Wkly Rep. 2010;57:1226–28. [PubMed] [Google Scholar]
- 23.Chen LS, Johnson EO, Breslau N, Hatsukami D, Saccone NL, et al. Interplay of genetic risk factors and parent monitoring in risk for nicotine dependence. Addiction. 2009;104:1731–40. doi: 10.1111/j.1360-0443.2009.02697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi IG, Son HG, Yang BH, Kim SH, Lee JS, et al. Scanning of genetic effects of alcohol metabolism gene (ADH1B and ADH1C) polymorphisms on the risk of alcoholism. Hum Mutat. 2005;26:224–34. doi: 10.1002/humu.20209. [DOI] [PubMed] [Google Scholar]
- 25a.Colombo G, Serra S, Vacca G, Carai MA, Gessa GL. Endocannabinoid system and alcohol addiction: pharmacological studies. Pharmacology, biochemistry, and behavior. 2005;81:369–80. doi: 10.1016/j.pbb.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 26.Comings DE, Muhleman D, Gade R, Johnson P, Verde R, et al. Cannabinoid receptor gene (CNR1): association with IV drug use. Mol Psychiatry. 1997;2:161–68. doi: 10.1038/sj.mp.4000247. [DOI] [PubMed] [Google Scholar]
- 27.Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B. 2004;129B:104–9. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 28.Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5′-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–48. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Covault J, Gelernter J, Kranzler H. Association study of cannabinoid receptor gene (CNR1) alleles and drug dependence. Mol Psychiatry. 2001;6:501–2. doi: 10.1038/sj.mp.4000925. [DOI] [PubMed] [Google Scholar]
- 30.Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137:1768–75. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 31.Degenhardt L, Hall W. Extent of illicit drug use and dependence, and their contribution to the global burden of disease. Lancet. 2012;379:55–70. doi: 10.1016/S0140-6736(11)61138-0. [DOI] [PubMed] [Google Scholar]
- 32.De Vries TJ, de Vries W, Janssen MC, Schoffelmeer AN. Suppression of conditioned nicotine and sucrose seeking by the cannabinoid-1 receptor antagonist SR141716A. Behav Brain Res. 2005;161:164–68. doi: 10.1016/j.bbr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 33.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 34.Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, et al. Variations in GABRA2, encoding the α 2 subunit of the GABAA receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–52. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–49. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 37.Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. Addict Biol. 2009;14:338–48. doi: 10.1111/j.1369-1600.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ehlers CL, Gizer IR, Vieten C, Wilhelmsen KC. Linkage analyses of cannabis dependence, craving, and withdrawal in the San Francisco family study. Am J Med Genet B. 2010;153B:802–11. doi: 10.1002/ajmg.b.31050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enoch MA. The role of GABAA receptors in the development of alcoholism. Pharmacol Biochem Behav. 2008;90:95–104. doi: 10.1016/j.pbb.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Enoch MA, Schwartz L, Albaugh B, Virkkunen M, Goldman D. Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. Am J Med Genet B. 2006;141B:599–607. doi: 10.1002/ajmg.b.30336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falvella FS, Galvan A, Colombo F, Frullanti E, Pastorino U, Dragani TA. Promoter polymorphisms and transcript levels of nicotinic receptor CHRNA5. J Natl Cancer Inst. 2010;102:1366–70. doi: 10.1093/jnci/djq264. [DOI] [PubMed] [Google Scholar]
- 42.Farres J, Moreno A, Crosas B, Peralba JM, Allali-Hassani A, et al. Alcohol dehydrogenase of class IV (σσ-ADH) from human stomach. cDNA sequence and structure/function relationships. Eur J Biochem. 1994;224:549–57. doi: 10.1111/j.1432-1033.1994.00549.x. [DOI] [PubMed] [Google Scholar]
- 43.Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, et al. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- 44.Forget B, Hamon M, Thiebot MH. Cannabinoid CB1 receptors are involved in motivational effects of nicotine in rats. Psychopharmacology. 2005;181:722–34. doi: 10.1007/s00213-005-0015-6. [DOI] [PubMed] [Google Scholar]
- 45.Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–45. [PubMed] [Google Scholar]
- 46.Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, et al. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–15. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B. 2005;136B:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- 49.Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–69. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gizer IR, Ehlers CL, Vieten C, Seaton-Smith KL, Feiler HS, et al. Linkage scan of alcohol dependence in the UCSF Family Alcoholism Study. Drug Alcohol Depend. 2011;113:125–32. doi: 10.1016/j.drugalcdep.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gizer IR, Ehlers CL, Vieten C, Seaton-Smith KL, Feiler HS, et al. Linkage scan of nicotine dependence in the University of California, San Francisco (UCSF) Family Alcoholism Study. Psychol Med. 2011;41:799–808. doi: 10.1017/S0033291710001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glatt SJ, Lasky-Su JA, Zhu SC, Zhang R, Zhang B, et al. Genome-wide linkage analysis of heroin dependence in Han Chinese: results from wave two of a multi-stage study. Drug Alcohol Depend. 2008;98:30–34. doi: 10.1016/j.drugalcdep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grucza RA, Wang JC, Stitzel JA, Hinrichs AL, Saccone SF, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64:922–29. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guze SB, Cloninger CR, Martin R, Clayton PJ. Alcoholism as a medical disorder. Compr Psychiatry. 1986;27:501–10. doi: 10.1016/0010-440x(86)90054-4. [DOI] [PubMed] [Google Scholar]
- 55.Haller G, Druley T, Vallania FL, Mitra RD, Li P, et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum Mol Genet. 2012 doi: 10.1093/hmg/ddr498. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammond D, Fong GT, Cummings KM, Hyland A. Smoking topography, brand switching, and nicotine delivery: results from an in vivo study. Cancer Epidemiol Biomark Prev. 2005;14:1370–75. doi: 10.1158/1055-9965.EPI-04-0498. [DOI] [PubMed] [Google Scholar]
- 57.Hanchar HJ, Chutsrinopkun P, Meera P, Supavilai P, Sieghart W, et al. Ethanol potently and competitively inhibits binding of the alcohol antagonist Ro15-4513 to α4/6β3δ GABAA receptors. Proc Natl Acad Sci USA. 2006;103:8546–51. doi: 10.1073/pnas.0509903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hansell NK, Agrawal A, Whitfield JB, Morley KI, Gordon SD, et al. Linkage analysis of alcohol dependence symptoms in the community. Alcohol Clin Exp Res. 2010;34:158–63. doi: 10.1111/j.1530-0277.2009.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashibe M, Morgenstern H, Cui Y, Tashkin DP, Zhang ZF, et al. Marijuana use and the risk of lung and upper aerodigestive tract cancers: results of a population-based case-control study. Cancer Epidemiol Biomark Prev. 2006;15:1829–34. doi: 10.1158/1055-9965.EPI-06-0330. [DOI] [PubMed] [Google Scholar]
- 60.Hatsukami DK, Morgan SF, Pickens RW, Champagne SE. Situational factors in cigarette smoking. Addict Behav. 1990;15:1–12. doi: 10.1016/0306-4603(90)90002-f. [DOI] [PubMed] [Google Scholar]
- 61.Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, et al. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry. 2011;70:513–18. doi: 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henningfield JE, Miyasato K, Jasinski DR. Abuse liability and pharmacodynamic characteristics of intravenous and inhaled nicotine. J Pharmacol Exp Ther. 1985;234:1–12. [PubMed] [Google Scholar]
- 63.Herman AI, Kranzler HR, Cubells JF, Gelernter J, Covault J. Association study of the CNR1 gene exon 3 alternative promoter region polymorphisms and substance dependence. Am J Med Genet B. 2006;141B:499–503. doi: 10.1002/ajmg.b.30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho MK, Tyndale RF. Overview of the pharmacogenomics of cigarette smoking. Pharmacogenomics J. 2007;7:81–98. doi: 10.1038/sj.tpj.6500436. [DOI] [PubMed] [Google Scholar]
- 65.Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, et al. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend. 2007;89:34–41. doi: 10.1016/j.drugalcdep.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopfer CJ, Young SE, Purcell S, Crowley TJ, Stallings MC, et al. Cannabis receptor haplotype associated with fewer cannabis dependence symptoms in adolescents. Am J Med Genet B. 2006;141B:895–901. doi: 10.1002/ajmg.b.30378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hukkanen J, Jacob P, III, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 68.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–37. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 69.Hurley TD, Perez-Miller S, Breen H. Order and disorder in mitochondrial aldehyde dehydrogenase. Chem-Biol Interact. 2001;130–32:3–14. doi: 10.1016/s0009-2797(00)00217-9. [DOI] [PubMed] [Google Scholar]
- 70.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–98. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 71.Ittiwut C, Listman J, Mutirangura A, Malison R, Covault J, et al. Interpopulation linkage disequilibrium patterns of GABRA2 and GABRG1 genes at the GABA cluster locus on human chromosome 4. Genomics. 2008;91:61–69. doi: 10.1016/j.ygeno.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kareken DA, Liang T, Wetherill L, Dzemidzic M, Bragulat V, et al. A polymorphism in GABRA2 is associated with the medial frontal response to alcohol cues in an fMRI study. Alcohol Clin Exp Res. 2010;34:2169–78. doi: 10.1111/j.1530-0277.2010.01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, et al. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res. 2011;35:963–75. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kendler KS, Schmitt E, Aggen SH, Prescott CA. Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch Gen Psychiatry. 2008;65:674–82. doi: 10.1001/archpsyc.65.6.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knight HM, Pickard BS, Maclean A, Malloy MP, Soares DC, et al. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression. Am J Hum Genet. 2009;85:833–46. doi: 10.1016/j.ajhg.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- 77.Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, et al. Association of ADH and ALDH genes with alcohol dependence in the Irish Affected Sib Pair Study of alcohol dependence (IASPSAD) sample. Alcohol Clin Exp Res. 2008;32:785–95. doi: 10.1111/j.1530-0277.2008.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lachman HM. An overview of the genetics of substance use disorders. Curr Psychiatry Rep. 2006;8:133–43. doi: 10.1007/s11920-006-0013-3. [DOI] [PubMed] [Google Scholar]
- 79.Lachman HM, Fann CS, Bartzis M, Evgrafov OV, Rosenthal RN, et al. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum Mol Genet. 2007;16:1327–34. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- 80.Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, et al. Association between alcoholism and γ-amino butyric acid α2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–98. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- 81.Levran O, Awolesi O, Linzy S, Adelson M, Kreek MJ. Haplotype block structure of the genomic region of the mu opioid receptor gene. J Hum Genet. 2011;56:147–55. doi: 10.1038/jhg.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li MD. The genetics of nicotine dependence. Curr Psychiatry Rep. 2006;8:158–64. doi: 10.1007/s11920-006-0016-0. [DOI] [PubMed] [Google Scholar]
- 83.Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10:225–31. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li MD, Yoon D, Lee JY, Han BG, Niu T, et al. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS ONE. 2010;5:e12183. doi: 10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li T, Liu X, Zhu ZH, Zhao J, Hu X, et al. No association between (AAT)n repeats in the cannabinoid receptor gene (CNR1) and heroin abuse in a Chinese population. Mol Psychiatry. 2000;5:128–30. doi: 10.1038/sj.mp.4000670. [DOI] [PubMed] [Google Scholar]
- 86.Lips EH, Gaborieau V, McKay JD, Chabrier A, Hung RJ, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int J Epidemiol. 2010;39:563–77. doi: 10.1093/ije/dyp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–21. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 89.Mahmoud S, Thorsell A, Sommer WH, Heilig M, Holgate JK, et al. Pharmacological consequence of the A118G μ opioid receptor polymorphism on morphine- and fentanyl-mediated modulation of Ca2+ channels in humanized mouse sensory neurons. Anesthesiology. 2011;115:1054–62. doi: 10.1097/ALN.0b013e318231fc11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maldonado RV, Berrendero OF. Involvement of the endocannabinoid system in drug addiction. Trends Neurosci. 2006;29:225–32. doi: 10.1016/j.tins.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 91.Merikangas KR, Stolar M, Stevens DE, Goulet J, Preisig MA, et al. Familial transmission of substance use disorders. Arch Gen Psychiatry. 1998;55:973–79. doi: 10.1001/archpsyc.55.11.973. [DOI] [PubMed] [Google Scholar]
- 92.Messina ES, Tyndale RF, Sellers EM. A major role for CYP2A6 in nicotine C-oxidation by human liver microsomes. J Pharmacol Exp Ther. 1997;282:1608–14. [PubMed] [Google Scholar]
- 93.Minematsu N, Nakamura H, Furuuchi M, Nakajima T, Takahashi S, et al. Limitation of cigarette consumption by CYP2A6*4, *7 and *9 polymorphisms. Eur Respir J. 2006;27:289–92. doi: 10.1183/09031936.06.00056305. [DOI] [PubMed] [Google Scholar]
- 94.Mizoi Y, Yamamoto K, Ueno Y, Fukunaga T, Harada S. Involvement of genetic polymorphism of alcohol and aldehyde dehydrogenases in individual variation of alcohol metabolism. Alcohol Alcohol. 1994;29:707–10. [PubMed] [Google Scholar]
- 95.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–45. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 96.Moore S, Montane-Jaime LK, Carr LG, Ehlers CL. Variations in alcohol-metabolizing enzymes in people of East Indian and African descent from Trinidad and Tobago. Alcohol Res Health. 2007;30:28–30. [PMC free article] [PubMed] [Google Scholar]
- 97.Mwenifumbo JC, Sellers EM, Tyndale RF. Nicotine metabolism and CYP2A6 activity in a population of black African descent: impact of gender and light smoking. Drug Alcohol Depend. 2007;89:24–33. doi: 10.1016/j.drugalcdep.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 98.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, et al. Characterization of CYP2A6 involved in 3′-hydroxylation of cotinine in human liver microsomes. J Pharmacol Exp Ther. 1996;277:1010–15. [PubMed] [Google Scholar]
- 99.Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, et al. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–17. [PubMed] [Google Scholar]
- 100.Nielsen DA, Ji F, Yuferov V, Ho A, He C, et al. Genome-wide association study identifies genes that may contribute to risk for developing heroin addiction. Psychiatr Genet. 2010;20:207–14. doi: 10.1097/YPG.0b013e32833a2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nikolov MA, Beltcheva O, Galabova A, Ljubenova A, Jankova E, et al. No evidence of association between 118A>G OPRM1 polymorphism and heroin dependence in a large Bulgarian case-control sample. Drug Alcohol Depend. 2011;117:62–65. doi: 10.1016/j.drugalcdep.2010.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oota H, Pakstis AJ, Bonne-Tamir B, Goldman D, Grigorenko E, et al. The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination. Ann Hum Genet. 2004;68:93–109. doi: 10.1046/j.1529-8817.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 103.Osier MV, Pakstis AJ, Kidd JR, Lee JF, Yin SJ, et al. Linkage disequilibrium at the ADH2 and ADH3 loci and risk of alcoholism. Am J Hum Genet. 1999;64:1147–57. doi: 10.1086/302317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002;71:84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palmer RH, Young SE, Hopfer CJ, Corley RP, Stallings MC, et al. Developmental epidemiology of drug use and abuse in adolescence and young adulthood: evidence of generalized risk. Drug Alcohol Depend. 2009;102:78–87. doi: 10.1016/j.drugalcdep.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Racz I, Bilkei-Gorzo A, Toth ZE, Michel K, Palkovits M, Zimmer A. A critical role for the cannabinoid CB1 receptors in alcohol dependence and stress-stimulated ethanol drinking. J Neurosci. 2003;23:2453–58. doi: 10.1523/JNEUROSCI.23-06-02453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ray LA, Hutchison KE. Associations among GABRG1, level of response to alcohol, and drinking behaviors. Alcohol Clin Exp Res. 2009;33:1382–90. doi: 10.1111/j.1530-0277.2009.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–15. [PubMed] [Google Scholar]
- 109.Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch Gen Psychiatry. 2003;60:1256–64. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- 110.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–17. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 111.Roh S, Matsushita S, Hara S, Maesato H, Matsui T, et al. Role of GABRA2 in moderating subjective responses to alcohol. Alcohol Clin Exp Res. 2011;35:400–7. doi: 10.1111/j.1530-0277.2010.01357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B. 2009;150B:453–66. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schmidt LG, Samochowiec J, Finckh U, Fiszer-Piosik E, Horodnicki J, et al. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend. 2002;65:221–24. doi: 10.1016/s0376-8716(01)00164-8. [DOI] [PubMed] [Google Scholar]
- 116.Schoedel KA, Hoffmann EB, Rao Y, Sellers EM, Tyndale RF. Ethnic variation in CYP2A6 and association of genetically slow nicotine metabolism and smoking in adult Caucasians. Pharmacogenetics. 2004;14:615–26. doi: 10.1097/00008571-200409000-00006. [DOI] [PubMed] [Google Scholar]
- 117.Schuckit MA. An overview of genetic influences in alcoholism. J Subst Abuse Treat. 2009;36:S5–14. [PubMed] [Google Scholar]
- 118.Shiraishi K, Kohno T, Kunitoh H, Watanabe S, Goto K, et al. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65–70. doi: 10.1093/carcin/bgn257. [DOI] [PubMed] [Google Scholar]
- 119.Soyka M, Preuss UW, Hesselbrock V, Zill P, Koller G, Bondy B. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 2008;42:184–91. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 119a.Subst. Abuse Mental Health Serv. Adm. (SAMHSA) NSDUH Ser. H-41, HHS Publ. (SMA) 11-4658. SAMHSA; Rockville, MD: 2011. Results from the 2010 national survey on drug use and health: summary of national findings. [Google Scholar]
- 120.Tan EC, Tan CH, Karupathivan U, Yap EP. Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport. 2003;14:569–72. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
- 121.Tanaka F, Yamamoto K, Suzuki S, Inoue H, Tsurumaru M, et al. Strong interaction between the effects of alcohol consumption and smoking on oesophageal squamous cell carcinoma among individuals with ADH1B and/or ALDH2 risk alleles. Gut. 2010;59:1457–64. doi: 10.1136/gut.2009.205724. [DOI] [PubMed] [Google Scholar]
- 123.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124a.Tob. Genet. Consort. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–47. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–84. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Truong T, Hung RJ, Amos CI, Wu X, Bickeboller H, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102:959–71. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: what we have learned. Harv Rev Psychiatry. 2001;9:267–79. [PubMed] [Google Scholar]
- 128.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–72. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 129.US Dep. Health Hum. Serv. Rep Surg Gen. US Dep. Health Hum. Serv., Cent. Dis. Control Prev., Natl. Cent. Chronic Dis. Prev. Health Promot., Off. Smok. Health; Atlanta, GA: 2010. How tobacco smoke causes disease: the biology and behavioral basis for smoking-attributable disease. [Google Scholar]
- 130.van der Stelt M, Di Marzo V. The endocannabinoid system in the basal ganglia and in the mesolimbic reward system: implications for neurological and psychiatric disorders. Eur J Pharmacol. 2003;480:133–50. doi: 10.1016/j.ejphar.2003.08.101. [DOI] [PubMed] [Google Scholar]
- 131.van der Zwaluw CS, Engels RC. Gene-environment interactions and alcohol use and dependence: current status and future challenges. Addiction. 2009;104:907–14. doi: 10.1111/j.1360-0443.2009.02563.x. [DOI] [PubMed] [Google Scholar]
- 132.Verweij KJ, Zietsch BP, Lynskey MT, Medland SE, Neale MC, et al. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105:417–30. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- 134.Volkow N, Rutter J, Pollock JD, Shurtleff D, Baler R. One SNP linked to two diseases---addiction and cancer: a double whammy? Nicotine addiction and lung cancer susceptibility. Mol Psychiatry. 2008;13:990–92. doi: 10.1038/mp.2008.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Volkow ND, Wang GJ, Fowler JS, Tomasi D. Addiction circuitry in the human brain. Annu Rev Pharmacol Toxicol. 2012;52:321–36. doi: 10.1146/annurev-pharmtox-010611-134625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wall TL, Carr LG, Ehlers CL. Protective association of genetic variation in alcohol dehydrogenase with alcohol dependence in Native American Mission Indians. Am J Psychiatry. 2003;160:41–46. doi: 10.1176/appi.ajp.160.1.41. [DOI] [PubMed] [Google Scholar]
- 137.Wallner M, Olsen RW. Physiology and pharmacology of alcohol: the imidazobenzodiazepine alcohol antagonist site on subtypes of GABAA receptors as an opportunity for drug development? Br J Pharmacol. 2008;154:288–98. doi: 10.1038/bjp.2008.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain. 2009;146:270–75. doi: 10.1016/j.pain.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 139.Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18:3125–35. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139a.Wang JC, Grucza R, Cruchaga C, Hinrichs A, Bertelsen S, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4:e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140a.Wellcome Trust Case Control Consort. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wessel J, McDonald SM, Hinds DA, Stokowski RP, Javitz HS, et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerström test for nicotine dependence. Neuropsychopharmacology. 2010;35:2392–402. doi: 10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Whitfield JB. Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet. 2002;71:1247–50. doi: 10.1086/344287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.World Health Organ. 2003 [Google Scholar]
- 144.World Health Organ. The global burden. 2010. [Google Scholar]
- 146.Wu C, Hu Z, Yu D, Huang L, Jin G, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–72. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- 147.Xie P, Kranzler HR, Krauthammer M, Cosgrove KP, Oslin D, et al. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biol Psychiatry. 2011;70:528–36. doi: 10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Xie P, Kranzler HR, Zhang H, Oslin D, Anton RF, et al. Childhood adversity increases risk for nicotine dependence and interacts with α5 nicotinic acetylcholine receptor genotype specifically in males. Neuropsychopharmacology. 2012;37:669–76. doi: 10.1038/npp.2011.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoshida A, Huang IY, Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc Natl Acad Sci USA. 1984;81:258–61. doi: 10.1073/pnas.81.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J. CNR1 variation modulates risk for drug and alcohol dependence. Biol Psychiatry. 2007;62:616–26. doi: 10.1016/j.biopsych.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 151.Zuo L, Kranzler HR, Luo X, Yang BZ, Weiss R, et al. Interaction between two independent CNR1 variants increases risk for cocaine dependence in European Americans: a replication study in family-based sample and population-based sample. Neuropsychopharmacology. 2009;34:1504–13. doi: 10.1038/npp.2008.206. [DOI] [PMC free article] [PubMed] [Google Scholar]