Abstract
How is a long strand of genomic DNA packaged into a mitotic chromosome or nucleus? The nucleosome fiber (beads-on-a-string), in which DNA is wrapped around core histones, has long been assumed to be folded into a 30-nm chromatin fiber, and a further helically folded larger fiber. However, when frozen hydrated human mitotic cells were observed using cryoelectron microscopy, no higher-order structures that included 30-nm chromatin fibers were found. To investigate the bulk structure of mitotic chromosomes further, we performed small-angle X-ray scattering (SAXS), which can detect periodic structures in noncrystalline materials in solution. The results were striking: no structural feature larger than 11 nm was detected, even at a chromosome-diameter scale (~1 μm). We also found a similar scattering pattern in interphase nuclei of HeLa cells in the range up to ~275 nm. Our findings suggest a common structural feature in interphase and mitotic chromatins: compact and irregular folding of nucleosome fibers occurs without a 30-nm chromatin structure.
Keywords: 30-nm chromatin fiber, chromatin, cryo-EM, fractal nature, interphase nuclei, irregular folding, mitotic chromosomes, X-ray scattering
Introduction
In current molecular biology textbooks, initial DNA packaging and organization into a chromosome or nucleus is often depicted as shown in Figure 1 (e.g., ref. 1). The current assumption is that DNA is wrapped around histones, forming “nucleosomes” (beads-on-a-string), followed by nucleosome folding into a 30-nm chromatin fiber (e.g., refs. 2–5). The famous "hierarchical helical folding" model asserts that the 30-nm chromatin fiber is folded progressively into larger fibers (i.e., ~100-nm and then ~200-nm fibers) to form the final mitotic chromosomes.6-9 A similar hierarchy is thought to exist in interphase nuclei (e.g., ref. 10).

Figure 1. In the textbook model, a long DNA molecule is wrapped around a basic core histone octamer that consists of H2A, H2B, H3 and H4 histone proteins, and forms a nucleosome with a diameter of 11 nm. The nucleosome has long been assumed to be folded into 30-nm chromatin fibers before the higher-order organization of mitotic chromosomes or interphase nuclei occurs. We show a typical one-start helix model between two well-known structural models for 30-nm chromatin fibers: one-start helix (solenoid) and two-start helix (zigzag). The images are reproduced with minor modifications fromreference 60 with permission from Elsevier.
To visualize mitotic chromosomes in a close-to-intact state, we performed cryoelectron microscopy (cryo-EM), in collaboration with Eltsov, Frangakis and Dubochet.11 For cryo-EM, mitotic HeLa cells were collected and frozen by high-pressure freezing. Thin sections were then prepared at low temperature and observed. Cryo-EM and subsequent computational image processing did not reveal 30-nm chromatin structures in the mitotic chromosomes, but rather a uniform disordered texture, strongly arguing against the current established hypothesis11(also see refs. 12 and 13). However, cryo-EM observations were limited to examining a portion of a chromosome because the section thickness was only ~70 nm, which may have prevented observation of hierarchical regular structures in the chromosomes.
Small-Angle X-ray Scattering (SAXS)
To investigate the bulk structure of mitotic chromosomes in solution, we performed small-angle X-ray scattering (SAXS). When X-rays are used to irradiate non-crystalline materials, scattering at small angles generally reveals periodic structures within samples (Fig. 2A) (e.g., ref. 14). A typical scattering pattern of SAXS composed of concentric rings is shown in Figure 2C. The signals at smaller angles (closer to the center) reflect larger periodic structures and vice versa.14
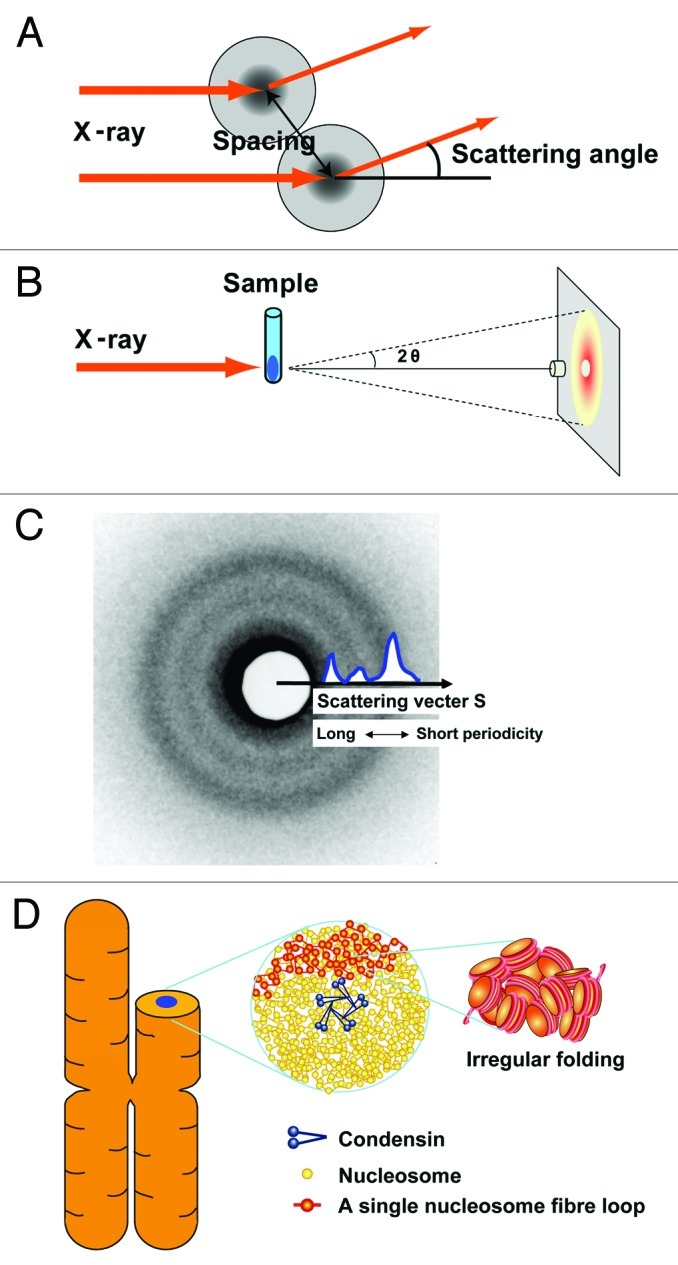
Figure 2. (A) When non-crystal materials are irradiated with X-rays, small-angle scattering generally reflects the size and spacing of internal structures. (B) Experimental setting: a chromosome pellet in a quartz capillary tube was exposed to a synchrotron X-ray beam and the scattering patterns were recorded with a CCD camera or imaging plate. The details of the setting are described in Nishino et al.15 (C) A typical scattering pattern is composed of concentric rings. The signals at smaller scattering angles [smaller size of the scattering vector (S) closer to the center] reflect larger periodic structures and vice versa. In Figure 3, the singles on the concentric rings are averaged and shown in a one-dimensional plot. The size of the scattering vector is defined by S = 2sin(θ)/λ, where λ is the wavelength and 2θ is the scattering angle. A periodic length is given by the inverse of S. Thus, "30-nm peak" refers to a scattering peak at S = 0.033 nm–1. (D) Chromosomes consist of irregularly folded nucleosome (beads on a string) fibers. Condensins (blue) hold the nucleosome fibers (red) globally around the chromosome center. Locally, the nucleosome fiber is folded in an irregular or disordered manner, forming loop structures that are collapsed toward the chromosome center (blue). The collapsed fiber (red) forms a domain that could be compatible with the large module observed by the Belmont group.8
At the SPring-8 synchrotron radiation facility in Japan, isolated human chromosomes at the bottom of a glass capillary were exposed to a synchrotron X-ray beam and the scattering patterns were recorded (Fig. 2B).15 Note that the chromosomes were not fixed or dehydrated to avoid possible artifacts caused by such treatments.16,17 The typical scattering pattern of mitotic chromosomes showed three peaks at 6, 11 and 30 nm,15 consistent with previous experiments by Langmore and Paulson.18,19 The 6- and 11-nm peaks are believed to come from edge-to-edge and face-to-face positioning of nucleosomes, respectively.18-20 The 30-nm peak is assumed to represent the side-by-side packing of 30-nm chromatin fibers,18-20 which has long been regarded as strong evidence for the existence of these fibers in chromosomes. However, based on cryo-EM, these 30-nm structures are not apparent in mitotic chromosomes.11-13
Mitotic Chromosome Organization
To examine the nature of the 30-nm peak observed by SAXS, the isolated chromosomes were examined by cryo-EM. As noted above, no 30-nm chromatin structures were observed inside the chromosomes.15 However, the cryo-EM images unexpectedly showed that the chromosome surface was coated with electron-dense granules the size of ribosomes.15 Western blotting and immunostaining with specific antibodies confirmed that there were contaminating ribosomes on the chromosome surface.15 The ribosomes were regularly stacked at ~30-nm spacing.15
We removed the ribosomes from the chromosome surface by washing with an isotonic buffer containing polyamine and EDTA (buffer A; see refs. 19, 21 and 22),15 while maintaining the size and shape of the chromosomes.15 Strikingly, in the chromosomes, no 30-nm peaks were detected by SAXS,15 although other peaks coming from an internal structure of nucleosomes remained.
Previous reports have suggested that chicken erythrocyte nuclei, which are almost completely transcriptionally silenced, contain 30-nm chromatin fibers23,24; therefore, these were used as positive controls. SAXS and cryo-EM analyses demonstrated 30-nm chromatin fiber structures in chicken erythrocyte nuclei, which lacked ribosomes.15 Therefore, we should have detected 30-nm fibers in the human mitotic chromosomes if they were present. We concluded from the combined SAXS and cryo-EM data that regularly folded 30-nm chromatin fibers are not present in human mitotic chromosomes.15
Next, we investigated an entire region of mitotic chromosomes using a newly developed apparatus for ultrasmall-angle X-ray scattering (USAXS).25 Again, we found no regular periodic structures between 50 and 1,000 nm.15 The cryo-EM, SAXS and USAXS data collectively indicate that irregular folding of nucleosome fibers is the bulk structure of human mitotic chromosomes.15
We then considered how the nucleosome fiber is organized into a mitotic chromosome. Because condensin26 and topoisomerase IIα,27 which are essential for chromosome condensation, form an axis in the chromosome22,28-32 (Fig. 2D), we assumed that they globally secure the nucleosome fibers around the chromosomal center. Locally, a nucleosome fiber is folded in an irregular manner toward the chromosome center (Fig. 2D). In addition, a genomic site is rather randomly incorporated into a wide, but not reproducibly specific, region in the chromosome during the condensation process. This view is consistent with the observation by the Belmont group that three-dimensional positions of GFP-labeled genomic loci showed intrinsic variability between sister chromatids in the chromosome.33
Interphase Chromatin Organization
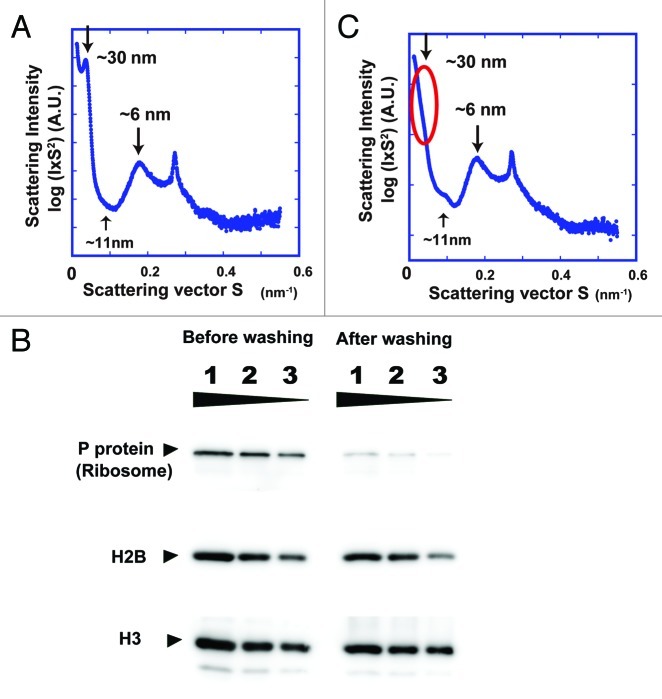
To examine the bulk periodic structure of interphase chromatin, we next focused on interphase nuclei of HeLa cells. Langmore and Paulson reported SAXS peaks at ~30, ~11 and ~6 nm in the nuclei of HeLa cells or mouse lymphocytes.18,19 These authors suggested that the 30-nm peak represents the side-by-side packing of 30-nm chromatin fibers,18,19 and this has long been considered strong evidence for the existence of these fibers in interphase chromatin. Consistent with their data, we also detected similar peaks at ~30, ~11 and ~6 nm in the HeLa nuclei, which were isolated using their procedure (Fig. 3A). Because we detected ribosome components in the nuclei sample by western blotting (Fig. 3B), in analogy with the case of the mitotic chromosomes, we again removed them by washing the nuclei with buffer A.19,21,22 This treatment removed most of the ribosome aggregates from the nuclear fractions (Fig. 3B). As expected, the 30-nm peak in the SAXS pattern disappeared almost completely (Fig. 3C), again suggesting the absence of the 30-nm structure. Note that the remaining peaks at larger angles, which came mainly from the internal structures of the nucleosomes, were unchanged (Fig. 3A and C). Similar to the case for mitotic chromosomes, the scattering profile of interphase chromatin also showed that the ~6-nm peak (face-to-face positioning) predominated over the ~11-nm peak (edge-to-edge positioning) (Fig. 3C). As formation of the 30-nm chromatin fiber requires similar frequencies of face-to-face (~6-nm peak) and edge-to-edge (~11-nm peak) positioning,15 the scattering profile in Figure 3C also supports the near absence of regular 30-nm chromatin fibers in interphase chromatins.
Figure 3. (A) SAXS profile of HeLa interphase nuclei, which were isolated using the Langmore and Paulson procedure.18,19 Three peaks at ~6, ~11 (faint) and ~30 nm were detected (arrows). In the plot of log(I × S2) vs. S, I is the measured average intensity and S is the size of the scattering vector, the inverse of the structure size or spacing (for details, see ref. 15). The 6- and 11-nm peaks are thought to come from edge-to-edge and face-to-face positioning of the nucleosomes, respectively. The 30-nm peak was assumed to represent the side-by-side packing of 30-nm chromatin fibers. (B) The presence and removal of ribosomes in the HeLa interphase nuclei were verified by western blotting. Nuclear lysates of ~2 × 104, 1 × 104, and 5 × 103 cells were loaded into lanes 1–3, respectively. Proteins were separated by SDS-PAGE and then transferred to membranes. Ribosomal P-protein64 and histone H2B (control) and histone H3 (control) were detected using specific antibodies (H2B, Upstate 07–371; H3 Abcom ab1791). P-protein was detected in the original nuclei, but much less in the nuclei after washing, whereas histones H2B and H3 were similarly observed in both nuclei. (C) Only the 30-nm peak disappeared after removal of ribosome aggregates, whereas the other peaks remained.
Consistent with our finding, interphase nuclei in most higher eukaryote cell types examined by cryo-EM reportedly contain no regular 30-nm chromatin fiber (e.g., refs. 16, 34 and 35). Using a combination of a chromosome conformation capture (3C) technique and polymer modeling, Dekker also found that the chromatin in a transcriptionally active domain in yeast did not form a compact 30-nm fiber, but instead was extended with a rather loose arrangement of nucleosomes.36 More recently, the Bazett-Jones group made a similar observation by electron-spectroscopic imaging (ESI), providing phosphorus and nitrogen mapping with sufficient contrast and resolution to visualize 10-nm nucleosome fibers.37,38 Although the cryo-EM and ESI, which are both EM-based methods, examine only a limited portion of nuclei, our SAXS study corroborates the notion that 30-nm chromatin fibers are absent in interphase chromatin.
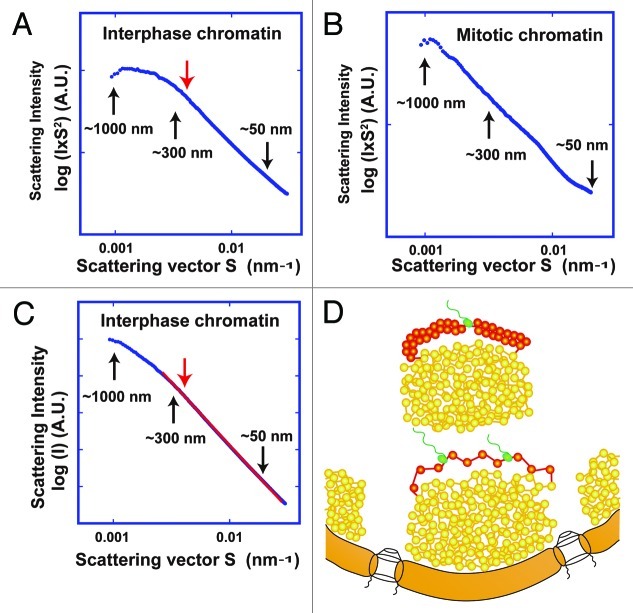
Furthermore, using the USAXS apparatus, we observed no periodic structures in the interphase chromatins in the range between ~50 and ~1,000 nm (Fig. 4A). Taken together with the SAXS data, we suggest that the bulk interphase chromatin also consists of irregularly folded nucleosome fibers (Fig. 4D).
Figure 4. (A) By ultrasmall-angle X-ray scattering (USAXS), no notable structures ~100–150 or 200–250 nm were detected in HeLa interphase nuclei. Beyond the ~275 nm range (red arrow), the slope in interphase chromatin decreased in magnitude. (B) For comparison, a USAXS profile of mitotic chromosomes is reproduced from Nishino et al.15 (C) The scattering intensity obeys the power law with respect to structure size or spacing. A plot of log (I) vs. log (S) on a straight line (red line) covers a wide range, extending over nearly three orders of magnitude. Least-squares fitting shows that I is proportional to S to the power of –3.36 (R = 0.999),39 suggesting that chromosomes do not possess notable regular structures over a very wide scale and exhibit a fractal nature of genome organization (see also ref. 15). (D) In the interphase nucleus, there are numerous compact chromatin domains like “chromatin liquid drops” (yellow balls).60 Red, transcribed nucleosome; Green, RNA polymerase and RNA. Formation of a 30-nm chromatin fiber might occur when nucleosome fibers are looped out from the chromatin domain or chromosome territory for transcription (top, see text). In our opinion, the transcriptional silencing can be established through dynamic capturing of transcriptional regions inside the compact chromatin domains. These domains can be considered as drops of viscous liquid, which could be formed by nucleosome-nucleosome interaction and a macromolecular crowding effect.60 Notably, this view is in line with predictions of the chromosome territory-interchromatin compartment (CT-IC) model65,66 and previous evidence for an interchromatin compartment as well as the perichromatin region (see ref. 35).
Similarity of Organization between Interphase and Mitotic Chromatins
Notably, a comparison of the USAXS scattering profiles of mitotic and interphase chromatins shows similarities in a range up to ~275 nm, suggesting a similar organization in this range (Fig. 4A and B). Beyond this range, the slope in interphase chromatin decreases in magnitude (Fig. 4A), and the curve becomes flat.
We found that both scattering patterns follow the power-law relationship between scattering intensity and scattering vector S over a wide range (Fig. 4C; ref. 15). This scattering property suggests a fractal nature,39 although we cannot rule out the possibility that other forms of organization might lead to a similar scattering pattern. The power-law relationship continues up to ~1,000 nm in mitotic chromosomes and ~275 nm in interphase chromatin (Fig. 4C and E in ref. 15). This suggests that mitotic and interphase chromatin have common structural features that are similar at many magnifications, at least in a range up to ~275 nm. Consistent with this finding, evidence for a fractal structure of human interphase chromatin was recently obtained40-43 (also see ref. 44).
The scattering similarity is also in good agreement with our notion that interphase and mitotic chromatin are locally indistinguishable17(also see refs. 45–46). Even in the interphase nuclei, numerous chromatin domains like “chromatin liquid drops” are already formed (Fig. 4D). For chromosome assembly, such chromatin domains are folded together, presumably by condensins and/or other protein factors, to create a rodlike chromosomal shape.17 The chromatin domains in interphase were originally identified as replication foci containing ~1 Mbp of the genome region by using pulse labeling.47-49 The domains have been further analyzed by super-resolution microscopy50 and Hi-C assay51 (a method to study the three-dimensional architecture of genomes). Recently it was reported that the domains are correlated with fractal globule structures.52 The fractal globule, which was identified by Lieberman-Aiden et al. using Hi-C assay,41 is an interesting structure, in which chromatin fibers are little entangled.41,42
Consistently, typical heterochromatin regions in plant or mammalian nuclei that have been visualized by cryo-EM have been reported to look very similar to mitotic chromosomes.34,35 Taken together, interphase chromatin and mitotic chromosomes have similar local organizations (e.g., chromatin domains, chromatin liquid drops or fractal globules), showing compact irregular folding of nucleosome fibers without a 30-nm chromatin fiber (Figs. 2D and 4D), although with different morphology at the micrometre scale.
30-nm Chromatin Fibers In Vitro and in Some Specific Cells
Although we suggested that 30-nm chromatin fiber is nearly absent in mitotic and interphase cells, there might be short stretches of 30-nm fibers or small amounts of other regularly folded hierarchies in the cells.15,53 In additon, clear “30-nm chromatin fibers” have been observed under the microscope (e.g., refs. 24 and 54–59). Why? A possible explanation is as follows: The formation of a 30-nm fiber requires the selective binding of nucleosomes, which are close neighbors on the DNA strand (intra-fiber nucleosomal association). A simple way to construct an intra-fiber nucleosomal association is “isolation of nucleosome fibers.” Such isolation could occur under dilute conditions, as in in vitro systems, in which interactions between nucleosome fibers are negligible. In particular, under low-salt conditions with 1–2 mM MgCl2, nucleosome fibers can gently repulse each other and easily form a 30-nm chromatin fiber (e.g., refs. 55 and 58). In conventional EM observations, such formation might be further stabilized through chemical cross-linking (e.g., glutaraldehyde fixation) and shrinkage resulting from alcohol dehydration during sample preparation.60 Isolation of nucleosome fibers might also occur when nucleosome fibers are looped out from the chromatin domain or chromosome territory for transcription (Fig. 4D, top).
In addition, specific cell types have nuclei containing apparent 30-nm chromatin fibers, including starfish sperm,24,61 chicken erythrocytes23,24,62 and mouse photoreceptor cells.63 Although the formation of a 30-nm fiber requires intra-fiber nucleosomal association, inter-fiber nucleosome associations are considered dominant in cells (Figs. 2D and 4D).11,60 Nucleosome fibers are highly interdigitated, such that they are prevented from forming 30-nm chromatin fibers, leading to irregular folding of the nucleosome fibers (polymer-melt-like structures) (Figs. 2D and 4D).11,15,60 In some specific cells that contain the regular 30-nm chromatin fibers, a unique mechanism may be present to increase intra-fiber nucleosome association, presumably via specific histone modifications or the binding of specific proteins.
In conclusion, interphase chromatin and mitotic chromosomes have similar local organizations (e.g., chromatin domains, chromatin liquid drops or fractal globules). The organizations show compact irregular folding of nucleosome fibers without a 30-nm chromatin fiber. Although the term “irregular” or “disordered” might provide an impression that the organizations are likely functionally irrelevant, the irregular folding implies less physical constraint and high dynamism, leading to a high degree of DNA accessibility.15 The irregular organization may thus have several advantages in template-directed biological processes in interphase nuclei, including RNA transcription and DNA replication, repair and recombination.15
Acknowledgments
We thank Drs M. Eltsov, K. Ito, Y. Takahashi, A.S. Frangakis and N. Imamoto, who contributed to the work published in the EMBO Journal. We also thank T. Fujisawa, T. Hayakawa and J. Dubochet for technical support and S. Tamura for technical assistance. We further thank T. Uchiumi for the anti-P antibody, and I. Hiratani, T. Cremer, A. Sasaki, S. Nozaki, M. Yamamoto, M. Nakasako, S. Nair and J. Hansen for helpful discussions and support. This work was supported by a grant-in-aid for a MEXT grant and JST CREST. S.H. and H.T. were JSPS fellows. The synchrotron radiation experiments were performed at BL29XU and BL45XU in SPring-8 with the approval of RIKEN (Proposal No. 20120023).
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/21222
References
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell, Fifth Edition 2007. [Google Scholar]
- 2.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4:809–14. doi: 10.1038/nrm1225. [DOI] [PubMed] [Google Scholar]
- 4.Robinson PJ, Rhodes D. Structure of the ‘30 nm’ chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol. 2006;16:336–43. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Tremethick DJ. Higher-order structures of chromatin: the elusive 30 nm fiber. Cell. 2007;128:651–4. doi: 10.1016/j.cell.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Sedat J, Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1978;42:331–50. doi: 10.1101/SQB.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Belmont AS, Sedat JW, Agard DA. A three-dimensional approach to mitotic chromosome structure: evidence for a complex hierarchical organization. J Cell Biol. 1987;105:77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strukov YG, Wang Y, Belmont AS. Engineered chromosome regions with altered sequence composition demonstrate hierarchical large-scale folding within metaphase chromosomes. J Cell Biol. 2003;162:23–35. doi: 10.1083/jcb.200303098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuda A, Shao L, Boulanger J, Kervrann C, Carlton PM, Kner P, et al. Condensed mitotic chromosome structure at nanometer resolution using PALM and EGFP- histones. PLoS One. 2010;5:e12768. doi: 10.1371/journal.pone.0012768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belmont AS, Bruce K. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eltsov M, Maclellan KM, Maeshima K, Frangakis AS, Dubochet J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc Natl Acad Sci U S A. 2008;105:19732–7. doi: 10.1073/pnas.0810057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDowall AW, Smith JM, Dubochet J. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986;5:1395–402. doi: 10.1002/j.1460-2075.1986.tb04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeshima K, Eltsov M. Packaging the genome: the structure of mitotic chromosomes. J Biochem. 2008;143:145–53. doi: 10.1093/jb/mvm214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roe R-J. Methods of X-Ray and Neutron Scattering in Polymer Science. 2000. [Google Scholar]
- 15.Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–53. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubochet J, Sartori Blanc N. The cell in absence of aggregation artifacts. Micron. 2001;32:91–9. doi: 10.1016/S0968-4328(00)00026-3. [DOI] [PubMed] [Google Scholar]
- 17.Maeshima K, Hihara S, Takata H. New insight into the mitotic chromosome structure: irregular folding of nucleosome fibers without 30-nm chromatin structure. Cold Spring Harb Symp Quant Biol. 2010;75:439–44. doi: 10.1101/sqb.2010.75.034. [DOI] [PubMed] [Google Scholar]
- 18.Langmore JP, Paulson JR. Low angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J Cell Biol. 1983;96:1120–31. doi: 10.1083/jcb.96.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulson JR, Langmore JP. Low angle x-ray diffraction studies of HeLa metaphase chromosomes: effects of histone phosphorylation and chromosome isolation procedure. J Cell Biol. 1983;96:1132–7. doi: 10.1083/jcb.96.4.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Widom J, Klug A. Structure of the 300A chromatin filament: X-ray diffraction from oriented samples. Cell. 1985;43:207–13. doi: 10.1016/0092-8674(85)90025-X. [DOI] [PubMed] [Google Scholar]
- 21.Lewis CD, Laemmli UK. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982;29:171–81. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 22.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Dev Cell. 2003;4:467–80. doi: 10.1016/S1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 23.Langmore JP, Schutt C. The higher order structure of chicken erythrocyte chromosomes in vivo. Nature. 1980;288:620–2. doi: 10.1038/288620a0. [DOI] [PubMed] [Google Scholar]
- 24.Woodcock CL. Chromatin fibers observed in situ in frozen hydrated sections. Native fiber diameter is not correlated with nucleosome repeat length. J Cell Biol. 1994;125:11–9. doi: 10.1083/jcb.125.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishino Y, Takahashi Y, Imamoto N, Ishikawa T, Maeshima K. Three-Dimensional Visualization of a Human Chromosome Using Coherent X-ray Diffraction. Physical Review Letters 2009; 102:18101 (4 pages) [DOI] [PubMed] [Google Scholar]
- 26.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–22. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 27.Wang JC. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–92. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli UK, Cheng SM, Adolph KW, Paulson JR, Brown JA, Baumbach WR. Metaphase chromosome structure: the role of nonhistone proteins. Cold Spring Harb Symp Quant Biol. 1978;42:351–60. doi: 10.1101/SQB.1978.042.01.036. [DOI] [PubMed] [Google Scholar]
- 29.Earnshaw WC, Heck MM. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985;100:1716–25. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986;188:613–29. doi: 10.1016/S0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- 31.Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci. 2004;117:6435–45. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- 33.Strukov YG, Belmont AS. Mitotic chromosome structure: reproducibility of folding and symmetry between sister chromatids. Biophys J. 2009;96:1617–28. doi: 10.1016/j.bpj.2008.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchet-Marquis C, Dubochet J, Fakan S. Cryoelectron microscopy of vitrified sections: a new challenge for the analysis of functional nuclear architecture. Histochem Cell Biol. 2006;125:43–51. doi: 10.1007/s00418-005-0093-x. [DOI] [PubMed] [Google Scholar]
- 35.Fakan S, van Driel R. The perichromatin region: a functional compartment in the nucleus that determines large-scale chromatin folding. Semin Cell Dev Biol. 2007;18:676–81. doi: 10.1016/j.semcdb.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem. 2008;283:34532–40. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fussner E, Ching RW, Bazett-Jones DP. Living without 30nm chromatin fibers. Trends Biochem Sci. 2011;36:1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Fussner E, Djuric U, Strauss M, Hotta A, Perez-Iratxeta C, Lanner F, et al. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011;30:1778–89. doi: 10.1038/emboj.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt PW. Use of scattering to determine the fractal dimension. The fractal approach to heterogeneous chemistry (edited by D Avnir) 1989:67-79. [Google Scholar]
- 40.Bancaud A, Huet S, Daigle N, Mozziconacci J, Beaudouin J, Ellenberg J. Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 2009;28:3785–98. doi: 10.1038/emboj.2009.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirny LA. The fractal globule as a model of chromatin architecture in the cell. Chromosome Res. 2011;19:37–51. doi: 10.1007/s10577-010-9177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell. 2012;148:908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi M. A fractal model of chromosomes and chromosomal DNA replication. J Theor Biol. 1989;141:117–36. doi: 10.1016/S0022-5193(89)80012-8. [DOI] [PubMed] [Google Scholar]
- 45.Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009;17:801–10. doi: 10.1007/s10577-009-9070-x. [DOI] [PubMed] [Google Scholar]
- 46.Cremer T, Markaki Y, Hübner B, Zunhammer A, Strickfaden H, Beichmanis S, et al. Chromosome Territory Organization within the Nucleus. Encyclopedia of Molecular Cell Biology and Molecular Medicine 2011. [Google Scholar]
- 47.Schermelleh L, Solovei I, Zink D, Cremer T. Two-color fluorescence labeling of early and mid-to-late replicating chromatin in living cells. Chromosome Res. 2001;9:77–80. doi: 10.1023/A:1026799818566. [DOI] [PubMed] [Google Scholar]
- 48.Berezney R, Malyavantham KS, Pliss A, Bhattacharya S, Acharya R. Spatio-temporal dynamics of genomic organization and function in the mammalian cell nucleus. Adv Enzyme Regul. 2005;45:17–26. doi: 10.1016/j.advenzreg.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 49.Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006;14:707–33. doi: 10.1007/s10577-006-1086-x. [DOI] [PubMed] [Google Scholar]
- 50.Markaki Y, Gunkel M, Schermelleh L, Beichmanis S, Neumann J, Heidemann M, et al. Functional nuclear organization of transcription and DNA replication: a topographical marriage between chromatin domains and the interchromatin compartment. Cold Spring Harb Symp Quant Biol. 2010;75:475–92. doi: 10.1101/sqb.2010.75.042. [DOI] [PubMed] [Google Scholar]
- 51.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20:761–70. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen JC. Human mitotic chromosome structure: what happened to the 30-nm fibre? EMBO J. 2012;31:1621–3. doi: 10.1038/emboj.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finch JT, Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976;73:1897–901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–58. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- 56.Woodcock CL, Frado LL, Rattner JB. The higher-order structure of chromatin: evidence for a helical ribbon arrangement. J Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Widom J. Physicochemical studies of the folding of the 100 A nucleosome filament into the 300 A filament. Cation dependence. J Mol Biol. 1986;190:411–24. doi: 10.1016/0022-2836(86)90012-4. [DOI] [PubMed] [Google Scholar]
- 58.Maeshima K, Eltsov M, Laemmli UK. Chromosome structure: improved immunolabeling for electron microscopy. Chromosoma. 2005;114:365–75. doi: 10.1007/s00412-005-0023-7. [DOI] [PubMed] [Google Scholar]
- 59.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci U S A. 2006;103:6506–11. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maeshima K, Hihara S, Eltsov M. Chromatin structure: does the 30-nm fibre exist in vivo? Curr Opin Cell Biol. 2010;22:291–7. doi: 10.1016/j.ceb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 61.Scheffer MP, Eltsov M, Bednar J, Frangakis AS. Nucleosomes stacked with aligned dyad axes are found in native compact chromatin in vitro. J Struct Biol. 2012;178:207–14. doi: 10.1016/j.jsb.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 62.Scheffer MP, Eltsov M, Frangakis AS. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc Natl Acad Sci U S A. 2011;108:16992–7. doi: 10.1073/pnas.1108268108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kizilyaprak C, Spehner D, Devys D, Schultz P. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One. 2010;5:e11039. doi: 10.1371/journal.pone.0011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uchiumi T, Traut RR, Kominami R. Monoclonal antibodies against acidic phosphoproteins P0, P1, and P2 of eukaryotic ribosomes as functional probes. J Biol Chem. 1990;265:89–95. [PubMed] [Google Scholar]
- 65.Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, Cremer M, et al. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Crit Rev Eukaryot Gene Expr. 2000;10:179–212. [PubMed] [Google Scholar]
- 66.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]