Abstract
Snakin/GASA proteins are widely distributed among plant species. They are expressed in different plant organs with high tissue and temporal specificity, and their subcellular localization varies among the different members. Interestingly, all of them maintain 12 cysteines of the C-terminus in highly conserved positions of the aminoacid sequences that are essential for their biochemical activity and probably responsible for their protein structure. Despite their common features, their functions are not completely elucidated and little is known about their mode of action. This review focuses on the current knowledge about this intriguing family of peptides and advances comprising gene regulation analyses, expression pattern studies and phenotypic characterization of mutants and transgenic plants. Furthermore, we discuss the roles of Snakin/GASA proteins in several aspects of plant development, plant responses to biotic or abiotic stress and their participation in hormone crosstalk and redox homeostasis.
Keywords: Abiotic stress, cell division, cell expansion, cysteine-rich domain, defense, hormone crosstalk, plant development, redox regulation, Snakin/GASA
Members of the Snakin/GASA protein family have been identi□ed in a wide range of plant species, such as tomato (Solanum lycopersicum) (GAST1, RSI-1),1,2 petunia (Petunia hybrida) (GIP1–5),3 Arabidopsis (Arabidopsis thaliana) (GASA 1–15),4-6 potato (Solanum tuberosum) (SN1–2),7,8 French bean (Phaseolus vulgaris) (FBCBP),9 rice (Oryza sativa) (OsGASR1–2, OsGSR1),10,11 gerbera (Gerbera hybrida) (GEG),12 strawberry (Fragaria ananassa) (FaGAST),13 beechnut (Fagus sylvatica) (FsGASA4),14 maize (Zea mays) (ZmGSL1–10),15 soybean (Glicine soja) (GsGASA1),16 and pepper (Capsicum annuum) (CaSn).17 Snakin/GASA genes encode small proteins in which three distinct domains can be de□ned: (1) a putative signal peptide of 18–29 residues, (2) a variable region displaying high divergence between family members both in aminoacid composition and sequence length, and (3) a C-terminal region of approximately 60 aminoacids containing 12 cysteine residues in conserved positions named GASA domain.5 Despite their common features, their functions are not completely elucidated and little is known about their mode of action. None of the Snakin/GASA protein sequences possess any known motif or active site, and even though many studies have attempted to determine their functions in numerous plant species, their biochemical activity is not fully understood yet. The fact that the number and positions of the cysteines have remained the same throughout evolution suggests that these residues play a central role and might be important to determine their biochemical/structural functions.18 This review focuses on the current knowledge about this intriguing family of peptides and discusses lines of evidence supporting the participation of Snakin/GASA genes in the interaction between different hormonal signaling pathways, in redox regulation and several aspects of plant biology.
Snakin/GASA Genes and Hormone Crosstalk
Most of Snakin/GASA genes are regulated by plant hormones and participate in hormonal signaling pathways modulating hormonal responses and levels. The tomato Gibberellic Acid-Stimulated Transcript 1 gene (GAST1) was the □rst member of this family to be identi□ed and was shown to be induced by gibberellic acid (GA) in shoots of the GA-deficient gib mutant.1 Since then, numerous Snakin/GASA genes have been described and most of them were reported to be transcriptionally regulated by this hormone. Exogenous application of GA has been shown to increase the transcript level of six out of 15 GASA genes (GASA1, GASA4, GASA6, GASA7, GASA8 and GASA13),4,19 GEG,12 GIP genes (GIP1, GIP2, GIP4 and GIP5),18 OsGASR1, OsGASR2 and OsGSR1,10,11 FaGAST,13 FsGASA4,14 and ZmGSL1, ZmGSL2, ZmGSL4, ZmGSL6, and ZmGSL9.15 In contrast, expression of GASA5, GASA9 and GASA11 genes as well as SN2 is inhibited by this hormone.8,19,20 Interestingly, the regulation of GASA4 expression by GA is tissue specific: it is upregulated in meristematic regions and repressed in cotyledons and leaves.5 Moreover, it has been recently reported that, under the influence of GA, the expression pattern of GsGASA1 in leaves and roots is more complex since it depends not only on the organ affected but also on the treatment length.16 Even though the gene regulation by this hormone was initially considered to be a defining feature of the whole family, this regulation could not be proven for all the members. Indeed, SN1, GASA10, GASA12, GASA14 and GASA15 expression is not affected by this hormone.7,19
In addition, differential responses to absicic acid (ABA) have been reported for several Snakin/GASA genes. GASA2/3 and GASA14 expression is induced after ABA treatment and is not affected by GA, whereas GASA9 expression is inhibited by both ABA and GA.19 Besides, ABA also acts as a GA antagonist in GAST1, GIP1, GASA4–7 and SN2 expression.1,3,8,19,20 In accordance with these reports, analyses of GASA promoters have revealed a high number of GA and ABA related cis-elements.19
The involvement of other hormones in modulating the expression of certain Snakin/GASA genes reveals an even more complex regulation pattern for these genes. For example, RSI-1 expression is induced by auxin2 and the GA-induced genes GASA1 and OsGSR1 are downregulated by brassinosteroids (BR).11,21 Moreover, OsGSR1 has been shown to regulate BR levels as a result of its direct interaction with a BR biosynthetic enzyme, and to modulate the GA response by downregulating the expression of the rice DELLA protein, a negative regulator of GA signaling.11 Furthermore, other Snakin/GASA genes participate in plant growth and stress tolerance through the modulation of DELLA ´s transcription.16,20,22
Finally, the overexpression of the GA-upregulated FsGASA4 has been reported to increase not only endogenous levels of salicylic acid (SA), but also the expression of genes involved in SA biosynthesis and action.14 In addition, SA signaling has been shown to be blocked in the GASA5-overexpressing plants and this blockage was partially rescued by exogenous GA.22 Taken together, these results suggest that Snakin/GASA proteins participate in the interaction between different hormonal signaling pathways; and therefore, they are involved in plant development and plant environmental responses.
Tissue and Developmental Specific Expression Patterns of Snakin/GASA Genes
Even though the expression of Snakin/GASA genes has been described in different plant organs, a detailed examination of the spatial localization by in situ hybridization or promoter::GUS histochemical staining revealed that the expression pattern of each family member is highly specific and restricted to defined tissues.2,5-7,11-13,15 For example, in strawberry, FaGAST expression is high in fruits and roots but very low in leaves and stolons. Within the fruit, its expression has only been detected in the receptacle but not in achenes, whereas in roots FaGAST expression is confined to the cells at the end of the elongation zone.13 In Arabidopsis, GASA8 and GASA10 were detected by RT-PCR in all the organs analyzed.19 Particularly in roots, pGASA8::GUS activity is strong in the cell-elongation zone and is absent from the root tip, while pGASA10::GUS activity has been detected in the vascular tissue and at the root tip.6
In addition, Snakin/GASA genes are mainly expressed in precise developmental stages. For instance, in gerbera, the expression pattern of GEG correlates with cessation of cell elongation both in the corolla and in the carpel,12 and in petunia, GIP2 is expressed in elongating stem and corollas, while GIP4 and GIP5 are expressed at earlier phases of development.3,18 Moreover, we have shown that SN1 promoter is active in young stages gradually decreasing as the plant ages.23
Expression analyses indicate that the spatial and temporal regulation of Snakin/GASA genes is highly specific. Even though partially overlapping localizations between different members of the family have been described, each one displays a distinct expression pattern. Interestingly, they all correlate with young tissues and actively growing organs suggesting that Snakin/GASA genes are involved in cellular processes such as cell division or expansion.
Subcellular Localization of Snakin/GASA Proteins
The subcellular localization of Snakin/GASA proteins varies among different family members. Even though the putative localization for some of these peptides has been proposed based on sequence analyses, the subcellular localization of only a few of them have been experimentally determined. The transient expression of a GIP1:: green fluorescent protein (GFP) fusion in tobacco bright yellow 2 (BY2) cells suggests an endoplasmic reticulum localization.18 OsGASR1::GFP, OsGASR2::GFP and OsGSR1::GFP fusion proteins transiently expressed in onion epidermal cells revealed that OsGASR peptides are restricted to the apoplasm or cell wall, while OsGSR1 protein localizes to the plasma membrane, cytoplasm and nucleus.10,11 Using the same experimental system, it has been demonstrated that GASA5 localizes to the cell wall and/or extracellular matrix, whereas GsGASA1 has been found throughout the onion epidermal cells.16,20 More recently, co-localization experiments performed by our group have recently shown that SN1::GFP is located in the plasma membrane of Nicotiana benthamiana agroinfiltrated leaves.24
Despite the presence of a putative signal peptide and the lack of other predicted targeting signals, not all the Snakin/GASA proteins are targeted to the cell wall/extracellular matrix. It is possible that post-translational modifications, electrostatic interactions, covalent bonds to membrane lipids or attachment/interaction with other proteins are responsible for their localization. Several examples of protein interactions involving snakin/GASA proteins have been reported. A SN2-like peptide from French bean has been shown to be associated with a proline-rich protein, resulting in a two-component protein complex (FBCBP).9 As it was mentioned before, it has been demonstrated that OsGSR1 interacts with the BR biosynthetic enzyme DIM/DWF1;11 and recently we have shown that SN1 self-interacts in vivo.24
Determining the appropriate subcellular localization of proteins is an important step toward elucidating their functions. However, more detailed research in this respect needs to be done to gain insights into the biological role of Snakin/GASA proteins.
Involvement of Snakin/GASA Genes in Plant Development and Stress Tolerance
The function assignment of some Snakin/GASA members has been mainly based on expression profile analyses and the phenotypic characterization of mutants and transgenic plants. Some of them have been involved in cellular processes such as promotion of cell elongation,1,3,18 cell elongation arrest,12,13 and cell division.5,10,18,24 Moreover, it has been demonstrated that Snakin/GASA proteins play important roles in development programs, including root formation, stem growth, flowering time and fruit ripening.2,13,15,18,20 In addition, members of this family have been also involved in biotic or abiotic stress responses like pathogen defense,7,8,17,25,26 heat, salt or oxidative stress tolerance.14,27
Interestingly, different family members could fulfill the same function or just the opposite one. Moreover, even though a specific function was initially assigned to a particular Snakin/GASA gene, later evidence established that a certain member could play different roles and might be involved in diverse plant cellular processes. For example, it was first reported that SN1 and SN2, the only snakin/GASA peptides that have been isolated up to now, exhibits a broad spectrum antimicrobial activity in vitro.7,8 Afterward, our group has demonstrated that the overexpression of SN1 gene in potato enhances resistance to commercially relevant pathogens suggesting that it also has in vivo antifungal and antibacterial activity.25 Even though SN1 constitutive overexpression does not seem to alter plant phenotype, we have recently demonstrated that SN1 silencing affects cell division, leaf metabolism and cell wall composition. Taken together, these findings suggest that it participates in plant growth and development beyond its already known involvement in defense.24 In Arabidopsis, GASA4 plays a role in regulating floral meristem identity and promotes seed size and weight.6 At the same time, it has been reported that GASA4 is involved in regulating hypocotyl elongation and 〉owering in response to the integration of light signaling and GA,28 and, equally important, the overexpression of this gene enhances tolerance to heat stress.27 Conversely, it has been suggested that GASA5 acts as a negative regulator not only in stem growth and 〉oral development but also in response to heat stress.20,22 Moreover, it has been demonstrated that GASA5 overexpression blocks SA signaling and reduces the antioxidant capacity of Arabidopsis and the accumulation of heat shock proteins.22 Remarkably, the overexpression of the GA-responsive gene FsGASA4 enhances salt, oxidative and heat stress tolerance in seed germination by means of increasing SA biosynthesis. These results demonstrate that GA may play a crucial role in early plant responses to adverse environmental conditions by modulating SA levels.14,29
Overall, Snakin/GASA proteins could act as integrators of internal and environmental cues participating in hormone homeostasis to modulate plant development and stress tolerance through adjusting the balance of cell growth promotion and cell growth inhibition.
Snakin/GASA Proteins and Redox Status
Computational analyses suggest that the oxidized cysteines in all Snakin/GASA proteins have the potentiality of creating up to □ve disul□de bridges.30 These conserved cysteines may be required for the generation of an essential 3D structure and/or for an interaction with other proteins.18 On the other hand, disulfide bonds may act catalytically if they can be reversibly reduced and oxidized (thiol/disulfide). As all Snakin/GASA proteins contain putative redox-active sites (i.e., pairs of cysteines separated by one or two aminoacids), it has been speculated that they may play a role in redox regulation.30 In fact, Weiss and coworkers have demonstrated that the expression of GIP2, GIP4 and GIP5 is induced by H2O2 and that the overexpression of GIP2 in transgenic petunia reduces H2O2 levels in leaves after wounding and in guard cells after osmotic stress or ABA treatments.30 Moreover, this group has recently demonstrated that the overexpression of GASA4 in transgenic Arabidopsis is able to suppress the accumulation of both H2O2 and nitric oxide in wounded leaves. Interestingly, expression of mutated versions of this protein in plants revealed that the conserved cysteines of GASA4 are essential for its redox activity and the promotion of GA responses such as flowering and seed germination.31 In addition, we observed that redox balance is altered in SN1 silencing lines since higher levels of reactive oxygen species (ROS) were detected in these plants (Nahirñak, unpublished results). Furthermore, scavengers of ROS such as ascorbate, galactinol and raffinose were significantly reduced in SN1 silencing lines with respect to wild type plants.24 The fact that these lines show affected cell division and reduced ascorbate levels supports the proposed role of this molecule in regulating that process and suggests that SN1 might play an important role in cell division by modulating ascorbate accumulation. In brief, these findings suggest that Snakin/GASA proteins participate in plant developmental processes and stress responses regulating redox homeostasis through its conserved cysteine-rich domain.
Conclusions
The identification of Snakin/GASA genes in distantly related species highlights the importance of these proteins and suggests that they fulfill essential functions in plants. Even though they have been involved in different aspects of plant growth and development, the exact role played by these proteins is still intriguing. It is possible that the specific biological function in diverse processes of the different members of this family is determined by their spatial and temporal expression during plant development and by their variable N-terminal sequence. On the other hand, the highly conserved C-terminus domain of Snakin/GASA proteins is essential for their biochemical activity and probably responsible for their protein structure and/or protein interactions. Although it has been demonstrated that these proteins act as antioxidants, it remains still unknown whether it is a direct or indirect effect and how this antioxidant activity affects physiological and developmental responses. Further exhaustive experiments concerning biochemical characterization of Snakin/GASA proteins are required to improve the understanding of the mechanism of action. Moreover, interaction studies with other proteins from the host and/or pathogens would provide more hints as to the function of this family.
Compelling evidence supports the involvement of Snakin/GASA genes in the complex crosstalk among different hormones at both levels biosynthesis and action. Relationships of these genes and GA responses have been the most studied to date; however, the existence of interactions between these genes and other hormones are becoming apparent. In this review, we propose a simplified model explaining the participation of Snakin/GASA proteins in developmental processes and stress tolerance through their involvement in redox and hormone homeostasis (Fig. 1). So far, several unresolved questions remain to be answered; i.e., Is Snakin/GASA gene regulation by internal/external signals direct or hormone-mediated? ; Which is the role of these genes in the different hormonal pathways? ; Are Snakin/GASA modulation of DELLA’s transcription, hormonal homeostasis and redox status (inter)related?
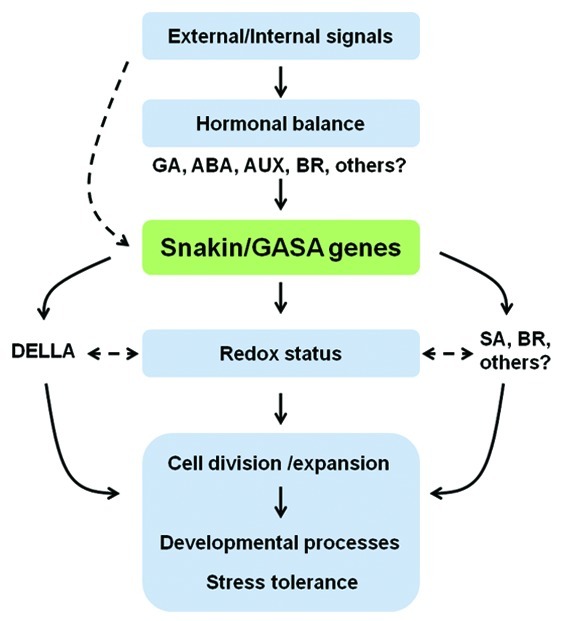
Figure 1. Simplified model of Snakin/GASA proteins’s role. Snakin/GASA gene expression is modulated in response to different hormones and/or external/internal signals. The encoded proteins regulate redox homeostasis through its conserved cysteine-rich domain and consequently participate in developmental processes and stress tolerance through adjusting the balance of cell growth promotion and cell growth inhibition. The involvement of Snakin/GASA genes in plant growth and stress responses was shown, for some members of the family, to be mediated by the modulation of DELLA’s transcription or biosynthesis of certain hormones. Nevertheless, the functional link between these changes and the redox status remains unclear.
Acknowledgments
This research was supported by PEAEBIO 244611 and 243532 (INTA) grants. We thank Mariana del Vas for critically reading this manuscript and Julia Sabio for her invaluable English assistance.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20813
References
- 1.Shi L, Gast RT, Gopalraj M, Olszewski NE. Characterization of a shoot-specific, GA3- and ABA-regulated gene from tomato. Plant J. 1992;2:153–9. [PubMed] [Google Scholar]
- 2.Taylor BH, Scheuring CF. A molecular marker for lateral root initiation: the RSI-1 gene of tomato (Lycopersicon esculentum Mill) is activated in early lateral root primordia. Mol Gen Genet. 1994;243:148–57. doi: 10.1007/BF00280311. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Nissan G, Weiss D. The petunia homologue of tomato gast1: transcript accumulation coincides with gibberellin-induced corolla cell elongation. Plant Mol Biol. 1996;32:1067–74. doi: 10.1007/BF00041390. [DOI] [PubMed] [Google Scholar]
- 4.Herzog M, Dorne AM, Grellet F. GASA, a gibberellin-regulated gene family from Arabidopsis thaliana related to the tomato GAST1 gene. Plant Mol Biol. 1995;27:743–52. doi: 10.1007/BF00020227. [DOI] [PubMed] [Google Scholar]
- 5.Aubert D, Chevillard M, Dorne AM, Arlaud G, Herzog M. Expression patterns of GASA genes in Arabidopsis thaliana: the GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol Biol. 1998;36:871–83. doi: 10.1023/A:1005938624418. [DOI] [PubMed] [Google Scholar]
- 6.Roxrud I, Lid SE, Fletcher JC, Schmidt ED, Opsahl-Sorteberg HG. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007;48:471–83. doi: 10.1093/pcp/pcm016. [DOI] [PubMed] [Google Scholar]
- 7.Segura A, Moreno M, Madueño F, Molina A, García-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 8.Berrocal-Lobo M, Segura A, Moreno M, López G, García-Olmedo F, Molina A. Snakin-2, an antimicrobial peptide from potato whose gene is locally induced by wounding and responds to pathogen infection. Plant Physiol. 2002;128:951–61. doi: 10.1104/pp.010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bindschedler LV, Whitelegge JP, Millar DJ, Bolwell GP. A two component chitin-binding protein from French bean -- association of a proline-rich protein with a cysteine-rich polypeptide. FEBS Lett. 2006;580:1541–6. doi: 10.1016/j.febslet.2006.01.079. [DOI] [PubMed] [Google Scholar]
- 10.Furukawa T, Sakaguchi N, Shimada H. Two OsGASR genes, rice GAST homologue genes that are abundant in proliferating tissues, show different expression patterns in developing panicles. Genes Genet Syst. 2006;81:171–80. doi: 10.1266/ggs.81.171. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Wang Z, Xu Y, Joo SH, Kim SK, Xue Z, et al. OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J. 2009;57:498–510. doi: 10.1111/j.1365-313X.2008.03707.x. [DOI] [PubMed] [Google Scholar]
- 12.Kotilainen M, Helariutta Y, Mehto M, Pollanen E, Albert VA, Elomaa P, et al. GEG participates in the regulation of cell and organ shape during corolla and carpel development in gerbera hybrida. Plant Cell. 1999;11:1093–104. doi: 10.1105/tpc.11.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Fuente JI, Amaya I, Castillejo C, Sánchez-Sevilla JF, Quesada MA, Botella MA, et al. The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J Exp Bot. 2006;57:2401–11. doi: 10.1093/jxb/erj213. [DOI] [PubMed] [Google Scholar]
- 14.Alonso-Ramírez A, Rodríguez D, Reyes D, Jiménez JA, Nicolás G, López-Climent M, et al. Evidence for a role of gibberellins in salicylic acid-modulated early plant responses to abiotic stress in Arabidopsis seeds. Plant Physiol. 2009;150:1335–44. doi: 10.1104/pp.109.139352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann R, Sakai H, Hochholdinger F. The Gibberellic Acid Stimulated-Like gene family in maize and its role in lateral root development. Plant Physiol. 2010;152:356–65. doi: 10.1104/pp.109.149054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li KL, Bai X, Li Y, Cai H, Ji W, Tang LL, et al. GsGASA1 mediated root growth inhibition in response to chronic cold stress is marked by the accumulation of DELLAs. J Plant Physiol. 2011;168:2153–60. doi: 10.1016/j.jplph.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 17.Mao Z, Zheng J, Wang Y, Chen G, Yang Y, Feng D, et al. The new CaSn gene belonging to the snakin family induces resistance against root-knot nematode infection in pepper. Phytoparasitica. 2011 doi: 10.1007/s12600-011-0149-5. [DOI] [Google Scholar]
- 18.Ben-Nissan G, Lee JY, Borohov A, Weiss D. GIP, a Petunia hybrida GA-induced cysteine-rich protein: a possible role in shoot elongation and transition to flowering. Plant J. 2004;37:229–38. doi: 10.1046/j.1365-313X.2003.01950.x. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Wang X. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chin Sci Bull. 2008;53:3839–46. doi: 10.1007/s11434-008-0525-9. [DOI] [Google Scholar]
- 20.Zhang S, Yang C, Peng J, Sun S, Wang X. GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol Biol. 2009;69:745–59. doi: 10.1007/s11103-009-9452-7. [DOI] [PubMed] [Google Scholar]
- 21.Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–8. doi: 10.1104/pp.010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Wang X. Overexpression of GASA5 increases the sensitivity of Arabidopsis to heat stress. J Plant Physiol. 2011;168:2093–101. doi: 10.1016/j.jplph.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Almasia NI, Narhirñak V, Hopp HE, Vazquez-Rovere C. Isolation and characterization of the tissue and development-specific potato snakin-1 promoter inducible by temperature and wounding. Electron J Biotechnol. 2010 doi: 10.2225/vol13-issue5-fulltext-12. [DOI] [Google Scholar]
- 24.Nahirñak V, Almasia NI, Fernandez PV, Hopp HE, Estevez JM, Carrari F, et al. Potato snakin-1 gene silencing affects cell division, primary metabolism, and cell wall composition. Plant Physiol. 2012;158:252–63. doi: 10.1104/pp.111.186544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almasia NI, Bazzini AA, Hopp HE, Vazquez-Rovere C. Overexpression of snakin-1 gene enhances resistance to Rhizoctonia solani and Erwinia carotovora in transgenic potato plants. Mol Plant Pathol. 2008;9:329–38. doi: 10.1111/j.1364-3703.2008.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faccio P, Vazquez-Rovere C, Hopp E, Gonzalez G, Decima-Oneto C, Favret E, et al. Increased Tolerance to Wheat Powdery Mildew by Heterologous Constitutive Expression of the Solanum chacoense snakin-1 Gene. Czech J Genet Plant Breed. 2011;47:135–41. [Google Scholar]
- 27.Ko CB, Woo YM, Lee DJ, Lee MC, Kim CS. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant physiology and biochemistry: PPB / Societe francaise de physiologie vegetale 2007; 45:722-8. [DOI] [PubMed]
- 28.Chen IC, Lee SC, Pan SM, Hsieh HL. GASA4, a GA-stimulated gene, participates in light signaling in Arabidopsis. Plant Sci. 2007;172:1062–71. doi: 10.1016/j.plantsci.2007.03.012. [DOI] [Google Scholar]
- 29.Alonso-Ramírez A, Rodríguez D, Reyes D, Jiménez JA, Nicolás G, López-Climent M, et al. Cross-talk between gibberellins and salicylic acid in early stress responses in Arabidopsis thaliana seeds. Plant Signal Behav. 2009;4:750–1. doi: 10.4161/psb.4.8.9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wigoda N, Ben-Nissan G, Granot D, Schwartz A, Weiss D. The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J. 2006;48:796–805. doi: 10.1111/j.1365-313X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- 31.Rubinovich L, Weiss D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. 2010;64:1018–27. doi: 10.1111/j.1365-313X.2010.04390.x. [DOI] [PubMed] [Google Scholar]


