Abstract
PLDZ2 is a member of the Arabidopsis phospholipase D gene family that is induced in both shoot and root in response to phosphate (Pi) starvation. Recently, through deletion and gain-of-function analyses of the PLDZ2 promoter, we identified a 65 bp region (denominated enhancer EZ2) capable of conferring tissue-specific and low-Pi responses to a minimal inactive promoter. The EZ2 element contains two P1BS motifs, each of which is the binding site for PHR1 and related transcription factors. This structural organization is evolutionarily conserved in orthologous promoters within the rosid clade. To determine whether EZ2 is significantly over-represented in Arabidopsis genes coexpressed with PLDZ2, we constructed a PLDZ2 coexpression network containing 26 genes, almost half of them encoding enzymes or regulatory proteins involved in Pi recycling. A variant of the P1BS motif was found to be highly enriched in the promoter regions of these coexpressed genes, showing an EZ2-like arrangement in seven of them. No other motifs were significantly enriched. The over-representation of the EZ2 arrangement of P1BS motifs in the promoters of genes coexpressed with PLDZ2, suggests this unit has a particularly important role as a regulatory element in a coexpression network involved in the release of Pi from phospholipids and other molecules under Pi-limiting conditions.
Keywords: Arabidopsis thaliana, Phosphate-responses, coexpression network, gene regulation, phosphate deprivation
Phosphorus is an essential element for all forms of life, being a constituent of vital molecules such as ATP, nucleic acids and phospholipids. Availability of orthophosphate (Pi), the only assimilable form of phosphorus, is often a limiting factor for plant growth in both natural and agricultural ecosystems, and deficiency of this nutrient has a direct negative effect on agronomic productivity. Phospholipids are an important reserve of Pi for the plant, and one of the many strategies that plants have evolved to cope with phosphorus limitation is the recycling of Pi from these membrane components. In plants subjected to Pi deprivation, the total content of diverse phospholipids such as phosphatidylcholine (PtdCho), phosphatidylethanolamine (PtdEtn), and phosphatidylglycerol decreases, and this reduction is coupled with an increased rate of synthesis of non-phosphorous lipids, such as the galactolipid digalactosyldiacylglycerol (DGDG) and the sulpholipid sulphoquinovosyldiacylglycerol (SQDG). This mechanism of lipid turnover allows the salvage of Pi for use in primary metabolism.1,2
Synthesis of DGDG and SQDG requires diacylglycerol (DAG), and the production of this compound from PtdCho and PtdEtn has been proposed to occur through two alternative pathways: the direct hydrolysis of phospholipids by phospholipase C (PLC) or a two-step reaction involving phospholipase D (PLD) and a phosphatidic acid phosphatase. It has been reported that phospholipase DZ2 (PLDZ2) actively participates in the hydrolysis of PtdCho and PtdEtn to release Pi and to provide DAG for the synthesis of galactolipids.3 The expression of PLDZ2 is highly induced in both shoots and roots under Pi starvation, and this increase in expression is regulated at the transcriptional level. We recently reported the characterization of the promoter region of PLDZ2.4 This analysis led to the identification of a 65-bp Pi-responsive enhancer (denominated EZ2) which confers gene expression properties similar to those of the PLDZ2 promoter in response to Pi starvation. EZ2 contains two P1BS motifs, separated by 26 bp, each of them corresponding to the consensus binding site for the transcription factor PHR1. Mutation analysis demonstrated the two P1BS motifs and adjacent sequences are important for the enhancer function of EZ2, which shows characteristics of a composite regulatory element. Moreover, analysis of orthologous promoters showed the structural organization of EZ2 is evolutionarily conserved within the rosid clade, suggesting that architectural features such as the distance between the two P1BS motifs are important for the regulatory properties of the enhancer element.
Since the EZ2 arrangement is highly conserved in the promoter regions of PLDZ2 orthologs, we wondered whether other Arabidopsis genes displaying expression patterns similar to that of PLDZ2 would show an enrichment of similar arrangements of P1BS motifs in their promoters or the enrichment of any other motif associated to this regulatory element.
Different microarray experiments have identified sets of genes differentially expressed in response to Pi deficiency, though only a small percentage of the selected genes overlap among these studies.5-7 This suggests that many of the genes initially described as responsive to low Pi represent secondary effects of the interactions between Pi availability and the experimental conditions employed in each particular instance, rather than a specific response to Pi starvation.
Transcriptome coexpression analysis has been successfully used to identify regulatory relationships from gene expression data in model organisms, leading to the discovery of cis-regulatory elements through their overrepresentation in promoters of genes coexpressed under different conditions and states.8,9 This analysis has also been employed to predict the function of genes under the assumption that a set of genes involved in the same or related metabolic pathway are under the control of a shared regulatory system and thus are coordinately expressed over a wide variety of different conditions.10
A large number of microarray data obtained under different experimental conditions are available in the public databases and similarity coefficients between individual genes have been calculated on the basis of these data sets. A higher coefficient indicates a constitutive or condition-independent coexpression relationship, allowing the identification of truly corregulated gene sets when the coexpression profile for a specific condition is compared with this “condition-independent” profile derived from public data sets. Coexpression networks constructed from pairwise correlation coefficients are a helpful method to visualize the regulatory relationships between genes and provide an efficient way to identify functional transcription modules associated with specific biological processes.
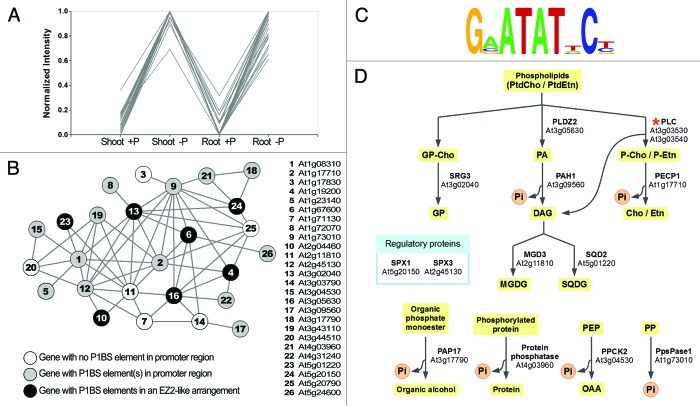
For the initial selection of genes coexpressed with PLDZ2 we used the data set from a transcriptomic analysis of Pi starvation responses in Arabidopsis plants7 (NCBI GEO accession GSE16722). Expression data from shoots and roots of wild-type plants grown under Pi-limiting or Pi-sufficient conditions, representing a total of 2739 probes called differentially expressed (1.5-fold change in at least one of the conditions) were used for the analysis. Normalization of ratio of expression intensity between Pi-deprived and Pi-supplied shoot samples was performed by subtracting the minimum value for each probe followed by dividing by the adjusted maximum value for that probe. This procedure scales all of the values for each probe so that they all fall in the range from 0 to 1 (Fig. 1A). This made it possible to classify the probes by the shape of the Pi-condition-dependent changing pattern alone and not by the absolute value of the expression level. When batch-learning-self-organizing mapping11 (BL-SOM) is applied to these data, the probes can be classified into the cells of a two-dimensional array (map) on the basis of the similarity of their expression patterns. Genes showing similar patterns are clustered into the same or neighboring cells. This procedure clusters genes that have similar expression patterns and might therefore be functionally related. Probes in the same cell as the probe corresponding to PLDZ2 and those in the immediate adjacent cells were selected for further analysis (100 probes). Since the probes have been classified based on their similar expression profile, those grouped in the selected cells can be considered as coexpressed in the experimental conditions of the microarray analysis. To establish whether these genes also have a condition-independent relationship, the presence of coexpression networks within the selected group was determined using ATTED-II (the Arabidopsis thaliana trans-factor and cis-element prediction database).12 This webtool uses Mutual Rank (MR) calculated from 1388 microarray experiment data as a measure of gene coexpression.13 Using the selected group of genes as input for the analysis in ATTED-II, a graph (network) is generated, where the pairs of coexpressed genes are represented with circles connected by lines that indicate the coexpression relationship between the genes. A threshold of MR < 10 was applied to retain only the most significant relationships. For this analysis, only 26 probes, corresponding to the same number of genes, form part of the PLDZ2 coexpression network, which was visualized using the software Pajek14 (Fig. 1B). The intergenic regions of the genes in the network were analyzed with the motif discovery tools Weeder and SCOPE15,16 to search for enriched sequence motifs that may correspond to functional regulatory elements. This analysis identified a variant of the P1BS motif (GVATATNCB; Fig. 1C) that is present in the promoter regions of 20 out of the 26 genes in the network, with an average content of 2.4 motifs per gene, and seven of the promoters have two of these motifs separated by less than 50 bp (denoted as an EZ2-like arrangement). No other obvious motifs were found to be significantly enriched in the regions analyzed. The degree of enrichment for the P1BS motif observed in this gene set is considerably higher than that previously reported for broader gene groups selected based on their response to Pi deficiency or their differential response to the conditional expression of PHR1.7
Figure 1. Coexpression analysis of PLDZ2 and other low-Pi-responsive genes. (A) Expression profiles of genes differentially expressed between plants grown under limiting (-P) and sufficient (+P) phosphate conditions showing a similar profile to that of PLDZ2 gene according to microarray data from Bustos et al. (2010). Each line represents the mean normalized expression value for each gene in the four samples. (B) DNA motif found to be enriched in the intergenic regions of the 26 genes in the PLDZ2 coexpression network. The motif was ilustrated using WebLogo.17 (C) PLDZ2 coexpression network comprising 26 genes showing a condition-independent relationship. Circles indicate genes and lines represent coexpression relationship as obtained by using ATTED-II. Lengths of the lines in this graph do not have any value. Tones of gray denote the presence of the P1BS motif reported in panel B in the upstream intergenic region of the corresponding gene. (D) Genes from the PLDZ2 coexpression network involved in processes related to the recycling of Pi. Asterisk indicates PLC genes (cross-hybridized to the same probe and thus indistinguishable), which are coexpressed with PLDZ2, but due to the lack of coexpression data for the corresponding probe, they are not included in the coexpression network. Both of them contain P1BS motifs in the intergenic region and At3g03530 has an EZ2-like arrangement. PtdCho, phosphatidylcholine; PtdEtn, phosphatidyethanolamine; P-Cho, phosphocholine; P-Etn, phosphoethanolamine; PA, phosphatidic acid; GP-Cho, glycerophosphatidylcholine; GP, glycerol-3-phosphate; DAG, diacylglycerol; Cho, choline; Etn, ethanolamine; MGDG, galactosyl-1,2-diacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; PEP, phosphoenolpyruvate; OAA, oxaloacetic acid; PP, pyrophosphate.
It is interesting to note that 12 of the genes in the network encode enzymes and regulatory proteins involved in the release of Pi from phospholipids and other molecules, demonstrating that in this case coexpression is correlated with functional relationship (Fig. 1D). This suggests that genes in the coexpression network that are annotated to encode proteins with unknown function may also be important players in the Pi recycling process. This and similar analyses may serve as a basis for experimental studies seeking to understand the regulatory mechanisms that occur during the response to low-Pi availability.
Since the motif found in this analysis is more restrictive than the canonical P1BS motif (GNATATNC), we wondered whether this motif variant might be associated with a specific gene expression profile. The self-organizing map (SOM) used to select the group of genes coexpressed with PLDZ2 was used to visualize the distribution of the genes containing this P1BS motif variant in their 1 kb proximal promoter regions, since genes are grouped in the map based on their expression profile (Fig. 2A). Even though the P1BS motif variant is significantly enriched in the promoters of genes with similar expression profile to PLDZ2, this element is also enriched in Pi-responsive genes displaying different profiles (Fig. 2B). For instance, the element is also enriched in the SOM cell with coordinates (19,7), which contains genes induced only in roots under low-Pi conditions. Interestingly, when searching for this motif in an EZ2-like organization, the arrangement shows significant association with genes coexpressed with PLDZ2 (Fig. 2C), suggesting this particular combination of motifs has an important role in determining the regulatory properties of the PLDZ2 coexpression network.
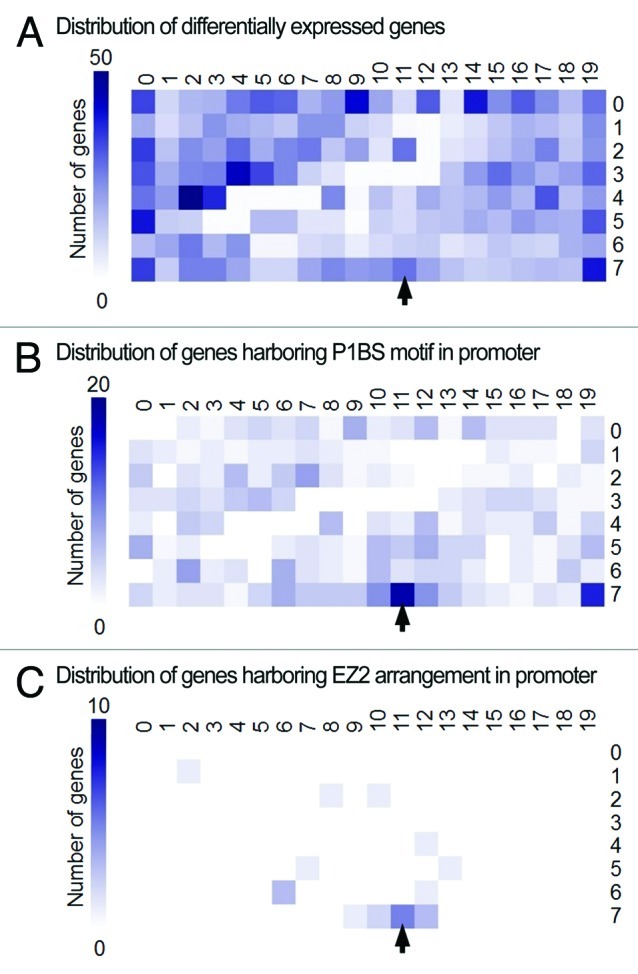
Figure 2. Distribution of the P1BS motif variant and the EZ2-like arrangement in Pi-responsive genes. (A) SOM displaying the distribution of 2739 probes called differentially expressed according to their signal values in shoots and roots of wild-type plants grown under Pi-limiting or Pi-sufficient conditions (data from experiment NCBI GEO accession GSE16722). Probes are organized based on the similarity of their expression profiles, thus probes showing similar patterns are clustered into the same or adjacent cells. (B) SOM displaying the distribution of 440 genes containing the P1BS motif variant (GVATATNCB) in their 1 kb proximal promoter regions. (C) SOM displaying the distribution of 21 genes containing an EZ2-like arrangement of P1BS motifs in their 1 kb proximal promoter regions. The gradient scale on the left of each map designates the number of genes in a given cell. Arrow indicates location of PLDZ2 in the map.
The results shown here raises the question whether distinct arrangements of the P1BS motif determine condition-specific gene expression patterns, or whether additional regulatory motifs are present in other coexpression networks. Similar analyses including other groups of genes responsive to Pi starvation would help answer this question.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20749
References
- 1.Tjellström H, Andersson MX, Larsson KE, Sandelius AS. Membrane phospholipids as a phosphate reserve: the dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ. 2008;31:1388–98. doi: 10.1111/j.1365-3040.2008.01851.x. [DOI] [PubMed] [Google Scholar]
- 2.Nussaume L, Maréchal E, Thibaud MC, Block MA. Plant plasma membrane and phosphate deprivation. In: The Plant Plasma Membrane. Plant Cell Monographs Vol 19, Murphy AS, Peer W, Schulz B (eds), Berlin Heidelberg: Springer Verlag, 2011; p. 237; 10.1007/978-3-642-13431-9_10 [Google Scholar]
- 3.Cruz-Ramírez A, Oropeza-Aburto A, Razo-Hernández F, Ramírez-Chávez E, Herrera-Estrella L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc Natl Acad Sci U S A. 2006;103:6765–70. doi: 10.1073/pnas.0600863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oropeza-Aburto A, Cruz-Ramírez A, Acevedo-Hernández GJ, Pérez-Torres CA, Caballero-Pérez J, Herrera-Estrella L. Functional analysis of the Arabidopsis PLDZ2 promoter reveals an evolutionarily conserved low-Pi-responsive transcriptional enhancer element. J Exp Bot. 2012;63:2189–202. doi: 10.1093/jxb/err446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci U S A. 2005;102:11934–9. doi: 10.1073/pnas.0505266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, et al. Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007;30:85–112. doi: 10.1111/j.1365-3040.2006.01608.x. [DOI] [PubMed] [Google Scholar]
- 7.Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:•••. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48:381–90. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- 9.Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2009;150:535–46. doi: 10.1104/pp.109.136028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci U S A. 2007;104:6478–83. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, et al. Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:10205–10. doi: 10.1073/pnas.0403218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, et al. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 2007;35(Database issue):D863–9. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obayashi T, Kinoshita K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009;16:249–60. doi: 10.1093/dnares/dsp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batagelj V, Mrvar A. Pajek- Program for large network analysis. Connections. 1998;21:47–57. [Google Scholar]
- 15.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32(Web Server issue):W199-203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson JM, Chakravarty A, DeZiel CE, Gross RH. SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35(Web Server issue):W259-64. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]