Abstract
In our previous study we applied the Agilent 44K tobacco gene chip to introduce and analyze the tobacco male gametophyte transcriptome in mature pollen and 4h pollen tubes. Here we extended our analysis post-pollen mitosis II (PMII) by including a new data set obtained from more advanced stage of the ongoing progamic phase – pollen tubes cultivated in vitro for 24 h. Pollen mitosis II marks key events in the control of male gametophyte development, the production of two sperm cells. In bicellular species covering cca 70% of angiosperms including Nicotiana tabacum, PMII takes place after pollen germination in growing pollen tube. We showed the stable and even slightly increasing complexity of tobacco male gametophyte transcriptome over long period of progamic phase–24 h of pollen tube growth. We also demonstrated the ongoing transcription activity and specific transcript accumulation in post-PMII pollen tubes cultivated in vitro. In all, we have identified 320 genes (2.2%) that were newly transcribed at least after 4h of pollen tube cultivation in vitro. Further, 699 genes (4.8%) showed over 5-fold increased accumulation after the 24h of cultivation.
Keywords: de novo pollen tube transcriptome, male gametophyte development, pollen tube growth, transcriptomics
In our original work,1 we applied the Agilent 44K tobacco gene chip to conduct the first thorough transcriptomic analysis of the tobacco male gametophyte development and the dynamics of gene expression throughout the first four hours of pollen germination and tube growth. We presented four independent data sets – two male gametophytic (mature pollen (MPG) and 4h pollen tubes (PT4) and two sporophytic (leaves (L) and roots (R) as a reference. In the meantime of the publication process of the original article, we extended our analyses and supplemented them with a data set from more advanced stage of the ongoing progamic phase – pollen tubes cultivated in vitro for 24 h (PT24). This extension allowed us to compare pollen tube transcriptomes pre- and post-pollen mitosis II (PMII). In tobacco, PMII occurs at the time window between 10–12 h of cultivation and is associated with changes in male germ unit (MGU) organization and pollen tube growth characteristics (ref. 1 and Breznenová, Čapková, Hafidh and Honys, unpublished data).
Mature pollen was isolated according to Petrů et al.2 and pollen tubes were in vitro germinated as described previously.3 RNA extraction, probe preparation and microarray hybridization were described in the original article1 as well as the microarray data normalization and analyses by freely available dChip 1.3 software4 and CLC Genomic Workbench v. 4.5.1 (CLC bio, Aarhus, Denmark). As before, two biological replicates were characterized.
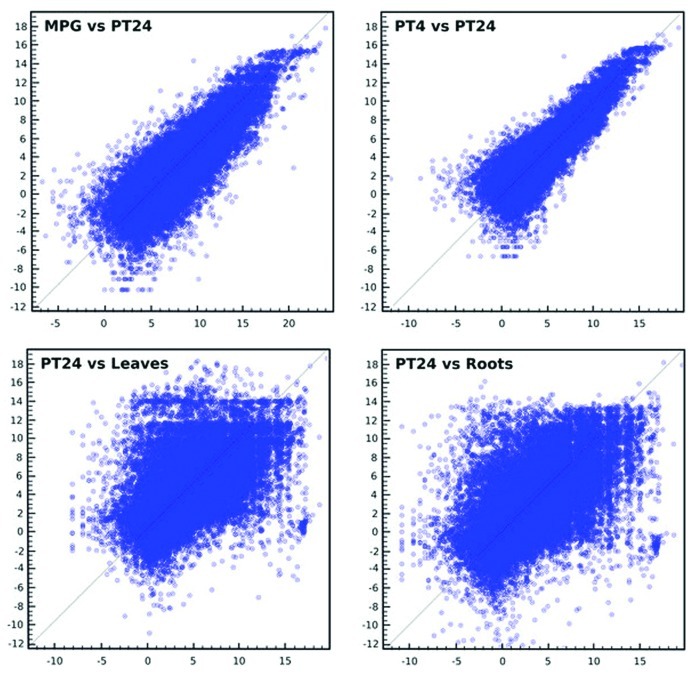
Principal component analysis (Fig. 1) confirmed the position of the new 24 h pollen tube transcriptome in the close vicinity to PT4 within the unique cluster of male gametophytic data sets clearly distinguished from sporophytic samples. Scatterplots were used to further visualize the distribution of p-values for individual probes. A close relationship between tissues of similar origin was confirmed as well as a great variability in relation to disparate tissues (Fig. 2).
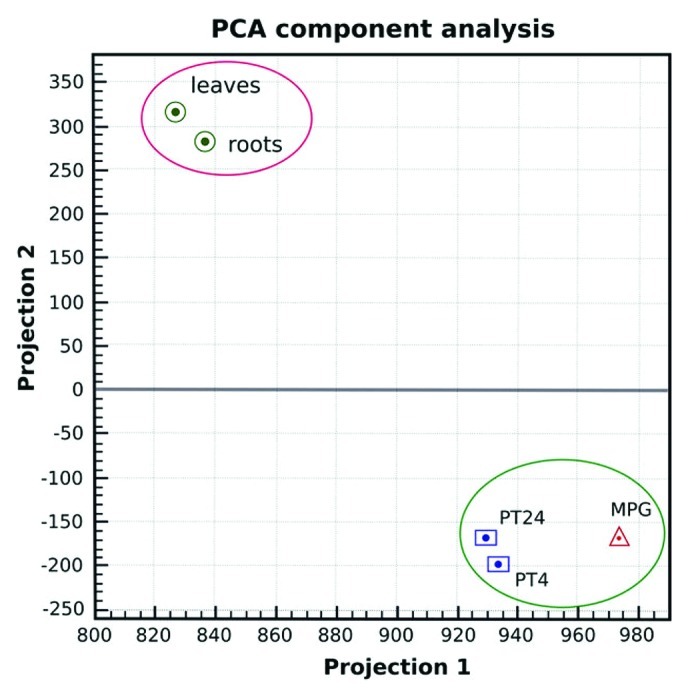
Figure 1. Output of the first and second principal component (PCA) analysis of the log2-transformed data sets. The largest and second-largest principal component (variability projection 1 and 2, respectively) are displayed in orthogonal directions, assessing the overall homogeneity between replicates and variability between samples of different tissue types as reflected in their grouping.
Figure 2. Scatterplots of log2-transformed microarray data showing a correlation of 24 h data set with other tissue arrays. A wider symmetrical scattering of the spots from the 'trendline' implicates higher variability between the tissues, with less dependency between variance and mean expression values, as compared with more tightly packed male gametophytic spots reflecting a closer relationship between these data sets.
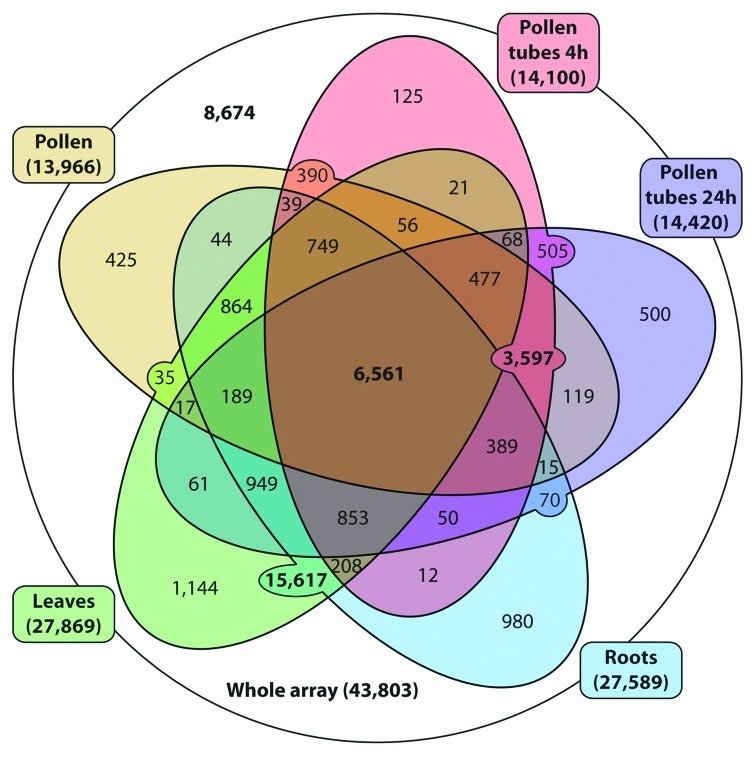
Of the 43,803 probes present on the array, 35,129 (80%) were reliably expressed (present call “well-above-background” in both replicates) in at least one tissue (Fig. 3). In the original article, we already demonstrated the reduced complexity of tobacco male gametophyte transcriptome in comparison to sporophytic tissues. In this respect tobacco followed the trend shown for plant species with more advanced tricellular pollen–Arabidopsis and rice.5-7 Here, the complexity of 24h pollen tube transcriptome was shown to be similar, though slightly higher than that of earlier male gametophytic data sets. There were 14,420 genes (41% of expressed genes) reliably expressed in 24h pollen tubes (Table S1), only 320 genes more than in 4h pollen tubes and 454 more than in mature pollen. Thus there is moderate but significant increase in transcription during pollen tube growth, particularly after PMII. Interestingly, this increase in transcriptional activity was reflected not only by an increase in the number of expressed genes but also in continuous accumulation of their respective transcripts. Of the 14,420 transcripts present in PT24, 6,934 (48.1% of genes active in PT24) have their expression signal in PT24 stronger than in other two male gametophytic data sets. Moreover, as many as 699 genes (4.8%) have their relative expression signal 5-fold higher than both MPG and PT4. In contrast to generally stable and relatively low proportion of male gametophyte-specific genes across various plant species,5-7 high proportion of these upregulated genes (227 genes, 32.5%) were putatively male gametophyte-specific. These results confirmed the ongoing high transcription activity and specific transcript accumulation in tobacco pollen tubes after PMII. Our finding also supplements results of a recent study by Qin et al.8 that revealed a specific subset of genes in Arabidopsis (semi-in-vivo (SIV)-enriched and SIV-exclusive) to be induced in pollen tubes grown through the pistil but not when grown in vitro. We rectify that among the SIV-enriched list (140 genes), we have identified at least eight tobacco homologs (47% of the total identified homologs) that had showed significant increase in accumulation, as much as 15-fold (Fig. S1, Table S2) after 24h of in vitro pollen tube cultivation. We supplement the previous findings in Arabidopsis that at least some of the identified SIV-enriched genes are expressed within a late progamic transcription program rather than as a response to the interaction with the pistil. However, the later still may have an influence on their expression. Among this set of genes there are potential receptor proteins that may respond to female cues for guidance of the pollen-tube tip growth. In the light of recent identification and biochemical characterization of long-term RNA-storage and transport of EPP particles,3,9 it is likely that these newly synthesized and accumulated transcripts are stored within these particles.
Figure 3. Five-way Venn diagram showing the quantification of all possible overlaps between three male gametophytic and two sporophytic data sets.
Taking together, here we present the extended tobacco male gametophytic transcriptomic data set that is now available to the scientific community. Moreover, we showed ongoing transcription activity and specific transcript accumulation in post-PMII pollen tubes cultivated in vitro.
Supplementary Material
Acknowledgment
The authors gratefully acknowledge the financial support from Czech Science Foundation (Grant No. P501/11/P321).
Note
Supplemental material can be found at: http://www.landesbioscience.com/journals/psb/article/20745/
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20745
References
- 1.Hafidh S, Breznenová K, Růžička P, Feciková J, Capková V, Honys D. Comprehensive analysis of tobacco pollen transcriptome unveils common pathways in polar cell expansion and underlying heterochronic shift during spermatogenesis. BMC Plant Biol. 2012;12:24. doi: 10.1186/1471-2229-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrů E, Hrabětová E, Tupý J. The technique of obtaining germinating pollen without microbial contamination. Biol Plant. 1964;6:68–9. doi: 10.1007/BF02930799. [DOI] [Google Scholar]
- 3.Honys D, Rĕnák D, Feciková J, Jedelský PL, Nebesárová J, Dobrev P, et al. Cytoskeleton-associated large RNP complexes in tobacco male gametophyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing, and localization. J Proteome Res. 2009;8:2015–31. doi: 10.1021/pr8009897. [DOI] [PubMed] [Google Scholar]
- 4.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honys D, Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pina C, Pinto F, Feijó JA, Becker JD. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005;138:744–56. doi: 10.1104/pp.104.057935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei LQ, Xu WY, Deng ZY, Su Z, Xue Y, Wang T. Genome-scale analysis and comparison of gene expression profiles in developing and germinated pollen in Oryza sativa. BMC Genomics. 2010;11:338. doi: 10.1186/1471-2164-11-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, et al. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet. 2009;5:e1000621. doi: 10.1371/journal.pgen.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hafidh S, Capková V, Honys D. Safe keeping the message: mRNP complexes tweaking after transcription. Adv Exp Med Biol. 2011;722:118–36. doi: 10.1007/978-1-4614-0332-6_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.