Abstract
Stomata are epidermal bi-celled structures that differentiate within special cell lineages initiated by a subset of protodermal cells. Recently, we showed that the Arabidopsis photomorphogenic repressor COP10 controls specific cell-lineage and cell-signaling developmental mechanisms in stomatal lineages. Loss-of-function cop10-1 mutant cotyledons and leaves produced (in the light and in the dark) abundant stomatal clusters, but nonlineage epidermal cells were not affected. Here we examine COP10 role in hypocotyls, cylindrical organs displaying a distinct epidermal organization with alternate files of protruding and non-protruding cells, with the latter producing a limited number of stomata. COP10 prevents stomatal clusters and restricts stomata production in hypocotyls; these roles are specific to lineage cells as in cotyledons, since COP10 loss of function does not elicit stomatal fate in nonlineage cells; COP10 also sustains the directional cell expansion of all hypocotyl epidermal cell types, and seems necessary for the differentiation between protruding and non-protruding cell files.
Keywords: constitutive photomorphogenic 10, tmm, epidermis development, hypocotyl, stoma, stomata lineage, stomatal cluster
The mature epidermes of Arabidopsis thaliana cotyledons and leaves are essentially composed of stomata and pavement cells (plus trichomes in some epidermes and organ sides). This neatly simple cell pattern is built from a similarly simple-patterned protodermis through a complex process where several short-lived cell types in stomatal lineages appear and rapidly evolve as the organ grows.1 Stomatal lineage development has been described in considerable depth in the last years using powerful genetic and molecular analyses in Arabidopsis thaliana.2,3 As we understand it now, a protodermal cell initiates a stomatal lineage undergoing an asymmetric division whose smaller product, the meristemoid, can experience up to three additional asymmetric divisions oriented in an inward spiral that places the smaller product in each division centrally with respect to the larger products, termed stomatal lineage ground cells (SLGCs); the late meristemoid becomes a guard mother cell, committed to divide symmetrically and produce the guard cell pair that constitutes the stoma. SLGCs can reiterate the behavior of protodermal cells, producing satellite stomatal lineages, but eventually differentiate into pavement cells. Thus, the developing epidermis of flat organs is composed of two main cell populations: stomatal lineage cells, and nonlineage cells. At the end of this dynamic process, most stomatal lineage cells and all nonlineage cells (except for those producing trichomes) differentiate into pavement cells.
Many genes involved in stomatal lineage development have been identified in the last years, including positive drivers of consecutive stomatal precursor cell stages and negative regulators that restrict cell transits and fates (for recent comprehensive reviews see refs. 2–4).
Since lineages initiate asynchronously in interspersed protodermal and stomatal lineage ground cells across the epidermis, and since only one cell in each lineage shall produce a stoma, the coordination of positive and negative regulators is essential to prevent the appearance of stomata in contact; they also contribute to establish the stomata abundance that better fits physiological and environmental conditions in the mature organs. Wild Arabidopsis genotypes encompass an ample variation for stomata and pavement cell abundance, suggestive of an adaptive value for these characters as the various populations evolved in natural environments.5 Environmental factors modulate stomatal abundance, although the mechanisms mediating the responses are largely unknown as yet. Light is one of these factors for which a few pieces of interesting information have been obtained.6 Photoreceptors (notably CRY1, CRY2, phyA and phyB) are required for full stomata differentiation in the light and mediate the increase in stomata abundance observed at high light intensity.7,8 COP1, a key component of the photomorphogenic switch that promotes skotomorphogenesis in the absence of light, was recently involved in preventing stomata formation in the dark and promoting stomata spacing,8 its lack of function leading to large stomatal clusters; elegant genetic evidence partly based on the null cop1–5 mutant indicated that COP1 conveys photoreceptor-perceived light information to the stomatal development gene circuits through YDA, a MAPK that restricts the function of downstream positive regulators of stomata development;8,9 however, the developmental mechanisms involved in these COP1 functions were not described.8 Recently,10 we provided further insight showing that COP10, another component of the photomorphogenic switch and part of the CDD complex that interacts with COP1,11,12 controls distinct events along the stomatal pathway.
Loss of function cop10–1 mutants display a final phenotype similar to cop1–5, but they have a more extended lifespan and a healthier growth; this allowed us to describe cell division and differentiation histories of individual cop10–1 stomatal lineages, by following the developmental course of living cotyledon epidermes through serial epidermal imprints. COP10 reduces stomatal lineage initiation in the dark and modulates it in light-grown plants. Regardless of the light conditions, COP10 extends meristemoid divisions and represses their premature stomatal fate, decreasing stomatal abundance; it also ensures the proper orientation of asymmetric divisions, contributing to a correct spacing. As the epidermis develops and fate errors accumulate in the cop10–1 cotyledon, the phenotype becomes increasingly severe and by 10 d after germination (dag) stomata proportion doubles the wild type one. In cop10–1, most lineage cells ended up differentiating stomata, what gave rise to abundant, large stomatal clusters; cop10–1 stomata were also morphologically and functionally impaired. In contrast, cotyledon and leaf epidermes also produced apparently normal (albeit smaller) pavement cells, and normal trichomes in the adaxial leaf epidermis. In fact, the cell distance between stomatal units (stomata or stomatal clusters) was statistically indistinguishable between the two genotypes.10
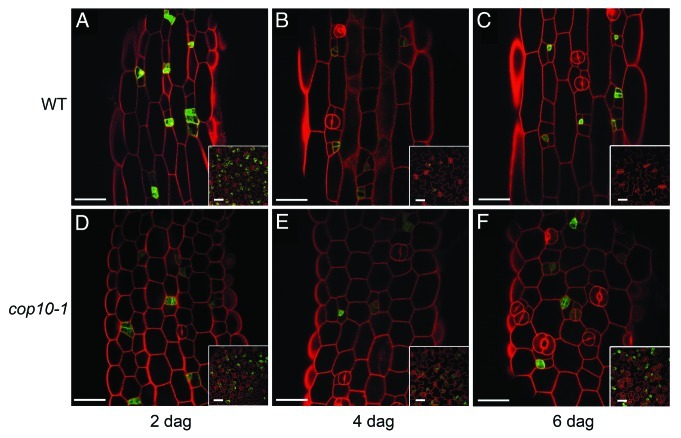
These findings prompted us to propose that COP10 acts by restricting stomata formation specifically on the lineage cell population, at least in flat organs. However, cylindrical organs were not examined. This is relevant because stems and hypocotyls show distinct features regarding stomata development in comparison with cotyledons, with a very limited stomata production. In fact, the great majority of epidermal cells in both cylindrical organs are nonlineage cells.13,14 In addition, hormones with significant impact on stomata development in hypocotyls show no or weak effects on cotyledons, and conversely.15-17 In this report, we provide a preliminary description of the qualitative hypocotyl phenotype of cop10–1 compared with wild-type seedlings (Fig. 1). Both genotypes carried a TMM::TMM-GFP transgene that marks proliferating stomatal lineage cells.18 In the wild type, and as previously described,13,14 non-protruding files showed cell divisions at 2 dag, some of them associated to stomatal lineages as shown by TMM-GFP accumulation (Fig. 1); at 4 and 6 dag lineages had progressed and stomata were present in non-protruding files; protruding cells remained large and undivided. In cop10–1 hypocotyls both file types contained un-elongated cells with no apparent divisions except for those linked to stomatal lineages marked by GFP, mostly in non-protruding files; along time, only lineage cells seemed to undergo division and differentiation and by 6 dag cop10–1 had more stomata than the wild type, and some of them were clustered.
Figure 1.Hypocotyl epidermal phenotype of cop10–1. Confocal images of light-grown wild type (A-C) and cop10–1 (D-F) seedlings carrying the TMM::TMM-GFP fusion that marks proliferating stomatal lineages (green). Plants were examined at 2, 4 and 6 dag after staining with propidium iodide to show cell contours (red). Insets show corresponding adaxial cotyledon epidermes. In cop10–1 hypocotyls, protruding and non-protruding cells are stunted and appear similar, contrasting with the wild type distinct cell patterns. Stomatal lineages are not overabundant at any time in cop10–1 hypocotyls, which show only occasional and small stomatal clusters at 6 dag in contrast with the equivalent cotyledons. Bars, 40 μm.
However, the striking stomatal clustering phenotype characteristic of cop10–1 cotyledons (insets in Figure 1) was not matched in hypocotyls, where stomata overproduction and clustering were very moderate. This agrees with the fact that cotyledon clusters built up progressively as the overabundant cop10–1 lineages divided and satellized, and cell fate and pattern errors accumulated;10 in hypocotyls most epidermal cells never enter the stomatal pathway as judged by the sparse cells showing TMM::TMM-GFP expression in both genotypes, and since the infrequent lineages in cylindrical organs satellize limitedly,14 few cells might be able to respond to loss-of-COP10 function. Similarly, sdd1–3 mutants produced few stomatal clusters in hypocotyls, and double sdd1–3/cop10–1 mutants, which showed an additive phenotype in cotyledons, with abundant large clusters,10 had a very mild hypocotyl phenotype, much like each single mutant (not shown). Therefore, the effect of COP10 on stomatal lineage development seems qualitatively similar in both embryonic organs. The fate identity loss in hypocotyls epidermal cells produced by ectopic overexpression of positive stomata-fate regulators such as MUTE,19 that transforms nearly all of them in stomata regardless of their lineage or nonlineage origin is thence not matched by COP10 loss of function, whose effects are exerted only in lineage cells. In addition, in cop10–1 hypocotyls epidermal cell expansion and differentiation between protruding and non-protruding files are also affected.
In hypocotyls, the sdd1–3 phenotype is similar to that of cop10–1 (see above), but both differ from tmm, which displays stomataless cylindrical organs. This puzzling tmm phenotype has been explained by the TMM-mediated inhibition of a stomatal development repression pathway exclusive of cylindrical organs that involves CHAL-type peptides.20,21 SDD1,20 and COP10 (this work) do not seem to be involved in this pathway, since loss of function of either gene do not prevent stomata development in hypocotyls. In cotyledons, however, the distinct stomatal developmental processes altered in cop10–1 closely match those produced by alterations on TMM and SDD1, two genes specifically involved in stomata development and whose epidermal expression is restricted to stomatal lineage cells.22-24 Mutants in both genes have phenotypes additive with those of either cop1–4 (TMM,8) or cop10–1 (SDD1,10), indicating that the two stomatal repressors act in routes genetically parallel to those involving COP genes.8,10 It might seem surprising that three parallel routes control precisely the same developmental processes; however, the molecular mechanisms involved might be completely different. For instance, a decreased COP10 function may over-stabilize cell-autonomous stomatal morphogenetic proteins, while a decreased SDD1 activity may sustain lower levels of stomatal inhibiting factors; in both cases, stomata-promoting proteins may over-accumulate ectopically and promote stomatal fate in several lineage cells, leading to apparently similar phenotypes through different lineage-based or cell-cell communication regulatory pathways.
Another way of looking at this issue is that the apparently similar phenotypes might simply mean that what seem to be well defined, discrete events are hiding un-described complexity comprising distinct cell identities and processes, each one involving (in part) different genes in parallel routes. Then, the apparent similarity in the processes controlled by these genes may be a consequence of looking at gross phenotypes (i.e., a shortened life of stomatal lineages, or incorrect placement of asymmetric division planes). Recently we unveiled cryptic natural diversity in stomata development hidden under similar final epidermal phenotypes, by examining satellite lineage proportions very early during cotyledon development.5 Dissecting developmental phenotypes more precisely in different genetic backgrounds may also contribute to understanding the genetic and environmental control of stomatal development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Arabidopsis Col-0 seeds containing the proTMM::TMM-GFP translational fusion were a kind gift from Fred Sack (University of British Columbia, Canada); the Nottingham Arabidopsis Stock Centre (UK) provided cop10–1 and Ws seeds. This work was supported by grants from the Government of Spain (BIO2007–60276 to C.F. and M.M., and CSD2007–00057 to C.F.) and the Junta de Comunidades de Castilla-la Mancha (PAI07–0036–3278 to M.M.)
Glossary
Abbreviations:
- COP10
CONSTITUTIVE PHOTPHOGENIC 10
- SLGCS
stomatal lineage ground cells
- CRY
cryptochrome
- PHY
phytochrome
- YDA
yoda
- TMM
too many mouths
- GFP
green fluorescent protein
- SDD1
stomatal density and distribution 1
Footnotes
D.D. and I.B. have contributed equally to this work
Previously published online: www.landesbioscience.com/journals/psb/article/20995
References
- 1.Nadeau JA, Sack FD. Stomatal Development in Arabidopsis. In: Somerville CR, Meyerowitz EM, eds. The Arabidopsis Book. Rockville: American Society of Plant Biologist, 2002; PMID: 22303215; 10.1199/tab.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann DC, Sack FD. Stomatal development. Annu Rev Plant Biol. 2007;58:163–81. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- 3.Pillitteri LJ, Torii KU. Mechanisms of stomatal development. Annu Rev Plant Biol. 2012;63:591–614. doi: 10.1146/annurev-arplant-042811-105451. [DOI] [PubMed] [Google Scholar]
- 4.Dong J, Bergmann DC. Stomatal patterning and development. Curr Top Dev Biol. 2010;91:267–97. doi: 10.1016/S0070-2153(10)91009-0. [DOI] [PubMed] [Google Scholar]
- 5.Delgado D, Alonso-Blanco C, Fenoll C, Mena M. Natural variation in stomatal abundance of Arabidopsis thaliana includes cryptic diversity for different developmental processes. Ann Bot. 2011;107:1247–58. doi: 10.1093/aob/mcr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casson SA, Hetherington AM. Environmental regulation of stomatal development. Curr Opin Plant Biol. 2010;13:90–5. doi: 10.1016/j.pbi.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM. phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol. 2009;19:229–34. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- 8.Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 2009; 21:2624-2641; PMID: 19794114; 10.1105/tpc.109.069765. [DOI] [PMC free article] [PubMed]
- 9.Bergmann DC, Lukowitz W, Somerville CR. Stomatal development and pattern controlled by a MAPKK kinase. Science. 2004;304:1494–7. doi: 10.1126/science.1096014. [DOI] [PubMed] [Google Scholar]
- 10.Delgado D, Ballesteros I, Torres-Contreras J, Mena M, Fenoll C. Dynamic analysis of epidermal cell divisions identifies specific roles for COP10 in Arabidopsis stomatal lineage development. Planta. 2012;in press doi: 10.1007/s00425-012-1617-y. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng X-W. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 2002;16:554–9. doi: 10.1101/gad.964602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J, et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004;18:2172–81. doi: 10.1101/gad.1229504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger F, Linstead P, Dolan L, Haseloff J. Stomata patterning on the hypocotyl of Arabidopsis thaliana is controlled by genes involved in the control of root epidermis patterning. Dev Biol. 1998;194:226–34. doi: 10.1006/dbio.1997.8836. [DOI] [PubMed] [Google Scholar]
- 14.Bhave NS, Veley KM, Nadeau JA, Lucas JR, Bhave SL, Sack FD. TOO MANY MOUTHS promotes cell fate progression in stomatal development of Arabidopsis stems. Planta. 2009;229:357–67. doi: 10.1007/s00425-008-0835-9. [DOI] [PubMed] [Google Scholar]
- 15.Saibo NJM, Vriezen WH, Beemster GTS, Van Der Straeten D. Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 2003;33:989–1000. doi: 10.1046/j.1365-313X.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- 16.Gudesblat GE, Schneider-Pizoń J, Betti C, Mayerhofer J, Vanhoutte I, van Dongen W, et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat Cell Biol. 2012;14:548–54. doi: 10.1038/ncb2471. [DOI] [PubMed] [Google Scholar]
- 17.Kim TW, Michniewicz M, Bergmann DC, Wang ZY. Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature. 2012;482:419–22. doi: 10.1038/nature10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadeau JA, Sack FD. Control of stomatal distribution on the Arabidopsis leaf surface. Science. 2002;296:1697–700. doi: 10.1126/science.1069596. [DOI] [PubMed] [Google Scholar]
- 19.Pillitteri LJ, Bogenschutz NL, Torii KU. The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in Arabidopsis. Plant Cell Physiol. 2008;49:934–43. doi: 10.1093/pcp/pcn067. [DOI] [PubMed] [Google Scholar]
- 20.Abrash EB, Bergmann DC. Regional specification of stomatal production by the putative ligand CHALLAH. Development. 2010;137:447–55. doi: 10.1242/dev.040931. [DOI] [PubMed] [Google Scholar]
- 21.Abrash EB, Davies KA, Bergmann DC. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell. 2011;23:2864–79. doi: 10.1105/tpc.111.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisler M, Nadeau J, Sack FD. Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 2000; 12:2075-86; PMID: 11090210; 10.1105/tpc.12.11.2075. [DOI] [PMC free article] [PubMed]
- 23.Geisler MJ, Deppong DO, Nadeau JA, Sack FD. Stomatal neighbor cell polarity and division in Arabidopsis. Planta. 2003;216:571–9. doi: 10.1007/s00425-002-0912-4. [DOI] [PubMed] [Google Scholar]
- 24.Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–31. doi: 10.1101/gad.14.9.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]