Abstract
Daylight UV-B (UV-B) radiation (280–315 nm) is, because of its photochemical effects and potential destructive impact, an important environmental factor for plants. After decades of fruitless attempts, a receptor molecule, UVR8, for sensing of ambient UV-B radiation by plants has been characterized, and the initial steps in signal transduction have been identified. There are, however, other signaling pathways, and there are apparent contradictions in the literature. There is still much to find out about the complex signaling network in plants for processing of information about the daylight surrounding them.
Keywords: Arabidopsis, DNA damage, UV-B, UV-B receptor, UVR8, action spectrum, signaling pathway
Biological responses to UV radiation have been studied for a long time, almost since the discovery of this shortwave type of electromagnetic radiation, in the following abbreviated as UV. For a long time the investigations were focused on the responses to UV-C radiation, which comprises the wavelength range from X-rays to 280 nm (1 nm = 1 nanometer = 10−9 m). This kind of radiation is not present in daylight, but is easily generated in the laboratory. Recently, more interest has been paid to the UV components of daylight, and in particular to UV-B (280 to 315 nm wavelength). The main reason for this interest has been the thinning of the stratospheric ozone layer that took place from about 1970 till about 2000, due to pollution of the atmosphere by halogen compounds and nitrogen oxides. This thinning caused an increase of UV-B radiation except near the equator, and in particular at high latitudes.1
Many reviews have been written on this topic, and the United Nations Environmental Program (UNEP) has published regular reports. The latest full report of the UNEP panel for assessment of environmental effects (Environmental effects of ozone depletion and its interactions with climate change: 2010 Assessment) can be downloaded from www.ozone.unep.org/Assessment_Panels/, where also other reports related to ozone depletion are available. Information for the general public2 is also available at this web page.
The interest of politicians, who have had to take actions to protect the stratospheric ozone, has focused on health issues, the effects on fisheries and agriculture, and on primary production in ecosystems. As explained more fully in the above-mentioned UNEP report, it has been found that in the year 2000 at high latitudes the decrease in dry matter accumulation in land plants due to ozone depletion was up to 6%. The corresponding value for the whole Earth is much less (Rozema et al., in preparation).
DNA Damage and Repair
One of the main molecular 'targets' in cells, which are hit by UV radiation and can be damaged is DNA, but destruction is to some extent compensated by repair. Because ozone depletion has been most severe at high latitudes, where temperatures are low, an important question is to what extent repair can proceed at low temperature. We found that one kind of damage called CPD is repaired almost equally fast at 12°C as at 24°C, but only very slowly at 0°C.3–5 Repair of another kind of damage to DNA, called 6-4 photoproducts,4–6 was considerably slower already at 12°C as compared with at 24°C. An unexpected result is that, in addition, the damage process under UV-B radiation in these cells was more rapid at 24°C as compared with at 0°C. This difference was not found if the damage was inflicted by UV-C radiation. There are also other, less frequent, kinds of DNA damage caused by UV-B radiation, and other repair mechanisms.
UV-B Perception and Regulatory Mechanisms
For a long time it has been known that UV-B radiation induces effects in plants which cannot be classified as damage. Probably the most studied of these effects is induction of flavonoid synthesis, but also, e.g., the ability to carry out the DNA repair mentioned above is stimulated by irradiation with low-level UV-B, and many effects on gene activities and growth, secondary metabolism and developmental processes have been noted. It has therefore been understood that plants possess systems for perception of UV-B, just as blue and violet light is sensed by phototropin and by cryptochromes, and red and far-red light is sensed by phytochromes. That more than one system is involved is evident on spectral grounds as well as based on evidence from molecular biology (see below).
We shall in the following paragraphs focus on recent information about these regulatory effects, on the perception of UV-B radiation by the plant, and on the signal transduction pathways involved. Signaling can, in fact, be induced by DNA damage. One example of this is the inhibition of cell cycle progression and of cell division that is induced by UV, and which gives cells time to repair damage to DNA before it is duplicated.6 Another example is induction of coumestrol synthesis in Phaseolus vulgaris.7 Transduction of UV signals can also take place via various pathways involving reactive oxygen species (ROS) and/or nitrogen monoxide (NO). Here we will focus on signaling by the newly characterized UV-B receptor UVR8.
Molecular Characterization of the UVR8 Protein
A review of UVR8 was published by Jenkins,8 and since then there has been rapid progress.9–12 The UVR8 protein in the non-irradiated state is a cytosol-localized homodimer with a large number of aromatic amino acid residues: Fourteen tryptophans, 6 phenylalanines, and 4 tyrosines.10 Eighteen of the 24 aromatic residues are located at the contact surface between the monomers, which consist of only charged aromatic residues. The monomers are held together by hydrogen-bonded salt bridges especially between arginine 286 (+) and aspartic acid 107 (-), and between arginine 146 (+) and glutamine 182 (-), but there are additional salt bridges as well. When a photon of UV radiation is absorbed by an aromatic amino acid the charges are redistributed and the protein monomerizes. Tryptophans 285 and 233 have been found to be particularly important for this process.10–12
Once the protein has been split into monomers it is rapidly translocated to the nucleus and binds to COP1 protein (see below), a process followed by activation or inactivation of a great number of genes.10–12
Molecular Signaling Downstream of UVR8
In the past few years, significant progress has been made in understanding of the signaling mechanisms of plant UV-B responses, particularly the low-fluence UV-B-induced photomorphogenesis. In addition to UVR8, several downstream components have also been discovered. These include COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC1) and HY5 (ELONGATED HYPOCOTYL 5), which positively regulate the UV-B signaling, RUP1/2 (REPRESSOR OF UV-B PHOTOMORPHOGENSIS1/2) and BBX24/STO (B-BOX ZINC FINGER PROTEIN24/SALT TOLERANCE) that negatively modulate the photomorphogenic UV-B responses.
COP1 is a key signaling component of plant responses to light and negatively regulates light signaling by functioning as an ubiquitin E3 ligase.13 It represses photomorphogensis in darkness by degrading positive light-responsive transcription factors (such as HY5), and releases its suppression function by moving out of nuclei upon light illumination.14 By contrast, COP1 is stabilized and accumulated after UV-B exposure in an UVR8-dependent manner, resulting in the inhibition of degradation of the HY5 transcription factor.15 Thus, COP1 is regarded as a positive regulator in UV-B signaling, and the function of E3 ubiquitin ligase may be compromised by UV-B.16 This notion is supported by the recent observation that COP1 is constantly accumulated in white light supplemented with UV-B, suggesting that COP1 is located in the nuclei under natural light conditions.17
HY5 functions as a positive regulator in both light and UV-B signaling. It is activated by accumulation of COP1 in response to UV-B,16 contrasting to the release of its function by degradation of COP1 under white light.14 HY5 plays an important role in UV-B signaling.18 It is transcriptionally activated in an UVR8- and COP1-dependent manner15 and its activation triggers the expression of a subset of UV-B-induced genes, including those associated with UV-B tolerance.18
Beside the two positive regulators downstream of UVR8, another two groups of factors, RUP1/2 and BBX24/STO that were discovered recently, are believed to fine-tune the UV-B responses by means of feedback regulation.19,20 Both RUPs and COP1 proteins contain the conserved WD40 domain, which is responsible for the interaction of RUPs and COP1 with UVR8.19 However, unlike COP1, RUPs negatively regulate UV-B responses as the rup1 rup2 double mutant was found to be extremely sensitive to UV-B radiation.19 Moreover, this hypersensitivity largely depends on the functional UVR8 and HY5 protein, which is consistent with the evidence that UV-B induced the expression of RUP1 and RUP2 is in an UVR8-, COP1- and HY5-dependent manner.19 Though RUPs are believed to act in a negative feedback loop downstream of UVR8-COP1, it is yet unknown how the UVR8-RUP interaction results in the repression of UV-B responses.
Recently we identified a new negative factor of UV-B signaling, BBX24/STO, through characterization of the Arabidopsis bbx24/sto mutant for its responses to UV-B.20 BBX24/STO was originally found to confer salt tolerance in yeast,21 but later found to negatively regulate light signaling in Arabidopsis.22 The bbx24/sto mutant is hypersensitive to all light conditions including UV-B, suggesting a negative role of BBX24/STO in these light responses. However, the underlying molecular mechanisms appear to be very different. For example, COP1 is believed to mediate BBX24 degradation in darkness through the 26S proteasome pathway, but move away from nuclei upon light exposure.23 However, COP1 is stabilized by UV-B treatment and physically interacts with BBX24 in vivo, which leads to the accumulation of BBX24 under UV-B.20 Furthermore, our genetic analyses demonstrate that BBX24, at least partly, functions downstream of COP1 in UV-B signaling, as the response of cop1–4 is remarkably suppressed by bbx2420 and cop1–4 is a null mutant in UV-B signaling
BBX24 also interacts with HY5, both biochemically and genetically.20 We have demonstrated that BBX24 acts antagonistically with HY5 in UV-B signaling by attenuating UV-B-induced HY5 accumulation and transcriptional activity, leading to the repression of UV-B responses.20 Based on these findings, we propose that BBX24 is a new negative regulator of photomorphogenetic UV-B response that may function as a key component of the feedback regulatory module in UV-B signaling. Whether RCD1 also plays a role in this feedback regulatory module is to be determined, since BBX24 was shown earlier to interact with RCD1 (RADICAL-INDUCED CELL DEATH1).
In conclusion, our knowledge on plant UV-B responses has been greatly advanced by recent identification of several important signaling components. Briefly, in the presence of UV-B, the homodimeric UVR8 is converted to the active monomer form, resulting in its interaction with COP1. The COP1-UVR8 interaction stabilizes and activates HY5, leading to UV-B regulated gene expression and other responses such as photomorphogenesis. These UV-B responses are fine-tuned by a set of negative regulators, including BBX24/STO, RCD1 and RUP1/2. These different factors highlight a signaling cascade that mediates plant UV-B responses. However, compared with the large number of regulators in light signaling, the number of identified components in the UV-B pathway is very small. To identify more signaling components in the UV-B pathway, more diversified approaches will be required in future studies.
Spectral Considerations
As we can see from Table 1, initiation of coumestrol synthesis24 and closing of stomata26, as well as production of hydrogen peroxide by irradiation of protein25 have action spectra with peaks in the UV-C region, but they have high effectiveness also in the adjacent UV-B part of the spectrum.
Table 1. Processes in plants that can be initiated by UV-B .
| Process | Species | Peak wavel., nm | Reference |
|---|---|---|---|
| coumestrol synthesis |
Phaseolus vulgaris |
< 270 |
[24] |
| H2O2 production in vitro |
horse polyIgG in vitro |
275 |
[25] |
| stomatal closing |
Eragrostis tef |
275 |
[26] |
| anthocyanin formation |
Daucus carota |
280 |
[27] |
| CHS gene transcription |
Daucus carota |
280 and > 330 |
[28] |
| PAL gene transcription |
Daucus carota |
280 |
[29] |
| cotyledon curling |
Brassica napus |
285 |
[30] |
| MEB5.2 and LHCB1*3 regulation |
Arabidopsis thaliana |
≈285 |
[31] |
| PAL gene transcription |
Daucus carota |
290 |
[28] |
| growth inhibition |
A. thaliana |
290 |
[32] |
| anthocyanin formation |
Zea mays |
294 |
[33] |
| flavonoid accumulation |
Petroselinum hortense |
294 |
[34] |
| CHS and PDX1.3 regulation |
A. thaliana |
≈300 |
[31] |
| anthocyanin formation |
Sorghum bicolor |
302 |
[35] |
| CsPHR transcription |
Cucumis sativus |
310 |
[36] |
| CsPHR promoter activation | Cucumis sativus | 310 | [36] |
We shall now turn to those processes in the remaining parts of Table 1, which have action peaks in the UV-B band. We can see that peak wavelengths are scattered over almost all this band. Despite all the difficulties associated with in vivo action spectroscopy this range is much wider than experimental uncertainty. We can therefore be almost certain that more than one photoreceptor, not only UVR8, is involved in the capture of the radiation signal.
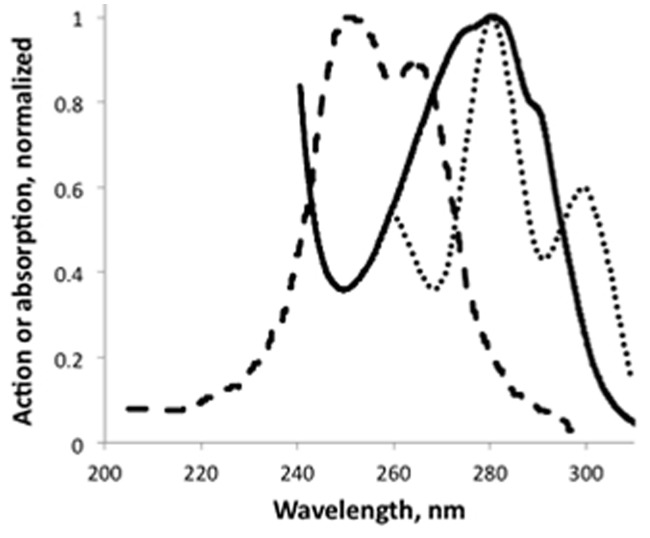
The spectral properties of the UVR8 receptor are still a bit uncertain. An experimental action spectrum37 (Fig. 1) for HY5 transcript accumulation (regarded as a UVR8 dependent process) showed a main peak near 280 nm and a smaller peak near 300 nm (uncertainty around ± 5 nm). In contrast to this, an absorption spectrum (supplementary information of Christie et al.,10) for purified UVR8 protein has a single UV-B maximum at 280 nm and two small shoulders, resulting in a much broader absorption around 280 nm. A theoretical (modeled) spectrum by Wu et al..12 is displaced to shorter wavelengths and has two maxima, as has the experimental action spectrum. Wu et al..12 comment their spectrum thus: “Taking also into account the 35-40 nm blue-shift at the current level of theory, for a single Trp residue relative to experiments, the two peaks seen for the full cluster are predicted to appear at approximately 275 and 300 nm, in very good agreement with that seen in the action spectrum of UVR8 dependent UV-B stimulation of HY5 transcription in A. thaliana leaves.”
Figure 1. UVR8 action spectrum,37 dotted, UVR8 absorption spectrum,10 solid line, and a modeled spectrum for UVR8,12 dashed.
One gets the impression that the absorption spectrum is some combination of original and wavelength-shifted action spectra, although we have not been able to verify this by a combination of only two spectra. It is not quite clear if the absorption spectrum of UVR8 published by Christie et al..10 in the supplementary material refers to the monomer or the dimer. The experimental description suggests the dimer, but perhaps it is a combination of the two.
The absorption spectrum of Christie et al...10 agrees better than the action spectrum of Brown et al..37 with in vivo action spectra for anthocyanin accumulation in carrot27 and for induction of PAL (phenyl ammonia lyase) promoter activity in carrot29 (Fig. 2). Also the action spectrum for accumulation of flavonoids in Petroselinum34 is single peaked, but more narrow than the spectrum by Brown et al..37 and displaced 14 nm to longer wavelengths. The spectrum for anthocyanin accumulation in leaves of Zea mays33 is single peaked, but more narrow than the spectrum of Brown et al., which37 is slightly two-peaked, but the minimum between the peaks is hardly significant. We regard all these action spectra related to the flavonoid pathway as compatible with the UVR8 absorption spectrum, although there seems to be some inactive absorption by UVR8 on the short-wavelength side. Possibly this latter divergence is in part due to internal screening in the plant, but this explanation is unlikely for the Petroselinum cell culture34.
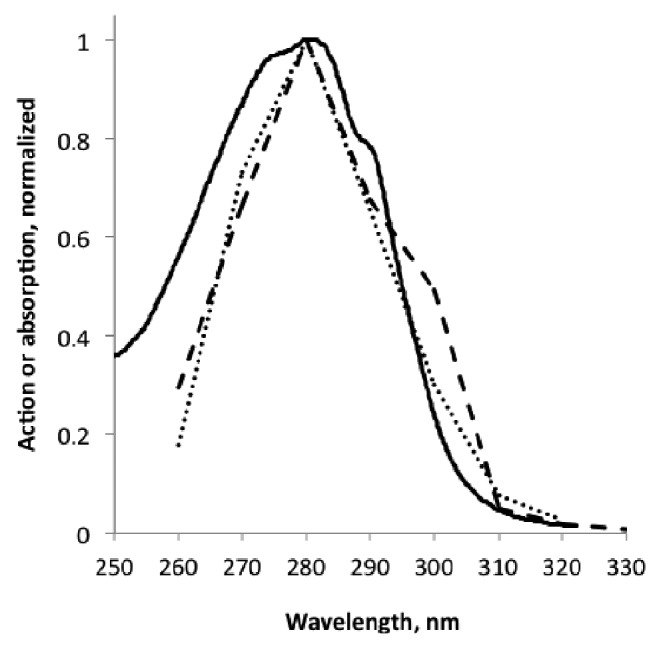
Figure 2. Comparison of the UVR8 absorption spectrum,10 solid line, with action spectra for anthocyanin accumulation in carrot,27 dashed, and for induction of PAL transcription,28 dotted.
One must not take for granted that UVR8 is the photoreceptor in all cases of UV-B signaling to this pathway. As we can see from Table 1 the action maximum for induction of anthocyanin accumulation in Sorghum35 is 302 nm. This corresponds to the long-wavelength peak in the spectrum Brown et al.,..37, and the short-wavelength peak could be hidden by screening pigments.
On the other hand, several processes not related to the flavonoid pathway also have action spectra compatible with UVR8 absorption, i.e., cotyledon curling in Brassica napus,30 growth inhibition in Arabidopsis thaliana,32 regulation of MEB5.2 and LHCB1*3 genes.31
The search for the UV-B receptors for the processes on the last four lines of Table 1 must continue. They are most likely outside the range for UVR8. UVR8 is unique among known photoreceptors in that it does not contain a non-amino acid chromophore. This property has certainly contributed to the delay in the characterization of this receptor molecule. It is also a property that reminds us of the yellow fluorescent protein38 and similar proteins of animal origin. The UV-B absorption band of UVR8 is due to 14 tryptophan, 6 phenylalanine, and 4 tyrosine residues per dimer, but tryptophans 285 are thought to have a special role in photoreception.10 This disagreement between absorbing and photofunctionally active amino acids could be thought of as an explanation for the difference between UVR8 absorption and action spectra, but this is not supported by the physiological action spectra. Neither is it supported by an attempt to decompose the absorption in components.
Green fluorescent protein has a chromophore containing one ring from a tyrosine residue, and one ring formed by a reaction between a glycine and a serine residue.38 The native green fluorescent protein can be modified in various ways to produce a range of spectra. Perhaps minor variations in UVR8 protein structure can also produce spectral variations that could account for action spectra peaking at 300 nm and greater wavelengths.
Acknowledgments
This work was supported by the grants to Prof. Shaoshan Li from the National Natural Science Foundation of China (NSFC, 31070242) and the Research Fund for the Doctoral Program of Higher Education of China (20114407110006).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/20815
References
- 1.McKenzie RL, Aucamp PJ, Bais AF, Björn LO, Ilyas M, Madronich S. Ozone depletion and climate change: impacts on UV radiation. Photochem Photobiol Sci. 2011;10:182–98. doi: 10.1039/c0pp90034f. [DOI] [PubMed] [Google Scholar]
- 2.Aucamp PJ, Björn LO, Lucas R. Questions and answers about the environmental effects of ozone depletion and its interactions with climate change: 2010 assessment. Photochem Photobiol Sci. 2011;10:301–16. doi: 10.1039/c0pp90045a. [DOI] [PubMed] [Google Scholar]
- 3.Li SS, Wang Y, Björn LO. Effects of temperature on UV-B-induced DNA damage and photorepair in Arabidopsis thaliana. J Environ Sci (China) 2004;16:173–6. [PubMed] [Google Scholar]
- 4.Li S, Paulsson M, Björn LO. Temperature-dependent formation and photorepair of DNA damage induced by UV-B radiation in suspension-cultured tobacco cells. J Photochem Photobiol B. 2002;66:67–72. doi: 10.1016/S1011-1344(01)00277-9. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Wang Y, Björn LO. Temperature effects on the formation of DNA damage in Nicotiana tabacum leaf discs induced by UV-B irradiation. Ecologic Sci. 2002;21:115–7. [Google Scholar]
- 6.Jiang L, Wang Y, Björn LO, Li S. UV-B-induced DNA damage mediates expression changes of cell cycle regulatory genes in Arabidopsis root tips. Planta. 2011;233:831–41. doi: 10.1007/s00425-010-1340-5. [DOI] [PubMed] [Google Scholar]
- 7.Beggs CJ, Stolzer-Jehle A, Wellmann E. Isoflavonoid formation as an indicator of UV stress in bean (Phaseolus vulgaris L.) Leaves: The significance of photorepair in assessing potential damage by increased solar UV-B radiation. Plant Physiol. 1985;79:630–4. doi: 10.1104/pp.79.3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–31. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 9.Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A. 2005;102:18225–30. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–6. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–6. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 12.Wu D, Hu Q, Yan Z, Chen W, Yan C, Huang X, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–9. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 13.Yi CL, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15:618–25. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–6. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 15.Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18:1975–90. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012;17:230–7. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:1397–402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:20132–7. doi: 10.1073/pnas.0914532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang L, Wang Y, Li QF, Björn LO, He JX, Li SS. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012 doi: 10.1038/cr.2012.34. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lippuner V, Cyert MS, Gasser CS. Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem. 1996;271:12859–66. doi: 10.1074/jbc.271.22.12859. [DOI] [PubMed] [Google Scholar]
- 22.Indorf M, Cordero J, Neuhaus G, Rodríguez-Franco M. Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 2007;51:563–74. doi: 10.1111/j.1365-313X.2007.03162.x. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, Wang Y, Björn LO, Li S. Arabidopsis radical-induced cell death1 is involved in UV-B signaling. Photochem Photobiol Sci. 2009;8:838–46. doi: 10.1039/b901187k. [DOI] [PubMed] [Google Scholar]
- 24.Beggs CJ, Stolzer-Jehle A, Wellmann E. Isoflavonoid formation as an indicator of UV stress in bean (Phaseolus vulgaris L.) Leaves. The significance of photorepair in assessing potential damage by increased solar UV-B radiation. Plant Physiol. 1985;79:630–4. doi: 10.1104/pp.79.3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negash L, Björn LO. Stomatal closure by ultraviolet radiation. Physiol Plant. 1986;66:360–4. doi: 10.1111/j.1399-3054.1986.tb05935.x. [DOI] [Google Scholar]
- 26.Wentworth P, Jr., Jones LH, Wentworth AD, Zhu X, Larsen NA, Wilson IA, et al. Antibody catalysis of the oxidation of water. Science. 2001;293:1806–11. doi: 10.1126/science.1062722. [DOI] [PubMed] [Google Scholar]
- 27.Takeda J, Abe S. Light-induced synthesis of anthocyanin in carrot cells in suspension- IV. the action spectrum. Photochem Photobiol. 1992;56:69–72. doi: 10.1111/j.1751-1097.1992.tb09604.x. [DOI] [Google Scholar]
- 28.Takeda J, Obi I, Yoshida K. Action spectra of phenylalanine ammonia-lyase and chalcone synthase expression in carrot cells in suspension. Physiol Plant. 1994;91:517–21. doi: 10.1111/j.1399-3054.1994.tb02982.x. [DOI] [Google Scholar]
- 29.Takeda J, Ozeki Y, Yoshida K. Action spectrum for induction of promoter activity of phenylalanine ammonia-lyase gene by UV in carrot suspension cells. Photochem Photobiol. 1997;66:464–70. doi: 10.1111/j.1751-1097.1997.tb03174.x. [DOI] [PubMed] [Google Scholar]
- 30.Gerhardt KE, Wilson MI, Greenberg BM. Ultraviolet wavelength dependence of photomorphological and photosynthetic responses in Brassica napus and Arabidopsis thaliana. Photochem Photobiol. 2005;81:1061–8. doi: 10.1562/2004-08-16-RA-276. [DOI] [PubMed] [Google Scholar]
- 31.Kalbina I, Li S, Kalbin G, Björn LO, Strid A. Two separate UV-B radiation wavelength regions control expression of different molecular markers in Arabidopsis thaliana. Funct Plant Biol. 2008;35:222–7. doi: 10.1071/FP07197. [DOI] [PubMed] [Google Scholar]
- 32.Gardner G, Lin C, Tobin EM, Loehrer H, Brinkman D. Photobiological properties of the inhibition of etiolated Arabidopsis seedling growth by ultraviolet-B irradiation. Plant Cell Environ. 2009;32:1573–83. doi: 10.1111/j.1365-3040.2009.02021.x. [DOI] [PubMed] [Google Scholar]
- 33.Beggs CJ, Wellmann E. Analysis of light-controlled anthocyanin formation in coleoptiles of Zea mays L.: The role of UV-B, blue, red, and far-red light. Photochem Photobiol. 1985;41:481–6. doi: 10.1111/j.1751-1097.1985.tb03515.x. [DOI] [Google Scholar]
- 34.Beggs CJ, Wellmann E. Photocontrol of flavonoid biosynthesis. In: Photomorphogenesis in plants, 2nd ed. (Kendrick RE, Kronenberg GHM, eds), Kluwer Academic Publishers, Dordrecht, Boston, London 1994; pp 733-51. [Google Scholar]
- 35.Yatsuhashi H, Hashimoto T, Shimizu S. Ultraviolet action spectrum for anthocyanin formation in broom sorghum first internodes. Plant Physiol. 1982;70:735–41. doi: 10.1104/pp.70.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioki M, Takahashi S, Nakajima N, Fujikura K, Tamaoki M, Saji H, et al. An unidentified ultraviolet-B-specific photoreceptor mediates transcriptional activation of the cyclobutane pyrimidine dimer photolyase gene in plants. Planta. 2008;229:25–36. doi: 10.1007/s00425-008-0803-4. [DOI] [PubMed] [Google Scholar]
- 37.Brown BA, Headland LR, Jenkins GI. UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochem Photobiol. 2009;85:1147–55. doi: 10.1111/j.1751-1097.2009.00579.x. [DOI] [PubMed] [Google Scholar]
- 38.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]