Abstract
Ayurvedic medicine plants continue to draw attention for the discovery of novel anticancer agents. Withaferin A (WA) is one such small-molecule constituent of the Ayurvedic medicine plant Withania somnifera with efficacy against cultured and xenografted human breast cancer cells. However, the mechanism underlying anticancer effect of WA is not fully understood. The present study was undertaken to determine the role of Notch signaling in anticancer effects of WA using human breast cancer cells as a model. Notably, Notch signaling is often hyperactive in human breast cancers. Exposure of MDA-MB-231 and MCF-7 human breast cancer cells to pharmacological concentrations of WA resulted in cleavage (activation) of Notch2 as well as Notch4, which was accompanied by transcriptional activation of Notch as evidenced by RBP-Jk, HES-1A/B, and HEY-1 luciferase reporter assays. On the other hand, WA treatment caused a decrease in levels of both transmembrane and cleaved Notch1. The WA -mediated activation of Notch was associated with induction of γ-secretase complex components Presenilin1 and/or Nicastrin. Inhibition of MDA-MB-231 and MDA-MB-468 cell migration resulting from WA exposure was significantly augmented by knockdown of Notch2 as well as Notch4 protein. Activation of Notch2 was not observed in cells treated with withanone or withanolide A, which are naturally-occurring structural analogues of WA. The results of the present study indicate that WA treatment activates Notch2 and Notch4, which impede inhibitory effect of WA on breast cancer cell migration.
Keywords: withaferin A, Notch2, Notch4, breast cancer, cell migration
Introduction
Breast cancer continues to be a leading cause of cancer-related deaths among women worldwide despite screening efforts for early detection of the disease as well as major chemotherapeutic advances involving targeted therapies [1-3]. Novel strategies for prevention and treatment of breast cancer are desirable to diminish disease-related cost, mortality, and morbidity associated with this disease. Constituents of Ayurvedic medicine, which has been practiced in India for thousands of years for the treatment of different ailments, continue to gain interest for the discovery of novel anticancer agents [4]. Withania somnifera (commonly known as Ashwagandha or Indian winter cherry) is one such medicinal plant with a broad spectrum pharmacological effects in experimental models, including cardioprotection from ischemia reperfusion injury [5], inhibition of 6-hydroxydopamine-induced Parkinsonism [6], antibacterial properties [7], immunomodulatory effects [8], and anticancer effects [9-12]. For instance, oral feeding of leaf extract of Ashwagandha resulted in growth retardation of HT1080 human fibrosarcoma in athymic mice [10]. Likewise, benzo[a]pyrene-induced forestomach tumor incidence and multiplicity were significantly inhibited by dietary administration of Withania somnifera root [11].
Anticancer effect of Withania somnifera is believed to be due to withanolides, including withaferin A (WA) [13-17] . WA treatment was shown to cause destruction of Ehrlich ascites tumor cells in vivo by causing immune activation [13]. Oral administration of WA for 14 weeks resulted in complete protection against 7,12-dimethylbenz[a]anthracene-induced oral carcinogenesis in hamsters [17]. The WA-mediated inhibition of human cancer cells implanted in athymic mice has also been reported [15, 16]. For example, studies from our own laboratory have shown WA-mediated inhibition of MDA-MB-231 human breast cancer xenograft growth in female athymic mice [16]. Moreover, the WA treatment inhibited breast cancer invasion and metastasis in vivo [18].
The mechanism underlying anticancer effect of WA is not fully understood, but the known effects following treatment with this agent in cultured cancer cells comprise of G2/M phase and mitotic arrest [19], reactive oxygen species-dependent apoptosis [20, 21], and suppression of multiple oncogenic pathways including signal transducer and activator of transcription 3 [22], estrogen receptor-α [23], and nuclear factor-κB [24]. Because pathogenesis of cancer is complex often involving activation of multiple oncogenes and deregulation of various checkpoints, targeting of multiple pathways with a single small-molecule is desirable for clinical management of cancer. Agent active against a single molecular target or pathway may have limited clinical utility as exemplified by estrogen receptor antagonists [25].
The Notch pathway regulates expression of genes involved in cell fate determination including proliferation and differentiation [26, 27]. The Notch pathway is implicated in mammary carcinogenesis [28-31]. The WA was previously shown to inhibit Notch1 activation in human colon cancer cells [32]. The Notch signaling is quite complex involving interplay between four receptors (Notch1-Notch4) and five ligands (Jagged 1, Jagged2, Delta-like ligands- 1,3, and 4). The primary objective of the present study was to determine the role of Notch2 and Notch4 in anticancer effects of WA using human breast cancer cells as a model.
Materials and methods
Reagents
The WA was purchased from Enzo Life Sciences (Plymouth Meeting, PA), whereas its naturally-occurring structural analogues (withanone and withanolide A) were purchased from ChromaDex (Irvine, CA). Dimethyl sulfoxide (DMSO), 4′,6-diamidino-2-phenylindole (DAPI), and anti-actin antibody were purchased from Sigma-Aldrich (St. Louis, MO). Reagents for cell culture, including media, antibiotic mixture, fetal bovine serum, and Alexa Flour-488 conjugated anti-rabbit antibody were purchased from Invitrogen-Life Technologies (Carlsbad, CA). The antibody against cleaved Notch1, Notch2, Jagged1, Jagged2, Presenilin1, and Nicastrin were from Cell Signaling Technology (Danvers, MA). An antibody specific for detection of cleaved Notch2 was from EMD Millipore (Billerica, MA). Anti-cleaved Notch1 antibody used for immunofluorescence microscopy was purchased from Abcam (Cambridge, MA). The antibodies against Notch1 and Notch4 (this antibody recognizes both transmembrane and cleaved forms of the protein), were from Santa Cruz Biotechnology (Santa Cruz, CA). Control non-specific small interfering RNA (siRNA) was purchased from Qiagen (Germantown, MD). The Notch2-targeted siRNA, Notch4-targeted siRNA, control small-hairpin RNA (shRNA), and Notch2-targeted shRNA were from Santa Cruz Biotechnology. Transwell™ chamber was purchased from Corning (Corning, NY).
Cell lines
The MDA-MB-231, MDA-MB-468, and MCF-7 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained as described by us previously [16] or recommended by the supplier. The MDA-MB-231 cells were stably transfected with Notch2 shRNA and control shRNA as described elsewhere [33]. Stock solutions of WA and its analogues were prepared in DMSO (final concentration <0.1%), and an equal volume of DMSO was added to the controls.
Western blotting
After desired treatment, the cells were collected and processed for immunoblotting as described by us previously [33, 34]. Immunoreactive bands were visualized using enhanced chemiluminescence reagent.
Immunocytochemistry analysis
The MDA-MB-231 or MCF-7 cells (1 × 105) were plated on coverslips and allowed to attach by overnight incubation. The cells were treated with different concentrations of WA for 24 h, fixed with 2% paraformaldehyde for 1 h at room temperature, permeabilized with 0.5% Triton X-100 for 10 min, and blocked with phosphate-buffered saline (PBS) supplemented with 0.5% bovine serum albumin and 0.15% glycine for 1 h. The cells were incubated overnight with antibody against cleaved Notch1 or cleaved Notch2 at 4°C, and then treated with 2 μg/ml of Alexa Fluor 488-conjugated secondary antibody for 1 h at room temperature. The cells were washed with PBS, counterstained with DAPI for 5 min at room temperature, mounted and observed under a Leica DC300F fluorescence microscope at ×100 objective magnification.
Luciferase reporter assay
RBP-Jk luciferase reporter kit (SABiosciences-Qiagen) was used to determine the effect of WA treatment on transcriptional activity of Notch. The HES-1A/B and HEY-1 luciferase reporter constructs were generously provided by Dr. Kimberly E. Foreman (Department of Pathology, Loyola University Medical Center, Maywood, IL) [35]. Luciferase activity was determined as described by us previously [33].
Reverse transcription (RT)-PCR and real-time quantitative PCR
Total RNA from control and WA-treated cells was isolated using RNeasy kit from Qiagen. cDNA was synthesized and reversed transcribed using Oligo(dT) primer and SuperScript III Reverse Transcriptase. The PCR was performed using GoTaq Green Master Mix (Promega, Madison, WI) and primers as described by us previously [36]. The RT-PCR primers and amplification conditions for Notch4 were: forward- 5′-TAGGGCTCCCCAGCTCTC-3′, reverse- 5′-GGCAGGTGCCCCCATT-3′ (95°C for 5 minutes followed by 35 cycles of 95°C for 30 seconds, 60°C for 1 minute, and 72°C for 30 seconds). Quantitative real-time PCR was performed using SYBR Green master mix (Applied Biosystems, Foster City, CA) on ABI StepOnePlus system (Applied Biosystems). Relative gene expression was normalized against GAPDH.
Cell growth assay
Cells were seeded into 96-well plates and allowed to attach. After 48 h of treatment with DMSO or WA, cell proliferation was determined using the CellTiter96 Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer's protocol.
Cell migration assay
The MDA-MB-231 or MDA-MB-468 cells were transfected with desired siRNA. Cell migration assay was performed as described by us previously [33, 36] using a Boyden chamber containing an 8 μm filter. Motile cells on the bottom face of the filter were fixed with methanol and stained with hematoxylin and eosin. At least 3 non-overlapping areas on each filter were scored for cell migration.
Wound healing assay
The MDA-MB-231 cells stably transfected with control shRNA or Notch2-targeted shRNA were seeded in 6-well plates. A monolayer of confluent cells was scratched with a pipette tip. The wounded cells were washed with PBS and incubated with RPMI-1640 medium containing 1% fetal bovine serum, puromycin (1 μg/mL), 1 mM thymidine, and desired concentrations of WA. The cells were allowed to migrate for 16 h, fixed with methanol, and stained with Giemsa staining solution. At least 3 non-overlapping areas were examined for wound healing.
Results
Effect of WA treatment on cleavage of Notch1, Notch2, and Notch4 in human breast cancer cells
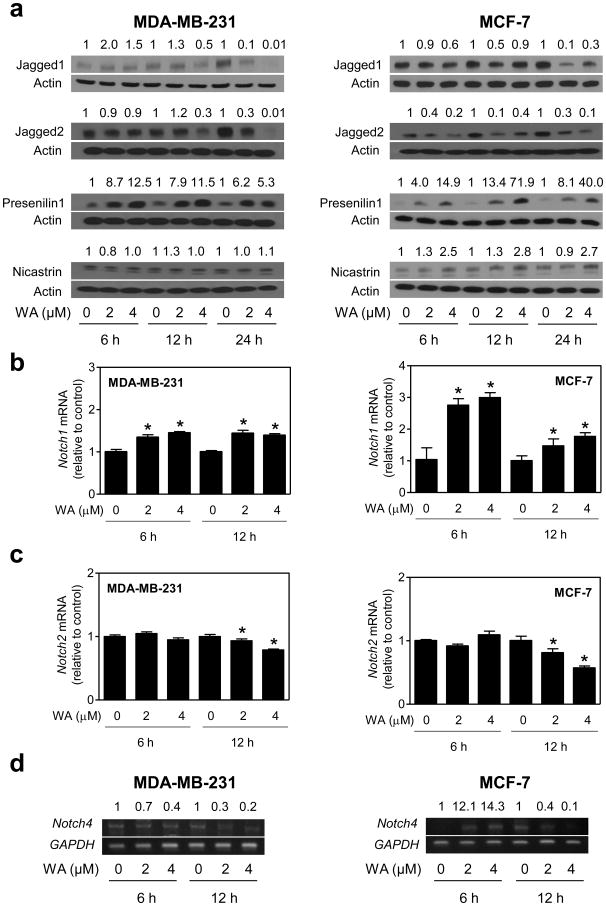
Final step of Notch activation involves cleavage of the receptor at a site located within the transmembrane domain mediated by the γ-secretase complex. Consistent with published results in colon cancer cells [32], the WA treatment resulted in a marked decrease in levels of transmembrane as well as cleaved (active) Notch1 in MDA-MB-231 and MCF-7 cells, which respectively are well-studied representatives of estrogen-independent and estrogen-responsive human breast cancer cells (Fig. 1a). On the other hand, levels of cleaved Notch2 as well as cleaved Notch4 were increased markedly upon a similar treatment with WA in both cell lines (Fig. 1a). Effect on cleaved Notch2 was relatively more pronounced in the MDA-MB-231 cell line compared with MCF-7, whereas an opposite trend was discernible on cleavage of Notch4. The WA-mediated cleavage of Notch2 and Notch4 was accompanied by a decrease in levels of respective transmembrane proteins (Fig. 1a). Opposing effects of WA treatment on levels of cleaved Notch1 (Fig. 1b) and cleaved Notch2 (Fig. 1c) was confirmed by immunocytochemistry. Thus, WA treatment elicited differential effects on cleavage of Notch1 (decrease) versus Notch2 and Notch4 (increase) in human breast cancer cells, and this response was neither cell line-specific nor influenced by the estrogen receptor status.
Fig. 1.
Effect of WA treatment on activation of Notch isoforms in human breast cancer cells. a Western blotting for transmembrane (uncleaved) and cleaved Notch isoforms using lysates from MDA-MB-231 or MCF-7 cells following treatment with DMSO (control) or WA (2 or 4 μM) for the indicated time periods. Blots were stripped and reprobed with anti-actin antibody. Numbers on top of the bands represent changes in protein levels relative to corresponding DMSO-treated controls. b Immunofluorescence microscopic analysis for cleaved Notch1 in MDA-MB-231 or MCF-7 cells after 24 hours of treatment with DMSO or 2 μM WA. c Immunofluorescence microscopic analysis for cleaved Notch2 in MDA-MB-231 or MCF-7 cells after 24 hours of treatment with DMSO or 2 μM WA. Each experiment was repeated at least twice and representative data from one such experiment are shown.
WA treatment caused transcriptional activation of Notch
Because WA treatment exhibited opposing effects on cleavage of Notch1 versus Notch2 and Notch4, it was of interest to determine overall impact on transcriptional activity of Notch. We approached this question by determining the effect of WA treatment on luciferase activities associated with RBP-Jk and downstream targets of Notch including HES-1A/B and HEY-1. Exposure of MDA-MB-231 and MCF-7 cells to WA resulted in a statistically significant increase in RBP-Jk-associated luciferase activity, which was accompanied by transcriptional activation of Notch downstream target HES-1A/B (Fig. 2). However, the effect of WA treatment on HEY-1 luciferase activity was different between MDA-MB-231 and MCF-7 cells. The WA-treated MDA-MB-231 cells exhibited a dose- and time-dependent increase in HEY-1 luciferase activity (Fig. 2). On the other hand, the WA-mediated increase in HEY-1 luciferase reporter activity was transient in MCF-7 cells with the increase seen only at 8 h time point. Mechanisms underlying regulation of HEY-1 gene expression are not fully understood but its expression can also be regulated by Wnt10b as exemplified in osteosarcoma cells [37]. It is possible that the decrease in HEY-1 luciferase activity seen at the 24 h time point in WA-treated MCF-7 cells is a reflection of Wnt10b suppression. Collectively, these data indicated that the WA -mediated cleavage of Notch2 and Notch4 resulted in transcriptional activation of Notch in both MDA-MB-231 and MCF-7 cells.
Fig. 2.
WA treatment causes transcriptional activation of Notch in human breast cancer cells. RBP-Jk-, HES-1A/B- and HEY-1-associated luciferase reporter activity in MDA-MB-231 and MCF-7 cells after 8 hours or 24 hours of treatment with DMSO (control) or the indicated concentrations of WA. Luciferase activity was normalized against protein concentration and renilla luciferase units. Results shown are mean ± SD (n = 3). *Significantly different (P < 0.05) compared with corresponding DMSO-treated control by one-way ANOVA with Dunnett's adjustment. The results are representative of at least two independent experiments.
Effects of WA treatment on Jagged1, Jagged2, Presenilin1, and Nicastrin protein levels
To further probe into the mechanism by which WA treatment increased cleavage of Notch2 and Notch4, we performed western blotting for Jagged1, Jagged2, and γ-secretase complex components Presenilin1 and Nicastrin using lysates from control (DMSO-treated) and WA-treated MDA-MB-231 and MCF-7 cells (Fig. 3a). Effect of WA treatment on protein levels of Jagged1 was different between MDA-MB-231 and MCF-7 cells. In MCF-7 cells, WA caused suppression of both Jagged1 and Jagged2 protein expression as early as 6 h after treatment, and this effect persisted for the duration of the experiment (Fig. 3a). A marked decrease in levels of Jagged1 and Jagged2 proteins in MDA-MB-231 cells at both concentrations of WA was evident only at the 24 h time point (Fig. 3a). At the same time, WA treatment resulted in a marked increase in Presenilin1 (both cell lines) and Nicastrin (MCF-7 cells) protein levels in MDA-MB-231 and MCF-7 cells and this effect generally persisted for the duration of the experiment (Fig. 3a). To our surprise the levels of Notch1 mRNA were increased after treatment with WA in both cell lines (Fig. 3b), whereas a decrease in Notch2 mRNA was discernible at 12 h time point (Fig. 3c). The level of Notch4 mRNA was decreased after treatment with WA in MDA-MB-231 in a dose- and time-dependent manner as revealed by RT-PCR (Fig. 3d). To the contrary, the WA-treated MCF-7 cells exhibited a biphasic response with an increase in Notch4 mRNA level at 6 h time point followed by its suppression after 12 h of treatment (Fig. 3d). Because of variability in mRNA expression of different Notch we conclude that induction of Presenilin1 and Nicastrin likely accounts for WA-mediated cleavage of Notch2 and Notch4. However, additional work is necessary to substantiate this mechanistic possibility.
Fig. 3.
Effect of WA treatment on protein levels of Notch ligands and γ-secretase complex components. a Western blotting for Jagged1, Jagged2, Presenilin1, and Nicastrin using lysates from MDA-MB-231 and MCF-7 cells following treatment with DMSO (control) or WA (2 or 4 μM) for the indicated time periods. Blots were stripped and reprobed with anti-actin antibody. Numbers on top of the bands represent changes in protein levels relative to corresponding DMSO-treated control. Real-time quantitative PCR for Notch1 (panel b) and Notch2 mRNA (panel c), and RT-PCR for Notch4 mRNA (panel d) in MDA-MB-231 and MCF-7 cells after 6 hours or 12 hours of treatment with DMSO (control) or the indicated concentrations of WA. Results shown are mean ± SD (n = 3). *Significantly different (P < 0.05) compared with corresponding DMSO-treated control by one-way ANOVA with Dunnett's adjustment. The results are representative of at least two independent experiments.
Notch2 knockdown augmented WA -mediated inhibition of cell viability and cell migration
We next proceeded to determine the role of Notch2 in WA-mediated inhibition of cell proliferation and cell migration by using MDA-MB-231 (a highly aggressive cell line with mesenchymal phenotype) and MDA-MB-468 cells (a pre-mesenchymal cell line). The MCF-7 cells were excluded from this experiment as they migrate poorly. The WA -mediated cleavage of Notch2 and Notch4 was also observed in the MDA-MB-468 cell line (results not shown). Level of transmembrane (uncleaved) Notch2 was decreased by >99% upon transfection of the MDA-MB-231 and MDA-MB-468 cells with a Notch2-trageted siRNA when compared with control siRNA transfected cells (Fig. 4a). Transient transfection with the Notch2-targeted siRNA alone resulted in a significant suppression of cell proliferation (Fig. 4b) as well cell migration (Fig. 4c) in both cell lines. Moreover, the WA-mediated inhibition of MDA-MB-231 and MDA-MB-468 cell proliferation (Fig. 4b) as well as cell migration (Fig. 4d) was significantly augmented by RNA interference of Notch2. However, this augmentation effect was much more pronounced on cell migration (Fig. 4d) than on cell proliferation (Fig. 4b).
Fig. 4.

Notch2 knockdown augments WA -mediated inhibition of cell migration. a Immunoblotting for transmembrane (uncleaved) Notch2 protein using lysates from MDA-MB-231 and MDA-MB-468 cells transiently transfected with control siRNA or Notch2-targeted siRNA. b Proliferation of MDA-MB-231 and MDA-MB-468 cells transiently transfected with control siRNA or Notch2-targeted siRNA and treated for 48 hours with DMSO (control) or different concentrations of WA. Results shown are mean ± S.D (n = 4). c Cell migration by MDA-MB-231 or MDA-MB-468 cells transiently transfected with control siRNA or Notch2-targeted siRNA and treated for 24 hours with DMSO or 2 μM WA (×100 magnification). d Quantitation of cell migration from the experimental data shown in panel c. Results shown are mean ± S.D (n = 3). *Significantly different (P<0.05) acompared with corresponding DMSO-treated control and bbetween control siRNA transfected cells and Notch2 siRNA transfected cells by one-way ANOVA with Tukey's (panel b) and/or Bonferroni's (panel d) multiple comparison tests. Similar results were observed in two independent experiments in each cell line. Results shown are relative to DMSO-treated control siRNA transfected cells (b and d).
We used MDA-MB-231 cells with stable transfection with a Notch2-targeted shRNA to further test its role in WA mediated inhibition of cell migration. Level of transmembrane Notch2 protein was decreased by >99% in MDA-MB-231 cells stably transfected with the Notch2-targeted shRNA compared with control shRNA-transfected cells (Fig. 5a). Similar to the results using siRNA, the WA -mediated inhibition of MDA-MB-231 cell proliferation was modestly increased upon stable knockdown of Notch2 at least at the 1 μM concentration (Fig. 5b). In addition, the MDA-MB-231 cells with stable transfection with Notch2-targeted shRNA were significantly more sensitive to WA-mediated inhibition of wound migration compared with control shRNA transfected cells (Fig. 5c,d). These results indicated that Notch2 activation by WA partially abrogated its inhibitory effect on cell migration.
Fig. 5.
Effect of WA treatment on cell viability and wound healing after stable knockdown of Notch2. a Western blotting for Notch2 in MDA-MB-231 cells stably transfected with control shRNA or Notch2-targeted shRNA. b Cell proliferation in MDA-MB-231 cells transfected with control shRNA or Notch2-targeted shRNA and treated for 48 hours with DMSO (control) or WA (1, 2 or 4 μM). Results shown are mean ± S.D (n = 4). c Representative microscopic images depicting wound healing in MDA-MB-231 cells transfected with control shRNA or Notch2-targeted shRNA and treated for 16 hours with DMSO (control) or WA (1 or 2 μM). d Quantitation of wound healing in MDA-MB-231 cells transfected with control shRNA or Notch2-targeted shRNA and treated for 16 hours with DMSO (control) and WA (1 or 2 μM) (× 100 magnification). Results shown are mean ± SD (n = 3). Significantly different (P < 0.05) compared with acorresponding DMSO-treated control, and bbetween cells transfected with control shRNA and Notch2 shRNA by one-way ANOVA followed by Tukey's multiple comparison test. The results are representative of at least two independent experiments. Results shown are relative to DMSO-treated control shRNA transfected cells (b and d).
Notch4 knockdown augmented WA-mediated inhibition of cell migration
We next studied the impact of Notch4 activation on WA's ability to inhibit cell proliferation and cell migration using MDA-MB-231 and MDA-MB-468 cells. Level of transmembrane Notch4 protein was decreased by 40% and >99% upon transient transfection of MDA-MB-231 and MDA-MB-468 cells with a Notch4-targeted siRNA, respectively (Fig. 6a). Unlike Notch2 siRNA (Fig. 4b), RNA interference of Notch4 alone had minimal effect on cell proliferation in either cell line (Fig. 6b). Furthermore, inhibition of cell proliferation resulting from WA exposure was not affected by RNA interference of Notch4 in either cell line (Fig. 6b). On the other hand, WA-mediated inhibition of cell migration was significantly augmented by Notch4 knockdown (Fig. 6c,d). This augmentation was relatively more pronounced in the MDA-MB-468 cell line than in the MDA-MB-231 cells possibly due to greater knockdown of the Notch4 protein in the former cell line. These results indicated that knockdown of Notch4 protein augmented WA-mediated inhibition of cell migration.
Fig. 6.

Knockdown of Notch4 augmented WA-mediated inhibition of cell migration. a Western blotting for transmembrane (uncleaved) Notch4 protein using lysates from the MDA-MB-231 and MDA-MB-468 cells transiently transfected with control siRNA or Notch4-targeted siRNA. b Proliferation of MDA-MB-231 and MDA-MB-468 cells transiently transfected with control siRNA or Notch4-targeted siRNA and treated for 48 hours with DMSO (control) or different concentrations of WA. Results shown are mean ± S.D (n = 4). c Cell migration by MDA-MB-231 or MDA-MB-468 cells transiently transfected with control siRNA or Notch4-targeted siRNA and treated for 24 hours with DMSO or 2 μM WA (×100 magnification). d Quantitation of cell migration from the experimental data shown in panel c. Results shown are mean ± S.D (n = 3). * Significantly different (P<0.05) acompared with corresponding DMSO-treated control, and bbetween control siRN A transfected and Notch4 siRNA transfected cells by one-way ANOVA with Tukey's (panel b) and/or Bonferroni's (panel d) multiple comparison test. Similar results were observed in two independent experiments in each cell line. Results shown are relative to DMSO-treated control siRNA transfected cells (b and d).
Effect of WA analogues on cell migration and Notch2 activation
We used a pair of WA analogues (structures shown in Fig. 7a) to test whether inhibition of cell migration and/or activation of Notch2 were unique to WA Initially, we compared WA, WE, and WLA for their efficacy against cell migration using MDA-MB-231 cells. The WE and WLA were practically ineffective against MDA-MB-231 cell migration (Fig. 7b). In addition, neither WE nor WLA was able to cause an increase in levels of cleaved Notch2 at least in the MDA-MB-231 and MCF-7 cells except for a modest increase seen with WE in MCF-7 cells at the 1 and 2 μM concentrations (Fig. 7c). These results indicated that even a subtle change in the withanolide structure could have a marked impact on its activity.
Fig. 7.
Effect of WA analogues on cell migration and Notch2 activation. a Chemical structures of withaferin A (WA), withanone (WE), and withanolide A (WLA). b Migration of MDA-MB-231 cells after 24 hours of treatment with DMSO (control) or the indicated concentrations of WA, WE, and WLA. Results shown are mean ± SD (n = 4). *Significantly different (P < 0.05) compared with corresponding DMSO-treated control by one-way ANOVA with Dunnett's adjustment. c Western blotting for cleaved Notch2 protein using lysates from MDA-MB-231 and MCF-7 cells following 24 hours of treatment with DMSO (control) or different concentrations of WA, WE, and WLA. Blots were stripped and reprobed with anti-actin antibody. Numbers on top of the bands represent changes in protein levels relative to corresponding DMSO-treated controls. Similar results were observed in two independent experiments in each cell line. Representative data from a single experiment are shown.
Discussion
Notch activation is linked to mammary carcinogenesis in both animal models and humans [28-31]. For example, overexpression of activated Notch1 and Notch3 in transgenic mice was shown to block mammary gland development but induce tumorigenesis of the breast [28]. In humans, high level expression of Notch1 and its ligands is associated with poor outcome [29-31]. Furthermore, Jagged 1 expression is associated with recurrence in lymph node-negative breast cancer [31]. Notch1 is involved in migration and invasion of human breast cancer cells [38], whereas Notch4 receptor signaling is implicated in regulation of breast cancer stem cell activity [39]. In addition, induction of Notch1 activity and function by ErbB2 as well as cross-talk between Notch and estrogen receptor has also been demonstrated [40,41]. Impetus to determine the effect of WA on Notch2 and Notch4 stemmed from the observations that this agent inhibits Notch1 activation in human colon cancer cells [32]. The present study indicates that while WA treatment inhibits cleavage of Notch1 consistent with the results observed in colon cancer cells [32], it is an activator of Notch2 as well as Notch4 in breast cancer cells. Whether or not WA activates Notch2 and/or Notch4 in colon or other cancer cells remains to be determined, but knockdown of these proteins clearly augments inhibitory effect of WA on cell migration. Importantly, the WA-mediated activation of Notch2 and Notch4 in breast cancer cells is evident at plasma achievable concentration of 2 μM [18].
The mechanism by which WA downregulates expression of Notch1 and inhibits its activation is unclear but this effect is not due to its transcriptional repression based on results shown on Fig. 3b. It is possible that WA treatment promotes proteasomal degradation of Notch1 and/or affects its translation. A role for prolyl-isomerase Pin1 in activation of Notch1 has also been proposed [42]. Pin1 has been shown to bind with Notch1 and promote its cleavage by the γ-secretase [42]. Interestingly, Pin1 is a direct target of Notch1 creating a positive feedback loop. Consistent with the Pin1-Notch1 cooperation, levels of these proteins correlate in breast cancer [42]. Interestingly, our unpublished studies indicate that WA treatment downregulates Pin1 protein expression at least in the MCF-7 cell line (Singh SV, unpublished results). It is possible that Pin1 downregulation contributes to the suppression of transmembrane and/or cleaved Notch1 after treatment with WA. Additional work is needed to probe into these mechanisms.
We observed cell line-specific differences in effect of WA on extent of Notch activation. For example, the WA-mediated increase in level of cleaved Notch2 is relatively more pronounced in the MDA-MB-231 cells (up to 16.7-fold increase compared with DMSO-treated control), which has mutant p53, than in the wild-type p53 expressing MCF-7 cell line (up to 3.4-fold increase over DMSO-treated control). In contrast, WA-mediated cleavage of Notch4 was relatively more pronounced in the MCF-7 cells compared with MDA-MB-231 cells. Similar cell line-specific differences are also discernible with respect to effect of WA on γ-secretase complex components. The molecular basis for these cell line-specific differences remains to be determined, but may be related to differences in p53 and/or Wnt/β-catenin status. The Wnt/β-catenin is constitutively active in the MCF-7 cells but not in the MDA-MB-231 cell line [43].
The present study reveals that Notch2 and Notch4 knockdown partially inhibits cell migration. Under these knockdown conditions, WA further inhibits cell migration. These results indicate that WA likely affects other pathways to inhibit breast cancer cell migration. Suppression of urokinase plasminogen activator (uPA) system consisting of the serine protease uPA and its glycolipid-anchored receptor (uPAR) is one such possibility to explain WA-mediated inhibition of cell migration. Previous studies have indicated that the uPA/uPAR system is causally involved not only in remodeling of the extracellular matrix but also enhancing cell proliferation and migration [44]. Our unpublished studies indicate that WA treatment suppresses uPA and uPAR protein levels in breast cancer cells (Lee J, Singh SV, unpublished results). The uPA is a direct transcriptional target of Jagged1-Notch signaling in breast cancer cells [45]. High uPA expression also correlates with breast cancer recurrence and metastasis [46]. Further studies are needed to experimentally address whether augmentation of WA-mediated inhibition of cell migration is related to suppression of the uPA/uPAR pathway.
Acknowledgments
Financial Disclosure: Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R01CA142604. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- WA
withaferin A
- WE
withanone
- WLA
withanolide A
- DMSO
dimethyl sulfoxide
- DAPI
4′,6-diamidino-2-phenylindole
- PBS
phosphate-buffered saline
- siRNA
small interfering RNA
- shRNA
small-hairpin RNA
- uPA
urokinase-type plasminogen activator
- uPAR
uPA receptor
Footnotes
Conflict of interest: None of the authors has any conflict of interest.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez RH. Present and future evolution of advanced breast cancer therapy. Breast Cancer Res. 2010;12(2):S1. doi: 10.1186/bcr2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins MJ, Baselga J. Breast cancer in 2010: Novel targets and therapies for a personalized approach. Nat Rev Clin Oncol. 2011;8:65–66. doi: 10.1038/nrclinonc.2010.217. [DOI] [PubMed] [Google Scholar]
- 4.Garodia P, Ichikawa H, Malani N, Sethi G, Aggarwal BB. From ancient medicine to modern medicine: ayurvedic concepts of health and their role in inflammation and cancer. J Soc Integr Oncol. 2007;5:25–37. doi: 10.2310/7200.2006.029. [DOI] [PubMed] [Google Scholar]
- 5.Gupta SK, Mohanty I, Talwar KK, Dinda A, Joshi S, Bansal P, Saxena A, Arya DS. Cardioprotection from ischemia and reperfusion injury by Withania somnifera: a hemodynamic, biochemical and histopathological assessment. Mol Cell Biochem. 2004;260:39–47. doi: 10.1023/b:mcbi.0000026051.16803.03. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad M, Saleem S, Ahmad AS, Ansari MA, Yousuf S, Hoda MN, Islam F. Neuroprotective effects of Withania somnifera on 6-hydroxydopamine induced Parkinsonism in rats. Hum Exp Toxicol. 2005;24:137–147. doi: 10.1191/0960327105ht509oa. [DOI] [PubMed] [Google Scholar]
- 7.Owais M, Sharad KS, Shehbaz A, Saleemuddin M. Antibacterial efficacy of Withania somnifera (ashwagandha) an indigenous medicinal plant against experimental murine salmonellosis. Phytomedicine. 2005;12:229–235. doi: 10.1016/j.phymed.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Rasool M, Varalakshmi P. Immunomodulatory role of Withania somnifera root powder on experimental induced inflammation: An in vivo and in vitro study. Vascul Pharmacol. 2006;44:406–410. doi: 10.1016/j.vph.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Devi PU, Sharada AC, Solomon FE. Antitumor and radiosensitizing effects of Withania somnifera (Ashwagandha) on a transplantable mouse tumor, Sarcoma-180. Indian J Exp Biol. 1993;31:607–611. [PubMed] [Google Scholar]
- 10.Widodo N, Kaur K, Shrestha BG, Takagi Y, Ishii T, Wadhwa R, Kaul SC. Selective killing of cancer cells by leaf extract of Ashwagandha: identification of a tumor-inhibitory factor and the first molecular insights to its effect. Clin Cancer Res. 2007;13:2298–2306. doi: 10.1158/1078-0432.CCR-06-0948. [DOI] [PubMed] [Google Scholar]
- 11.Padmavathi B, Rath PC, Rao AR, Singh RP. Roots of Withania somnifera inhibit forestomach and skin carcinogenesis in mice. Evid Based Complement Alternat Med. 2005;2:99–105. doi: 10.1093/ecam/neh064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leyon PV, Kuttan G. Effect of Withania somnifera on B16F-10 melanoma induced metastasis in mice. Phytother Res. 2004;18:118–122. doi: 10.1002/ptr.1378. [DOI] [PubMed] [Google Scholar]
- 13.Shohat B, Joshua H. Effect of withaferin A on Ehrlich ascites tumor cells. II. Target tumor cell destruction in vivo by immune activation. Int J Cancer. 1971;8:487–496. doi: 10.1002/ijc.2910080317. [DOI] [PubMed] [Google Scholar]
- 14.Devi PU, Kamath R, Rao BS. Radiosensitization of a mouse melanoma by withaferin A: in vivo studies. Indian J Exp Biol. 2000;38:432–437. [PubMed] [Google Scholar]
- 15.Srinivasan S, Ranga RS, Burikhanov R, Han SS, Damodaran C. Par-4-dependent apoptosis by the dietary compound withaferin A in prostate cancer cells. Cancer Res. 2007;67:246–253. doi: 10.1158/0008-5472.CAN-06-2430. [DOI] [PubMed] [Google Scholar]
- 16.Stan SD, Hahm ER, Warin R, Singh SV. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res. 2008;68:7661–7669. doi: 10.1158/0008-5472.CAN-08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manoharan S, Panjamurthy K, Menon VP, Balakrishnan S, Alias LM. Protective effect of Withaferin-A on tumour formation in 7,12-dimethylbenz[a]anthracene induced oral carcinogenesis in hamsters. Indian J Exp Biol. 2009;47:16–23. [PubMed] [Google Scholar]
- 18.Thaiparambil JT, Bender L, Ganesh T, Kline E, Patel P, Liu Y, Tighiouart M, Vertino PM, Harvey RD, Garcia A, Marcus AI. Withaferin A inhibits breast cancer invasion and metastasis at sub-cytotoxic doses by inducing vimentin disassembly and serine 56 phosphorylation. Int J Cancer. 2011;129:2744–2755. doi: 10.1002/ijc.25938. [DOI] [PubMed] [Google Scholar]
- 19.Stan SD, Zeng Y, Singh SV. Ayurvedic medicine constituent withaferin a causes G2 and M phase cell cycle arrest in human breast cancer cells. Nutr Cancer. 2008;60:51–60. doi: 10.1080/01635580802381477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahm ER, Moura MB, Kelley EE, Van Houten B, Shiva S, Singh SV. Withaferin A-induced apoptosis in human breast cancer cells is mediated by reactive oxygen species. PLoS One. 2011;6:e23354. doi: 10.1371/journal.pone.0023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayola E, Gallerne C, Esposti DD, Martel C, Pervaiz S, Larue L, Debuire B, Lemoine A, Brenner C, Lemaire C. Withaferin A induces apoptosis in human melanoma cells through generation of reactive oxygen species and down-regulation of Bcl-2. Apoptosis. 2011;16:1014–1027. doi: 10.1007/s10495-011-0625-x. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Hahm ER, Singh SV. Withaferin A inhibits activation of signal transducer and activator of transcription 3 in human breast cancer cells. Carcinogenesis. 2010;31:1991–1998. doi: 10.1093/carcin/bgq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahm ER, Lee J, Huang Y, Singh SV. Withaferin A suppresses estrogen receptor-α expression in human breast cancer cells. Mol Carcinog. 2011;50:614–624. doi: 10.1002/mc.20760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaileh M, Vanden Berghe W, Heyerick A, Horion J, Piette J, Libert C, DeKeukeleire D, Essawi T, Haegeman G. Withaferin A strongly elicits IkappaB kinase beta hyperphosphorylation concomitant with potent inhibition of its kinase activity. J Biol Chem. 2007;282:4253–4264. doi: 10.1074/jbc.M606728200. [DOI] [PubMed] [Google Scholar]
- 25.Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, Daly M, Wieand S, Tan-Chiu E, Ford L, Wolmark N. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 26.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 27.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 28.Hu C, Diévart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 30.Dickson BC, Mulligan AM, Zhang H, Lockwood G, O'Malley FP, Egan SE, Reedijk M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- 31.Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, Zhang H, Bull SB, O'Malley FP, Egan SE, Andrulis IL. JAG1 expression is associated with a basal phenotype and reccurence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;111:439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 32.Koduru S, Kumar R, Srinivasan S, Evers MB, Damodaran C. Notch-1 inhibition by Withaferin-A: a therapeutic target against colon carcinogenesis. Mol Cancer Ther. 2010;9:202–210. doi: 10.1158/1535-7163.MCT-09-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SH, Sehrawat A, Singh SV. Notch2 activation by benzyl isothiocyanate impedes its inhibitory effect on breast cancer cell migration. Breast Cancer Res Treat. 2012;134:1067–1079. doi: 10.1007/s10549-012-2043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao D, Srivastava SK, Lew KL, Zeng Y, Hershberger P, Johnson CS, Trump DL, Singh SV. Allyl isothiocyanate, a constituent of cruciferous vegetables, inhibits proliferation of human prostate cancer cells by causing G2/M arrest and inducing apoptosis. Carcinogenesis. 2003;24:891–897. doi: 10.1093/carcin/bgg023. [DOI] [PubMed] [Google Scholar]
- 35.Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest. 2006;86:842–852. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- 36.Kim SH, Sehrawat A, Sakao K, Hahm ER, Singh SV. Notch activation by phenethyl isothiocyanate attenuates its inhibitory effect on prostate cancer cell migration. PLoS One. 2011;6:e226615. doi: 10.1371/journal.pone.0026615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mödder UI, Oursler MJ, Khosla S, Monroe DG. Wnt10b activates Wnt, notch, and NFκB pathways in U2OS osteosarcoma cells. J Cell Biochem. 2011;112:1392–1402. doi: 10.1002/jcb.23048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Fu L, Gu F, Ma Y. Notch1 is involved in migration and invasion of human breast cancer cells. Oncol Rep. 2011;26:1295–1303. doi: 10.3892/or.2011.1399. [DOI] [PubMed] [Google Scholar]
- 39.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, Bundred NJ, Clarke RB. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010;70:709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindsay J, Jiao X, Sakamaki T, Casimiro MC, Shirley LA, Tran TH, Ju X, Liu M, Li Z, Wang C, Katiyar S, Rao M, Allen KG, Glazer RI, Ge C, Stanley P, Lisanti MP, Rui H, Pestell RG. ErbB2 induces Notch1 activity and function in breast cancer cells. Clin Transl Sci. 2008;1:107–115. doi: 10.1111/j.1752-8062.2008.00041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzo P, Miao H, D'Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, Hao L, Yao K, Rajan P, Hicks C, Siziopikou K, Selvaggi S, Bashir A, Bhandari D, Marchese A, Lendahl U, Qin JZ, Tonetti DA, Albain K, Nickoloff BJ, Miele L. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP, Del Sal G. The prolyl-isomerase Pin1 is Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol. 2009;11:133–142. doi: 10.1038/ncb1822. [DOI] [PubMed] [Google Scholar]
- 43.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Autocrine WNT signaling contributes to breast cancer cell proliferation via the canonical WNT pathway and EGFR transactivation. Breast Cancer Res. 2007;9:R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu M, Cohen B, Goldvasser P, Berman H, Virtanen C, Reedijk M. Plasminogen activator uPA is a direct transcriptional target of the JAG1-Notch receptor signaling pathway in breast cancer. Cancer Res. 2011;71:277–286. doi: 10.1158/0008-5472.CAN-10-2523. [DOI] [PubMed] [Google Scholar]
- 46.Annecke K, Schmitt M, Euler U, Zerm M, Paepke D, Paepke S, von Minckwitz G, Thomssen C, Harbeck N. uPA and PAI-1 in breast cancer: review of their clinical utility and current validation in the prospective NNBC-3 trial. Adv Clin Chem. 2008;45:31–45. doi: 10.1016/s0065-2423(07)00002-9. [DOI] [PubMed] [Google Scholar]