Summary
A faster semi-automated 96-well microtiter plate assay to determine viral infectivity titers, or viral focal units (vfu), of equine infectious anemia virus (EIAV) stocks is described. Optimization of the existing method modernizes a classic virological technique for viral titer determination by quantitating EIAV in experimentally infected cells via a cell-based ELISA. To allow for automation, multiple parameters of the current assay procedures were modified resulting in an assay that required only one quarter the original amount of virus and/or serum for infectivity or neutralization assays, respectively. Equivalent reductions in the required volumes of tissue culture, cell processing, and protein detection reagents were also achieved. Additionally, the new assay decreased the time required from start to finish from 10 days to 6 days (viral titer) or 7 days (viral neutralization), while increasing the number of samples that can be processed concurrently by transition to a 96-well microtiter plate format and by automated counting.
Keywords: Infectious Center Assay, viral titer, neutralization
1. Introduction
Equine Infectious Anemia Virus (EIAV), a lentivirus of the family Retroviridae, causes a persistent and potentially fatal infection in equids. Infection with this macrophage-tropic virus produces a chronic disseminated disease that is of worldwide importance in veterinary medicine (reviewed in Leroux et al. 2004; Craigo and Montelaro 2008). Natural and experimental infection with EIAV results in a rapid and dynamic disease process that differs distinctly from the slowly progressive degenerative diseases associated with other lentiviral infections, including HIV infection of humans. EIAV can be transmitted via iatrogenic sources such as contaminated syringe needles, but predominantly is spread by blood-feeding insect vectors (mainly horseflies and deerflies). Consequently, disease is most problematic in regions with warmer climates (Issel and Foil, 1984; Leroux et al., 2004). The actual number of infected animals in various geographical regions is not precisely known due to a lack of routine testing. Since its inception, testing in the United States has increased on an annual basis (APHIS, 2006), but the number of animals tested still represents a small proportion of the total equine population.
EIA disease in equids manifests as a progression through three stages: acute, chronic, and inapparent. The acute and chronic stages of EIA are defined by episodes of clinical disease triggered by waves of viremia and distinguished by fever, anemia, thrombocytopenia, edema, diarrhea, lethargy, and various wasting signs. Horses typically progress to life-long (long-term) inapparent carriers by 8-12 months post-infection, presumably due to the development of enduring protective host immunity (Craigo and Montelaro, 2008). Inapparent carriers, however, remain infected for life with the maintenance of markedly different levels of steady state virus replication in monocyte-rich tissue reservoirs (Montelaro et al., 1993; Hammond et al., 2000; Harrold et al., 2000). Stress or immune suppression of inapparent carriers can induce an increase in viral replication and potentially a recrudescence of disease (Kono et al., 1976; Montelaro et al., 1993; Craigo et al., 2002). Thus, EIAV offers a unique model for characterizing natural immunologic control of lentivirus replication and disease, for elucidating the nature and role of viral variation in persistence and pathogenesis, and ultimately for developing and modeling lentiviral vaccines.
Current EIAV research is focused on diverse basic and applied projects of relevance to veterinary and human medical fields. Basic research on the virus as a whole, its undefined gene products (the S2 gene), viral pathogenesis, and interactions with the host immune system are all areas of veterinary basic research programs. Applied research not only encompasses these same areas of veterinary interest, but also involves EIAV research as an animal model for HIV that include studies on viral entry, assembly, pathogenesis, and vaccine development. A common requirement for these varied disciplines studying EIAV is the in vitro determination of virus titer, yet the existing methods for virus quantification of infectious EIAV stocks are remarkably limited. Present assay methods are typically performed in large well formats such as 12-well and 24-well plates, use chemicals such as polybrene that can be a source of artifacts, or require primary macrophage cells to calculate TCID50 based on reverse transcriptase activity (Payne et al., 1989; Carpenter et al., 1990; Grund et al., 1996; Hammond et al., 1997; Zheng et al., 1997a; Zheng et al., 1997b; Sponseller et al., 2007; Craigo et al., 2009; Jiang et al., 2011). These methods, while locally standardized and used routinely over the years, have various aspects that could be improved, such as: eliminating the need for laborious techniques (e.g. macrophage isolation); reducing the required volumes of test virus and serum; eliminating the need for manual visualization of stained viral protein units that can cause significant error between assays; and the general streamlining of the method to utilize state-of-the-art automated processing and counting instrumentation.
To improve experiment precision while simplifying the determination of infectious viral titers, a new viral titer assay utilizing the colony counting technology employed on ELISpot readers was developed by strategic modification of our current EIAV infectivity assay. That assay, a classic virology protocol, detects viral proteins in cellular foci of EIAV infected fibroblastic cells in 24-well plates via a cell-based ELISA (Payne et al., 1989; Grund et al., 1996; Hammond et al., 1997; Craigo et al., 2009). Utilizing 96-well microtiter plates, new colorimetric detection chemicals, and automated counting on a CTL Immunospot, the new and improved assay achieves substantially higher levels of sensitivity, higher throughput, and reduces the artifact of visualizing stained plaques manually.
Materials and methods
2.1 Virus strains and cell lines
EIAV laboratory strains (EIAVPV and EIAVD9) utilized in this method development have been described previously (Rwambo et al., 1990; Grund et al., 1994; Leroux et al., 1997; Craigo et al., 2006; Craigo et al., 2007a; Craigo et al., 2007b; Craigo et al., 2010). Both strains, which infect fibroblastic cells in a non-lytic manner, were grown and harvested from infected equine dermal (ED) cells, and stored at −80°C as described previously (Li et al., 2000; Craigo et al., 2005; Craigo et al., 2007b). Fetal equine kidney (FEK) cells for infectivity and neutralization assays were cultured at 37°C and 5% CO2 in modified essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum, glutamine (2mM) and penicillin-streptomycin (100ug/ml) (Mediatech, Manassas, VA, USA).
2.2 Cell infections
FEK cells were seeded 24 hours pre-infection at a density of 1.5×104 cells/well in a volume of 100ul on 96-well Opaque White Microtest™ plates (BD Biosciences, San Jose, CA., USA) and cultured overnight to allow adherence. For viral titer determinations, two-fold serial dilutions of EIAV viral aliquots were performed in FEK culture medium, and 12.5ul of diluted virus inoculum added to FEK cells in triplicate. For viral neutralization assays, 50 viral focal units (vfu) of EIAV was incubated at 37°C, 5% CO2 for one hour with two-fold serial dilutions of heat-inactivated horse serum, and 50μl of the pre-incubated serum:virus mix added to cell wells in triplicate and returned to 37°C, 5% CO2 for 4-6 hours. For infectivity titer determinations and neutralization assays, 24 hours post-infection a 100μl overlay of prewarmed carboxymethylcellulose-sodium salt (CMC) was added to each well, and the plates incubated at 37°C, 5% CO2 for an additional 72 or 96 hours, respectively.
2.3 Preparation of cells for viral protein detection
Following 72 or 96 hours of incubation, cells were fixed by adding approximately 200μl/well of 3.7% formaldehyde in 1X PBS to the side of each well and incubated at room temperature for 10 minutes. This step was then repeated twice more to insure complete removal of CMC overlay. (Neutralization assay plates were washed three times with approximately 200μl 1X PBS prior to fixing.) Cell membranes were washed briefly with 1% Triton X-100 in 1X PBS and then disrupted by a 30 minute incubation at room temperature of approximately 200μl/well of the same 1% Triton X-100 solution. Plates were washed two times with 100ul/well of 5% Blotto (5% nonfat dry milk, 5% FBS, 0.25% Tween-20) and blocked at room temperature for one hour with 100μl/well 5% Blotto.
2.4 Detection of EIAV proteins in infected cell foci
Following blocking, EIAV proteins were detected by incubation at room temperature for one hour with 100μl/well of an antigen-specific primary antibody, or EIAV polyclonal serum (Lady, (Grund et al., 1996)), diluted 1:3000 in 5% blotto. Plates were washed four times with 1X PBS containing 0.025% Tween-20, after which 100μl/well of HRP-conjugated goat anti-horse IgG, (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), diluted 1:3000 in 5% blotto, was added and incubated at room temperature for one hour. Plates were then washed four times with 1X PBS containing 0.025% Tween-20, and 50μl/well of room temperature TrueBlue peroxidase substrate (KPL, Gaithersburg, MD) was added. Following a 10-minute room temperature incubation in the dark, the reaction was stopped by rinsing with distilled water and allowed to air-dry overnight. Fourteen to sixteen hours later, the plates were covered with parafilm and placed in dark to completely dry before counting.
2.5 Determination of viral focal units using automated counting instrumentation
Stained plates were scanned and counted with CTL BioSpot® software on an ImmunoSpot S4 UV Analyzer (C.T.L. Analyzers, LLC, Cleveland, OH). Focal units appeared as dark blue/green spots on a light blue background. Overlapping vfu were distinguished by appropriate manual setting of the instrumentation software separation tolerance parameter. Minimum viral focal unit size was adjusted for counting depending on cell growth, viral dispersion, background focal artifact, etc. For viral titer assays, determined virus focal units (spots) for all dilutions were plotted as a function of the reciprocal dilution of the stock using Prism software (GraphPad, La Jolla, CA). Linear plots of the reciprocal dilutions were log transformed, non-linear fit of the dose-response fitted, and statistically analyzed for linearity of the curve. Curves with R squares below 0.9 are discarded, and the assay repeated. Once the linearity of the assay was confirmed, titers were determined from the linear phase of the plot using the following formula:
Titer (vfu/ml) = viral focal units x dilution factor (vfu) / volume of added virus (ml) Calculations of 50% neutralization titers were performed by standard procedures, as described previously (Grund et al., 1996; Leroux et al., 1997; Hammond et al., 1999; Howe et al., 2002; Howe et al., 2005; Craigo et al., 2009).
3. Results
3.1 Infectious center assay (ICA) parameter optimization
The first step in generating an updated assay method was to determine the key steps of the current assay that could be modified to increase sensitivity, reproducibility, and importantly, to streamline the overall procedure. In order to utilize automated counting techniques, the cell-based ELISA needed to be transitioned from a manually counted 24-well assay to a 96-well format. Cell densities and volumes of trypsin, culture media, CMC, and serially diluted inoculum were adjusted accordingly from the standard 24-well plate format (Table 1). These initial changes in reagent and consumable materials were initially evaluated in the manually visualized titer assay (Grund et al., 1996; Howe et al., 2002; Craigo et al., 2009), using the indicated changes in the ICA reagents, and a BD-Falcon 96-well flat bottom microtiter plate. The reagent and consumable material modifications allowed for accurate titer determination by the standard ICA method (data not shown), thus validating these parameters as the basis for conversion of the assay to an automated microtiter format, as addressed below.
Table 1. Parameter modifications between the original and optimized ICA formats.
| 24-well, manual visualization |
96-well, automatic counting |
|
|---|---|---|
|
|
||
| FEK cell density | 6.0 × 104 | 1.5 × 104 |
| Culture media (MEM) volume | 800ul | 200ul |
| CMC overlay volume | 400ul | 100ul |
| Test inoculum (Titer) volume | 50ul | 12.5ul |
| Fixative incubations (number) | 2 | 3 |
| Block/diluent solution | 1X PBS/0.25% Tween-20 |
5% Blotto/5% FBS/0.25% Tween-20 |
| Block/primary & secondary antibody volumes | 200ul | 50ul |
| Wash steps between solutions (number) | 3 | 4 |
| 1X PBS wash prior to fixation (neutralization assay only) | 0 | 1 |
| Colorimetric reagent/volume/incubation time | AEC/200ul/45min | TrueBlue/50ul/15min |
| Microtiter plate | BD-Falcon 24-well | BD-Falcon 96-well white flat bottom |
| Neutralization assay inoculum volumes (diluted serum:50vfu) | 200ul | 50ul |
| Total days for assay (titer/neutraliztion) | 10/10 | 6/7 |
Initial experiments revealed intrinsic differences in the requirements for optimizing assays in 96-well plates compared to 24-well plates. Higher surface area to volume ratios encountered when transitioning from a 24-well plate to a 96-well plate assay led to decreased efficiency of the removal of liquids especially in the case of the highly viscous CMC. These problems created high background levels of cellular staining during the multiple steps involved in cell fixing and processing procedures. It was well established in the original titer assay that residual CMC led to increased background staining. The methods for ensuring complete removal of the CMC in the 24-well assay did not work completely for the 96-well plates. To decrease background attributed to residual CMC left in the 96-well plate wells, the fixative incubation was repeated twice following the initial ten-minute incubation instead of once.
Other well-known problems that contribute to increased background staining are insufficient blocking and inefficient removal of antibodies. In the EIAV assay, it has been observed that the use of equine immune serum as a primary antibody and anti-horse Ig as secondary antibody can lead to high backgrounds on equine cell substrate unless careful optimization of all staining parameters is performed. Evaluations of primary and secondary antibody concentrations revealed that reductions in both reagents equally reduced specific and background levels of reactivity. Thus, these antibody concentrations were not modified from the standard ICA assay method. Initial attempts to reduce the level of background staining included the use of an automated plate washer. However, the force used by the machine displaced the fixed cells and therefore was not a viable option. Therefore, to address the increased background that was still evident from the antibodies, a higher stringency blocking and antibody dilution reagent was utilized (see Table 1 and Materials and Methods section 2.3). This modified solution for blocking and antibody dilutions markedly decreased the levels of background staining as compared to the original solution. Additionally, to reduce further the background staining of the cell membranes, the number of washes at each step were increased (Table 1).
The colorimetric indicator utilized in the 24-well ICA to visualize EIAV infectious centers was a peroxidase substrate 3-amino-9-ethylcarbazole in a sodium acetate buffer (pH 5.5) supplemented with H2O2 (AEC). AEC staining of EIAV-infected FEK cells is typically faint, but visible, by use of a dissecting microscope for manual counting. This stain did not have sufficient colorimetric density for detection in automated counting and needed to provide a higher signal to noise ratio to work efficiently for the new ICA. The reagent that strongest reactivity with the highest signal to noise ratio was the TrueBlue peroxidase substrate by KPL. TrueBlue stained viral focus units more intensely, comes as a pre-formulated HRP stain reducing preparation time, and requires a shorter incubation time on plates as compared to AEC staining (Table 1). Once the colorimetric reagent was resolved, it was possible to test one last consumable reagent to increase the sensitivity of the staining and to optimize automated counting. Clear and white bottom microtiter plates from multiple vendors were assessed for their effect on staining sensitivity. BD Biosciences 96-well clear flat bottom plates and white flat bottom plates (BD Biosciences, San Jose, CA., USA), MIDSCI 96-well white plates (MIDSCI, St. Louis, MO., USA), and Corning 96-well clear bottom/white side plates (Corning Inc., Corning, NY, USA) were all compared in parallel assays. The results revealed that the BD white flat bottom plates produced the clearest resolution of stained cells.
Once the indicated parameters were adjusted, one final optimization became achievable. The previous ICA required an incubation period of 168 hours (7 days) post CMC-overlay in order to reach a staining level that was detectable manually with a dissecting microscope. The sensitivity of the new staining procedure coupled with the automated counting allowed for a remarkable decrease in incubation time. Through empirical determination this incubation period was reduced from the original 168 hours to 72 hours, thus achieving a 2.3-fold decrease in the necessary incubation time post CMC-overlay, and 40% less time required for the assay from start to finish.
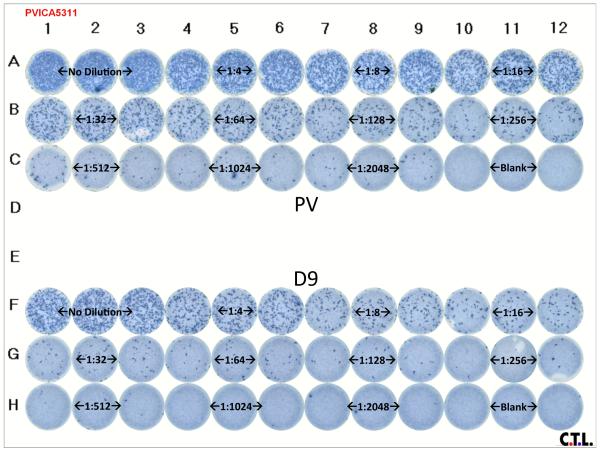
The new automated assay was evaluated simultaneously with the standard manual assay in a final test ICA with two viral stocks, EIAVPV and EIAVD9 (both having known titers). Utilizing the above described methods, the stained plates were scanned and enumerated as described in Materials and Methods (Figure 1). The new assay results were read 72 hours post CMC-overlay and the standard assay plates was read 168 hours as previously described. (Grund et al., 1996; Howe et al., 2002; Craigo et al., 2009). Figure 1 displays the stained plate of the new assay; stained plates from the old ICA are not presented, as the foci are not readily documented for visualization. Both the old and new ICAs resulted in approximately the same viral titer for the respective viral stocks: EIAVPV, 1.2 × 106 and 1.8 × 106, old and new ICA respectively, and EIAVD9, 3.2 × 105 and 2.4 × 105, old and new, respectively. These numbers matched what was expected within error from the previously calculated titers of both stocks.
Figure 1. ICA titer plate of EIAVPV and EIAVD9.
Reference stocks of EIAVPV and EIAVD9 were thawed and diluted in MEM as indicated in Materials and Methods. The ICA methods detailed in sections 2 and 3.1-3.2 were followed for infection, processing, and detection of viral focal units. Rows are designated A-H and columns are designated 1-12. Rows A-C contain the EIAVPV samples and rows F-H contain the EIAVD9 samples. Triplicate dilution levels are indicated over the wells in which they were plated. PV, EIAVPV; D9, EIAVD9; Blank, no virus control.
3.2 Assay Quantitation and Variability
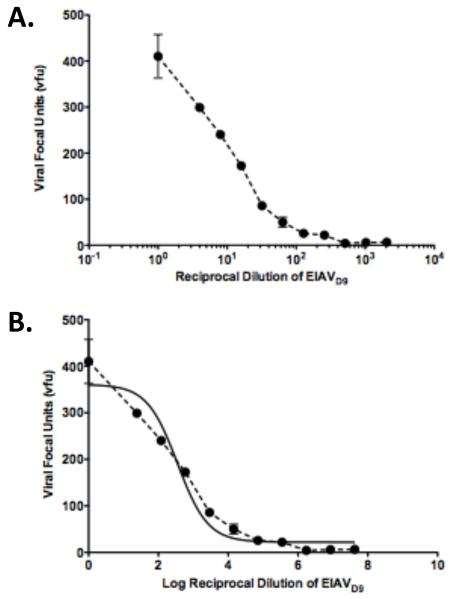
The new automated ICA allows for a faster and more consistent determination of viral titer. To increase further that standardization, linear regression plots that evaluate the relationship of all viral dilutions to their respective viral titers were incorporated to the automated ICA methods. Incorporating this validation of the assay verifies the linear nature of the data from all dilutions of the stock and also guides the selection of which dilutions to choose for titer determination. The previous methodology only selected 1-2 dilutions to verify the titer and did not have an average evaluation of all infected wells. The automated counting reduced the labor involved in counting the lower viral dilutions (higher numbers of vfu) and thus facilitated a more rigorous level of assay validation. The viral titer is determined from the plate only if the R square value of the linear regression analysis is > 0.9. The advantages of using this calculation in the automated ICA was demonstrated with the data generated from the EIAVD9 titer determination illustrated in Figure 1. Following spot enumeration (Figure 1), the vfu values of the EIAVD9 stock were plotted as a function of their reciprocal dilution (Figure 2A). That plot was log transformed (Figure 2B), and a non-linear fit determined. Multiple assays with both EIAVPV (data not shown) and EIAVD9 strains revealed that the CTL Immunospot would count reliably between 5 and 400 vfu (Figure 2). The linear range for the assay in Figure 2 is between 50 and 300 vfu and is representative of a typical linear range for the viral strains assayed during method development. The dilutions chosen for titer determination from the linear phase of the plot for EIAVD9 hence were dilutions 1:128 and 1:256 (Figures 1, 2).
Figure 2. Linear fitting of titer data.
(A) Enumerated viral focal units were plotted as a function of their reciprocal dilution. (B) Log transformed plots of the vfu:dilution pairs were plotted and fit for linearity. Non-linear fits of the dose-response curve were determined in Prism (GraphPad, La Jolla, CA). R square of the curve = 0.94. vfu, viral focal units.
The original ICA had an expected yet acceptable level of error between experiments, generally within a half log. To observe how much variation was actually apparent in the new ICA compared to the standard assay, comparative analysis between viral titer determinations from the two assays was performed. Several titer assays were randomly chosen from both the new and old ICA titer evaluations of EIAVPV that were performed during the optimization and validation of the new method (Table 2). Additionally included in the old ICA titers were calculations performed prior to the current assay development as part of reference viral stock maintenance. In general, the observed differences in the calculated titers between the old and new methods were low, approximately less than or equal to a half log error between the old and new assay titers. Statistical analysis of the differences in the means of the two assays confirmed that the differences were insignificant (P = 0.272). However, analysis of the variance within an assay revealed that the new assay had significantly lower levels of variation (P = 0.024) with a standard deviation of 0.3 ×106 as opposed to the 1.1 × 106 of the old assay (Table 2).
Table 2. Titers of EIAVPV.
| Sample* | Automated (vfu) | Manual (vfu) |
|---|---|---|
| 1 | 2.5 × 106 | 1.2 × 106 |
| 2 | 2.0 × 106 | 8.5 × 105 |
| 3 | 2.7 X106 | 2.8 × 106 |
| 4 | 1.8 × 106 | 9.3 × 105 |
| 5 | 2.1 × 106 | 9.9 × 105 |
| 6 | 2.2 × 106 | 3.2 × 106 |
|
| ||
| Standard deviation# | 0.3 × 106 | 1.1 × 106 |
P = 0.0241, unpaired t-test, Welch's correction (GraphPad,LaJolla, Ca)
Samples are not paired, but are chosen from random assays
3.3 Neutralization Assay Modifications
One of the more common uses of the titer assay is to detect and quantitate antibody neutralization of EIAV. The original ICA methodology for this assay included a pre-incubation of 50 vfu of the EIAV strain (to be neutralized) with serially-diluted polyclonal serum for one hour prior to overlay onto the cells, 24 hours post seeding. Initial attempts at transitioning this method included similar proportionate reductions in reagent volumes as described in section 3.1, but also included modifications in the volume of inoculum:serum pre-incubation mixture (Table 1). However, the inclusion of the test serum in the overlay of the cells increased the level of background staining, decreasing the signal to noise ratio that had been overcome by all of the modifications detailed in section 3.1. This increase in signal was also observed in negative test serum controls (data not shown). To address these technical issues, the following modifications to the automated ICA protocol were incorporated for neutralization assays specifically. It was determined that leaving the serum:viral inoculum mix on the cell layer prior to CMC overlay was a major problem for background staining; hence, after a 4-6 hour adsorption, the serum:virus inoculum was removed and replaced with MEM. CMC was overlaid the next day per the titer ICA and the original neutralization assay. Prior to fixing, the plates were rinsed with 1X PBS to assist in complete CMC removal. No changes were made to the blocking, antibody dilution reagents, or the colorimetric indicator. While the described modifications reduced the background noise in the cell wells, the modified procedures also reduced the signal. Therefore, the protocol was modified to include an additional 24 hours of incubation prior to staining, or 96 hours post CMC overlay as opposed to the 72 hours of the new titer ICA.
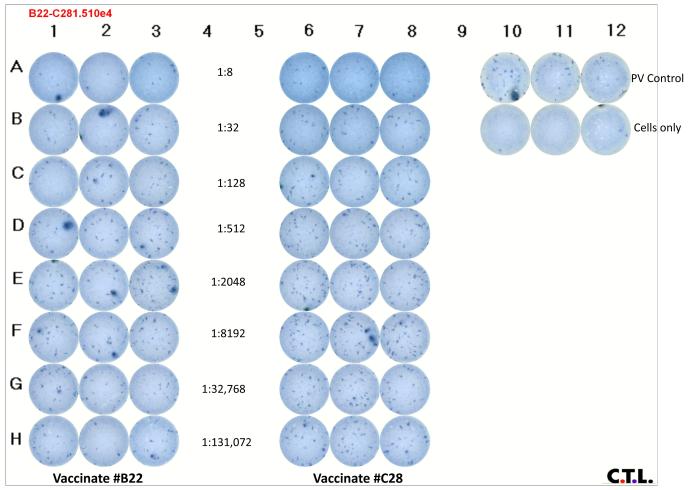
A final test neutralization ICA of the viral stock (EIAVPV) with reference immune serum (with defined 50% neutralization titers) from two horses experimentally inoculated with an engineered live-attenuated EIAV strain (Craigo et al., 2007b) was performed. Utilizing these modified methods, stained plates were scanned and enumerated as described in Materials and Methods section 2.5 (Figure 3). Reciprocal neutralization titers (50%) were calculated as described in Materials and Methods section 2.5. The reciprocal neutralization titers for the tested sera from horse C28 and B22 were calculated to be 220 and 350, respectively, consistent with previous neutralization determinations.
Figure 3. ICA neutralization titer (50%) plate of EIAVPV by sera from vaccinated horses.
A laboratory stock of EIAVPV was thawed, 50 vfu mixed with serially-diluted serum from EIAV vaccine studies (Craigo et al., 2007b) as indicated in section 3.3. The ICA methods detailed in sections 2, 3.3 and 3.1 were followed for infection, processing, and detection of viral focal units. Rows are designated A-H and columns are designated 1-12. Columns 1-3 have the ICA analysis of the indicated (columns 4-5) serial dilutions (rows A-H) of animal #B22 and columns 6-8 of animal #C28. Columns 10-12 have the control wells of 50 vfu of EIAVPV (row A) or no virus control (row B).
4. Discussion
One central technique that spans numerous avenues of research in multiple viral systems is the quantification of viral infectivity. Calculating the titer of a virus stock to elucidate the infectious doses per volume of the virus suspension for such things as experimental infections, attenuated vaccine inoculations, or for the analysis of neutralizing antibodies all start with the same question: how do we efficiently and sensitively determine the quantity? Classic virology historically utilizes cell-based assays, whether staining cell layers and utilizing plaque formation with a lytic virus infection or performing cell-based ELISAs when the viral infections are nonlytic. Many current methods have moved to reporter-based assays such as the TZM-cell luciferase assay utilized for infectious dose determinations of HIV viruses (Mascola et al., 2005). While these methods are highly sensitive, usually take less reagents and time, and have proven to be repeatable, it is still a measure of normalization instead of a direct counting of virus-infected cells. The goal of the current protocol was to achieve a faster, more sensitive titer assay that allowed for higher throughput while still capturing the classic virological assay of enumerating infectious centers. Therefore, this report details the development of a semi-automated viral titer assay that maintained the central characteristics of this classic assay while updating it to be a more sensitive, faster, higher throughput, and cost-effective microtiter plate assay with inherently lower levels of assay-to-assay error.
The transition from a 24-well manually enumerated assay to a 96-well semi-automated assay required some obvious and other not-so-obvious modifications. Reducing the cell number and volumes of viral stocks and reagents to physically fit into the well of a 96-well microtiter plate seemed obvious. However, the background issues that had to be overcome from the transition to a 96-well plate were less expected. Physically removing the CMC from the cell layer became a much larger problem than it had been in a 24-well plate. Additionally, the higher levels of background staining from the primary and secondary antibody incubations, which in the past were of little consequence, were an unexpected challenge. Both these issues seemed to stem from higher surface area to volume ratios in the transition from the square area of a well on the 24-well plates versus the well size on a 96-well plate. These issues were addressed effectively by increasing the amount of exposure to fixing reagents (in the case of CMC removal) and increasing the blocking ability of the antibody buffer by using 5% blotto instead of PBS. Increasing the number of washes between all processing steps also lowered the levels of background staining. The inefficiency of the original wash protocol originated from the same issues created by being in a smaller surface area.
Equally unanticipated was the additional background issues of the neutralization ICA. The test serum:virus mixture that is preincubated prior to inoculation on the cell layer created high levels of background such that the spots were unable to be differentiated from cell background at the lowest dilutions of serum (highest serum concentrations). It can easily be presumed that the test serum of the serum:virus inoculum was physically being trapped by the CMC overlay and consequently fixed to the cell membranes upon the first fixation step of the plate processing. The pre-rinse of wells with PBS prior to fixation alone did not overcome this issue completely, hence the modification to the protocol of removing the test serum:virus mix prior to CMC overlay post-adsorption. Likewise, merely removing this inoculum did not completely resolve the increased background, which is why both of these new steps were incorporated into the neutralization ICA.
Signal to noise ratios in general become a larger problem when shifting from manually visualizing viral focal units to training software to distinguish a positive spot from background noise. What appears visible and distinguishable to the eye may not be so clear to the machine. If the operator has to spend a large amount of time working with the machine to recognize the differences, the timesaving of the automated assay becomes less, especially when considering multi-plate assays. Switching the assay to the new colorimetric reagent, TrueBlue made a significant difference in the observed signal to noise ratio. Changing the plates helped with the resolution as well. These two specific changes for automatic counting enabled the software to distinguish readily the spots on the scans.
The final step added to this assay was the linearity plot of all dilutions (titer ICA only). While this seems like a minor check of the system, it is a very important parameter that reduces the variability of the assay by individuals and between different users in general. The control provided by the addition of this simple calculation to the assay is two fold. First, the plot ensured that the dilution series occurred in a dose-responsive manner to provide confidence in calculating the viral titer from one or two dilutions. Second, the plot demonstrated at which dilutions the assay is in a linear phase, allowing for a more guided selection of the best dilutions from which to calculate the titer. In the previous ICA not all dilutions were counted. Due to manual visualization the cut off for “too numerous to count” was much lower than what the machine is able to count. The dilutions that were used for titer determinations were often higher dilutions (lower levels of virus) and thus were likely not in the linear phase, as seen in Figure 2B starting at approximately the sixth dilution (1:128) in the series. This is likely one of the reasons that new ICA has a significantly lower level of inter-assay error as compared to the old method (Table 2).
The data presented in this report demonstrated modifications of a classic virology assay for the quantification, or titering, of viral suspensions. The transition of the previous assay from a 24-well manually counted ICA to a 96-well automatic count ICA allowed for a higher number of assays/per plate, per day, at significant reductions in the costs of reagents and volumes of virus and serum required. Additionally, the assay had significant reductions in the amount of time the ICA takes from start to finish, reducing from 10 days to 6 (titer) or 7 (neutralization) days, which is a timesaving of 40% and 30% for titer and neutralization assays, respectively. Ultimately, the faster, higher throughput, and more economical assay demonstrated significantly lower variation between experimental determinations.
Highlights.
We describe a semi-automated microtiter assay to determine viral titers.
Optimization of the existing method modernizes a classic virological technique.
Our new assay reduces required assay time while increasing assay capabilities.
Acknowledgments
The authors would like to thank Jonathan D. Steckbeck for editing the manuscript. This work was supported by the National Institutes of Health grant number R01 AI25850 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- APHIS . In: Equine Infectious Anemia (EIA) USDA, editor. Veternairy Services; 2006. [Google Scholar]
- Carpenter S, Evans LH, Sevoian M, Chesebro B. In vivo and in vitro selection of equine infectious anemia virus variants. In: Kurstak E, Marusyk RG, Murphy FA, Van Regenmortel MHV, editors. Applied Virology Research II: Virus Variation. Epidemiology, and Control, Plenum Publishing; New York: 1990. pp. 99–115. [Google Scholar]
- Craigo JK, Barnes S, Cook SJ, Issel CJ, Montelaro RC. Divergence, not diversity of an attenuated equine lentivirus vaccine strain correlates with protection from disease. Vaccine. 2010;28:8095–104. doi: 10.1016/j.vaccine.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigo JK, Barnes S, Zhang B, Cook SJ, Howe L, Issel CJ, Montelaro RC. An EIAV field isolate reveals much higher levels of subtype variability than currently reported for the equine lentivirus family. Retrovirology. 2009;6:95. doi: 10.1186/1742-4690-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigo JK, Durkin S, Sturgeon TJ, Tagmyer T, Cook SJ, Issel CJ, Montelaro RC. Immune suppression of challenged vaccinates as a rigorous assessment of sterile protection by lentiviral vaccines. Vaccine. 2007a;25:834–45. doi: 10.1016/j.vaccine.2006.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigo JK, Leroux C, Howe L, Steckbeck JD, Cook SJ, Issel CJ, Montelaro RC. Transient immune suppression of inapparent carriers infected with a principal neutralizing domain-deficient equine infectious anaemia virus induces neutralizing antibodies and lowers steady-state virus replication. J Gen Virol. 2002;83:1353–1359. doi: 10.1099/0022-1317-83-6-1353. [DOI] [PubMed] [Google Scholar]
- Craigo JK, Li F, Steckbeck JD, Durkin S, Howe L, Cook SJ, Issel CJ, Montelaro RC. Discerning an Effective Balance between Equine Infectious Anemia Virus Attenuation and Vaccine Efficacy. J Virol. 2005;79:2666–2677. doi: 10.1128/JVI.79.5.2666-2677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigo JK, Montelaro RC. Equine Infectious Anemia Virus (Retroviridae), Encyclopedia of Virology. Vol. 3. Elsevier; Oxford: 2008. pp. 167–174. [Google Scholar]
- Craigo JK, Sturgeon TJ, Cook SJ, Issel CJ, Leroux C, Montelaro RC. Apparent elimination of EIAV ancestral species in a long-term inapparent carrier. Virology. 2006;344:340–353. doi: 10.1016/j.virol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Craigo JK, Zhang B, Barnes S, Tagmyer TL, Cook SJ, Issel CJ, Montelaro RC. Envelope variation as a primary determinant of lentiviral vaccine efficacy. Proc Nat Acad Sci USA. 2007b;104:15105–15110. doi: 10.1073/pnas.0706449104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund CH, Lechman ER, Issel CJ, Montelaro RC, Rushlow KE. Lentivirus cross-reactive determinants present in the capsid protein of equine infectious anaemia virus. J.Gen.Virol. 1994;75(Pt 3):657–662. doi: 10.1099/0022-1317-75-3-657. [DOI] [PubMed] [Google Scholar]
- Grund CH, Lechman ER, Pezzuolo NA, Issel CJ, Montelaro RC. Fine specificity of equine infectious anaemia virus gp90-specific antibodies associated with protective and enhancing immune responses in experimentally infected and immunized ponies. J.Gen.Virol. 1996;77(Pt 3):435–442. doi: 10.1099/0022-1317-77-3-435. [DOI] [PubMed] [Google Scholar]
- Hammond SA, Cook SJ, Lichtenstein DL, Issel CJ, Montelaro RC. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J.Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Li F, McKeon BM, Sr., Cook SJ, Issel CJ, Montelaro RC. Immune responses and viral replication in long-term inapparent carrier ponies inoculated with equine infectious anemia virus. J.Virol. 2000;74:5968–5981. doi: 10.1128/jvi.74.13.5968-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Raabe ML, Issel CJ, Montelaro RC. Evaluation of antibody parameters as potential correlates of protection or enhancement by experimental vaccines to equine infectious anemia virus. Virology. 1999;262:416–430. doi: 10.1006/viro.1999.9939. [DOI] [PubMed] [Google Scholar]
- Harrold SM, Cook SJ, Cook RF, Rushlow KE, Issel CJ, Montelaro RC. Tissue sites of persistent infection and active replication of equine infectious anemia virus during acute disease and asymptomatic infection in experimentally infected equids. J.Virol. 2000;74:3112–3121. doi: 10.1128/jvi.74.7.3112-3121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe L, Craigo JK, Issel CJ, Montelaro RC. Specificity of serum neutralizing antibodies induced by transient immune suppression of inapparent carrier ponies infected with a neutralization-resistant equine infectious anemia virus envelope strain. J Gen Virol. 2005;86:139–149. doi: 10.1099/vir.0.80374-0. [DOI] [PubMed] [Google Scholar]
- Howe L, Leroux C, Issel CJ, Montelaro RC. Equine Infectious Anemia Virus Envelope Evolution In Vivo during Persistent Infection Progressively Increases Resistance to In Vitro Serum Antibody Neutralization as a Dominant Phenotype. J Virol. 2002;76:10588. doi: 10.1128/JVI.76.21.10588-10597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issel CJ, Foil LD. Studies on equine infectious anemia virus transmission by insects. J.Am.Vet.Med.Assoc. 1984;184:293–297. [PubMed] [Google Scholar]
- Jiang CG, Gao X, Ma J, Lin YZ, Wang XF, Zhao LP, Hua YP, Liu D, Zhou JH. C-terminal truncation of the transmembrane protein of an attenuated lentiviral vaccine alters its in vitro but not in vivo replication and weakens its potential pathogenicity. Virus Res. 2011;158:235–45. doi: 10.1016/j.virusres.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Kono Y, Hirasawa K, Fukunaga Y, Taniguchi T. Recrudesence of equine infectious anemia by treatment with immunosuppressive drugs. Nat Inst Anim Hlth Quart. 1976;16:8–15. [PubMed] [Google Scholar]
- Leroux C, Cadore JL, Montelaro RC. Equine Infectious Anemia Virus (EIAV): what has HIV’s country cousin got to tell us? Vet Res. 2004;35:485–512. doi: 10.1051/vetres:2004020. [DOI] [PubMed] [Google Scholar]
- Leroux C, Issel CJ, Montelaro RC. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J.Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Leroux C, Craigo JK, Cook SJ, Issel CJ, Montelaro RC. The S2 gene of equine infectious anemia virus is a highly conserved determinant of viral replication and virulence properties in experimentally infected ponies. J.Virol. 2000;74:573–579. doi: 10.1128/jvi.74.1.573-579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, D’Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79:10103–7. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelaro RC, Ball JM, Rushlow K. Equine retroviruses. In: Levy JA, editor. The Retroviridae. Vol. 2. Plenum Press; New York, N.Y.: 1993. pp. 257–360. [Google Scholar]
- Payne SL, Rushlow K, Dhruva BR, Issel CJ, Montelaro RC. Localization of conserved and variable antigenic domains of equine infectious anemia virus envelope glycoproteins using recombinant env-encoded protein fragments produced in Escherichia coli. Virology. 1989;172:609–615. doi: 10.1016/0042-6822(89)90203-1. [DOI] [PubMed] [Google Scholar]
- Rwambo PM, Issel CJ, Hussain KA, Montelaro RC. In vitro isolation of a neutralization escape mutant of equine infectious anemia virus (EIAV) Arch.Virol. 1990;111:275–280. doi: 10.1007/BF01311062. [DOI] [PubMed] [Google Scholar]
- Sponseller BA, Sparks WO, Wannemuehler Y, Li Y, Antons AK, Oaks JL, Carpenter S. Immune selection of equine infectious anemia virus env variants during the long-term inapparent stage of disease. Virology. 2007;363:156–165. doi: 10.1016/j.virol.2007.01.037. [DOI] [PubMed] [Google Scholar]
- Zheng YH, Nakaya T, Sentsui H, Kameoka M, Kishi M, Hagiwara K, takahashi H, Kono Y, Ikuta K. Insertions, duplications and substitutions in restricted gp90 regions of equine infectious anemia virus during febrile episodes in an experimentally infected horse. J Gen Virol. 1997a;78:807–820. doi: 10.1099/0022-1317-78-4-807. [DOI] [PubMed] [Google Scholar]
- Zheng YH, Sentsui H, Nakaya T, Kono Y, Ikuta K. In vivo dynamics of equine infectious anemia viruses emerging during febrile episodes: insertions/duplications at the principal neutralizing domain. J.Virol. 1997b;71:5031–5039. doi: 10.1128/jvi.71.7.5031-5039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]