Abstract
Particulate methane monooxygenase (pMMO) is an integral membrane metalloenzyme that oxidizes methane to methanol in methanotrophic bacteria, organisms that live on methane gas as their sole carbon source. Understanding pMMO function has important implications for bioremediation applications and for the development of new, environmentally friendly catalysts for the direct conversion of methane to methanol. Crystal structures of pMMOs from three different methanotrophs reveal a trimeric architecture, consisting of three copies each of the pmoB, pmoA, and pmoC subunits. There are three distinct metal centers in each protomer of the trimer, mononuclear and dinuclear copper sites in the periplasmic regions of pmoB and a mononuclear site within the membrane that can be occupied by copper or zinc. Various models for the pMMO active site have been proposed within these structural constraints, including dicopper, tricopper, and diiron centers. Biochemical and spectroscopic data on pMMO and recombinant soluble fragments, denoted spmoB proteins, indicate that the active site involves copper and is located at the site of the dicopper center in the pmoB subunit. Initial spectroscopic evidence for O2 binding at this site has been obtained. Despite these findings, questions remain about the active site identity and nuclearity and will be the focus of future studies.
Keywords: copper, methanotroph, membrane protein, dioxygen activation, ammonia monooxygenase, Cu-ZSM-5 zeolite, hemocyanin, tyrosinase
Introduction
Importance of biological methane oxidation
Methanotrophic bacteria oxidize methane to methanol in the first step of their metabolic pathway (Hanson & Hanson, 1996). Methane is a potent greenhouse gas, with a global warming potential more than 20 times that of carbon dioxide, and as the only biological methane sink, methanotrophs play a critical role in the global carbon cycle (Jiang et al., 2010). In addition, harnessing biological mechanisms of methane oxidation is becoming a pressing goal in the development of “green” catalysts. The notion of a “methanol economy” (Olah, 2005) could perhaps be realized if more efficient and environmentally benign catalytic processes existed for the direct and selective oxidation of methane to methanol. Current catalysts that activate the 104 kcal mol−1 C-H bond in methane require harsh conditions and produce waste (Arakawa et al., 2001). By contrast, methanotrophs perform this chemistry under ambient conditions, and thus their methane metabolizing enzymes may provide inspiration for the design of new catalysts.
Methanotrophs
Methanotrophs are divided into three phylogenetic groups on the basis of their carbon assimilation pathways, cell morphology, membrane arrangement, 16S RNA sequences, and other metabolic characteristics (Hanson & Hanson, 1996; Semrau et al., 2010). The type I methanotrophs, including the genera Methylomicrobium and Methylobacter, assimilate formaldehyde as a carbon source via the ribulose monophosphate pathway (RuMP) whereas the type II methanotrophs, including the genera Methylocystis, Methylosinus, and Methylocella, utilize the serine pathway. The type X methanotrophs, such as Methylococcus, use the RuMP, but also possess low levels of the serine pathway enzyme ribulose-bisphosphate carboxylase. Beyond the proteobacterial methanotrophs, filamentous methane oxidizers Crenothrix polyspora (Stoecker et al., 2006) and Clonothrix fusca (Vigliotta et al., 2007)) and acidophilic methanotrophs belonging to the Verrucomicrobia phylum (Op den Camp et al., 2009; Pol et al., 2007; Dunfield et al., 2007; Islam et al., 2008) have been discovered recently.
Methanotrophs are found in a wide variety of environments, and can oxidize a wide variety of other substrates, including halogenated hydrocarbons (Semrau et al., 2010), and thus represent a target for bioremediation applications. They grow at temperatures ranging from 4 °C to 72 °C and at extremes of pH and salt levels (Semrau et al., 2008; Semrau et al., 2010). Although most methanotrophs are obligate (only grow on one-carbon compounds), some strains are facultative and can grow on multicarbon compounds as well (Dedysh & Dunfield, 2011). The first methantroph genome to be sequenced was that of Methylococcus capsulatus (Bath) in 2004 (Ward et al., 2004). Since then, representative genomes of a wide variety of methanotrophs have been completed, including those of Methylosinus trichosporium OB3b (Stein et al., 2010), Methylocystis species strain Rockwell (Stein et al., 2011), Methylocella silvestris BL2 (Chen et al., 2010), Methylomonas methanica MC09 (Boden et al., 2011), Methylobacter tundripaludum SV96 (Svenning et al., 2011), Methylomicrobium alcaliphilum 20Z (Vuilleumier et al., 2012), and Methylacidiphilum infernorum (Hou et al., 2008). The combination of genomic data with the growing phylogenetic distribution of methanotrophs suggests that methanotrophs are more diverse in the environment than previously imagined (Jiang et al., 2010) and may produce unusual forms of metabolic enzymes, including methane monooxygenases (MMOs).
Methane monooxygenases
The oxidation of methane to methanol, the first step in the metabolic pathway of methanotrophs, is catalyzed by MMO enzymes. Methanol is further converted to formaldehyde by methanol dehydrogenase, and formaldehyde is then oxidized to formate and carbon dioxide by formaldehyde and formate dehydrogenases, respectively. Formaldehyde is also assimilated for biosynthesis of cell material composed of multicarbon compounds (Hanson & Hanson, 1996). There are two types of MMOs. The soluble form (sMMO) (Merkx et al., 2001) is expressed under conditions of copper limitation, and is only present in a few methanotroph strains. The membrane-bound form, referred to as particulate MMO (pMMO) (Lieberman & Rosenzweig, 2004; Balasubramanian & Rosenzweig, 2007; Hakemian & Rosenzweig, 2007), is expressed under copper-replete conditions and is present in all known methanotrophs with the exception of Methylocella silvestris BL2 (Theisen et al., 2005). In those organisms that possess both sMMO and pMMO, the switch in expression is copper-dependent (Ali & Murrell, 2009) (Choi et al., 2003; Prior & Dalton, 1985; Stanley et al., 1983). Although there is some information on the genes and gene products involved (Csáki et al., 2003; Stafford et al., 2003; Ukaegbu et al., 2006; Ukaegbu & Rosenzweig, 2009), the specific mechanism of this switch has not been determined.
The two MMO systems are completely different in sequence, overall structure, substrate selectivity, and active site composition. sMMO is a multicomponent monooxygenase composed of a hydroxylase (MMOH), which houses the active site, a reductase (MMOR), which shuttles electrons from NADH to the active site of MMOH, and a regulatory protein (MMOB) that is required for activity (Merkx et al., 2001). MMOH consists of three polypeptides arranged in an α2β2γ2 dimer (Rosenzweig et al., 1993). Methane and O2 bind at a carboxylate-bridged diiron center, and the catalytic mechanism has been studied intensively (Tinberg & Lippard, 2011). sMMO has a wide substrate range that includes aromatic and heterocyclic compounds (Jiang et al., 2010; Colby et al., 1977). pMMO is composed of three subunits, α, β and γ, also known as pmoB, pmoA, and pmoC, respectively (Zahn & DiSpirito, 1996; Lieberman & Rosenzweig, 2004), arranged in an α3β3γ3 trimer (Lieberman & Rosenzweig, 2005a; Lieberman & Rosenzweig, 2005b). Ammonia monooxygenase (AMO), a pMMO homolog, is the only other enzyme besides pMMO and sMMO known to oxidize methane and is believed to have a similar polypeptide composition as pMMO (Arp et al., 2002; Holmes et al., 1995). The physiological reductant for pMMO has not been identified definitively, but might involve quinones from the quinone pool reduced by a type 2 NADH:quinone oxidoreductase (Choi et al., 2003; Shiemke et al., 2004; Cook & Shiemke, 2002) or by methanol dehydrogenase (Basu et al., 2003). Unlike sMMO, pMMO has a narrow substrate specificity, only oxidizing alkanes containing up to five carbons and alkenes containing up to four carbons (Jiang et al., 2010; Burrows et al., 1984; Smith & Dalton, 1989; Nguyen et al., 1996). The nature of the pMMO metal active site has been controversial (Chan et al., 2004; Lieberman & Rosenzweig, 2004; Balasubramanian & Rosenzweig, 2007; Rosenzweig, 2008; Chan & Yu, 2008; Semrau et al., 2010), and is critically important since pMMO is the predominant methane oxidation catalyst in nature, although it should be noted that other systems oxidize methane in anerobic environments (Thauer, 2011). Major advances toward understanding the pMMO active site have been made over the past few years and are the focus of this review.
Structure of pMMO
Overall architecture
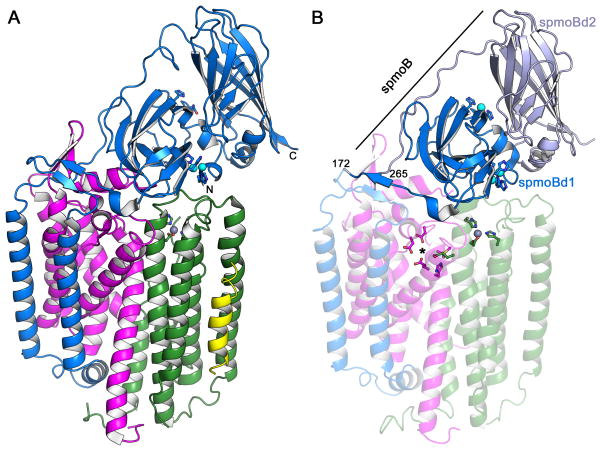
Crystal structures of pMMOs from three different organisms have been determined, the M. capsulatus (Bath) pMMO to 2.8 Å resolution (Lieberman & Rosenzweig, 2005a; Lieberman & Rosenzweig, 2005b), the M. trichosporium OB3b pMMO to 3.9 Å resolution (Hakemian et al., 2008), and the Methylocystis sp. strain M pMMO to 2.68 Å resolution (Smith et al., 2011b). Of the three, the Methylocystis sp. strain M pMMO structure is the best quality model (Smith et al., 2011b). The overall architecture of pMMO is the same in all three structures, an α3β3γ3 trimer with three copies each of the pmoB, pmoA, and pmoC subunits. The 100 kDa complex consisting of one copy each of pmoB, pmoA, and pmoC is typically referred to as the pMMO protomer (Figure 1A). The pmoB subunit consists of an N-terminal cupredoxin domain followed by two transmembrane helices, a long linker region, and a C-terminal cupredoxin domain. These cupredoxin domains, located in the periplasm, are the only soluble domains of pMMO. The pmoA and pmoC subunits are primarily composed of transmembrane helices. The recent determination of the Methylocystis sp. strain M pMMO structure revealed several discrepancies in these chains with the original M. capsulatus (Bath) pMMO crystallographic model, and the M. capsulatus (Bath) pMMO structure was re-refined accordingly, leading to a much improved structure (Smith et al., 2011b).
Figure 1.
Overall architecture of pMMO. (A) Structure of Methylocystis sp. strain M pMMO protomer (PDB accession code 3RGR). The pmoB, pmoA, and pmoC subunits are shown in blue, magenta, and green, respectively. The N- and C-termini of pmoB are labeled. An exogenous helix is shown in yellow. Copper ions are shown as cyan spheres and a zinc ion is shown as a gray sphere. Ligands are shown as ball-and-stick representations. (B) Structure of M. capsulatus (Bath) pMMO protomer (PDB accession code 3RGB). The amino terminal domain of pmoB (spmoBd1) is shown in blue, the carboxy terminal domain of pmoB is shown in gray (spmoBd2), and the two transmembrane helices are shown in transparent blue. In the recombinant spmoB protein, spmoBd1 and spmoBd2 are linked by a GKLGGG sequence connecting residues 172 and 265 (labeled). The pmoA and pmoC subunits are shown in transparent magenta and transparent green, respectively. A hydrophilic patch of residues proposed to house a tricopper active site is denoted with an asterisk. The mononuclear copper site at the interface of the two spmoB domains is not present in the Methylocystis sp. strain M pMMO structure. [A color version of this figure is available online.]
Both the M. trichosporium OB3b (Hakemian et al., 2008) and Methylocystis sp. strain M (Smith et al., 2011b) pMMO models include an additional transmembrane helix, not found in the M. capsulatus (Bath) structure, modeled as polyalanine, adjacent to the pmoC subunit (Figure 1A). The electron density maps and chain tracings clearly indicate that this helix is not a part of either pmoC or pmoA, but the limited resolution has precluded assigning the side chains. This helix is definitely not present in the M. capsulatus (Bath) pMMO structure (Figure 1B) and thus may be specific to pMMO from the type II methanotrophs. Unidentified, exogenous helices have been observed other membrane protein structures (Buschmann et al., 2010), and the significance of this helix for pMMO remains unclear.
Crystallographic metal centers
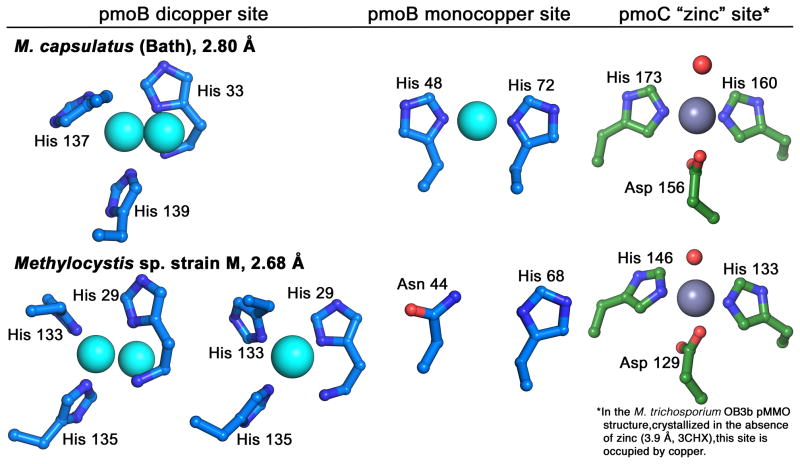
Three metal centers were observed in the original M. capsulatus (Bath) pMMO structure and identified using anomalous difference electron density maps (Lieberman & Rosenzweig, 2005a; Sommerhalter et al., 2005). Two copper centers were modeled in the soluble regions of pmoB, a mononuclear site at the interface between the two cupredoxin domains and a dinuclear site close to the membrane interface (Figure 1B). The mononuclear site is coordinated by His 48 and His 72 (Figure 2). The residue equivalent to His 48 is an asparagine in both the M. trichosporium OB3b and Methylocystis sp. strain M pMMOs, and neither contains copper at this site (Hakemian et al., 2008; Smith et al., 2011b) (Figures 1A, 2). The dinuclear site is coordinated by His 33, His 137, and His 139. Residue His 33 is the amino terminal residue of pmoB (the first 32 residues are a signaling sequence that is removed in vivo), and its amino group is within coordinating distance of the copper (Figure 2). These three histidine residues are conserved in all methanotroph pmoB sequences except those from the Verrucomicrobia (Op den Camp et al., 2009) (Figure 3). Since the crystallographic resolution is not sufficient to distinguish two copper ions so close together, this model relies heavily on extended X-ray absorption fine structure (EXAFS) data indicating the presence of a Cu-Cu interaction at 2.5–2.6 Å (Lieberman et al., 2006; Lieberman et al., 2003). This model is also consistent with the stoichiometry of 2–3 copper ions per 100 kDa M. capsulatus (Bath) pMMO protomer reported by several laboratories (Lieberman & Rosenzweig, 2004; Lieberman et al., 2003; Basu et al., 2003). However, in the structure of Methylocystis sp. strain M pMMO, two of the three putative dinuclear sites were modeled as mononuclear (Smith et al., 2011b) (Figure 2). This may be due to lability of the copper sites observed during isolation of this pMMO.
Figure 2.
Metal centers modeled in the pMMO crystal structures. The pmoB site modeled as dicopper in the M. capsulatus (Bath) pMMO structure is modeled as dicopper in one protomer and monocopper in two protomers in the Methylocystis sp. strain M pMMO structure. The monocopper site in pmoB from M. capsulatus (Bath) pMMO does not contain copper in the other pMMO structures. In both structures, zinc is found in the transmembrane site as a result of including zinc in the crystallization buffer. Copper occupies this site in M. trichosporium OB3b pMMO. [A color version of this figure is available online.]
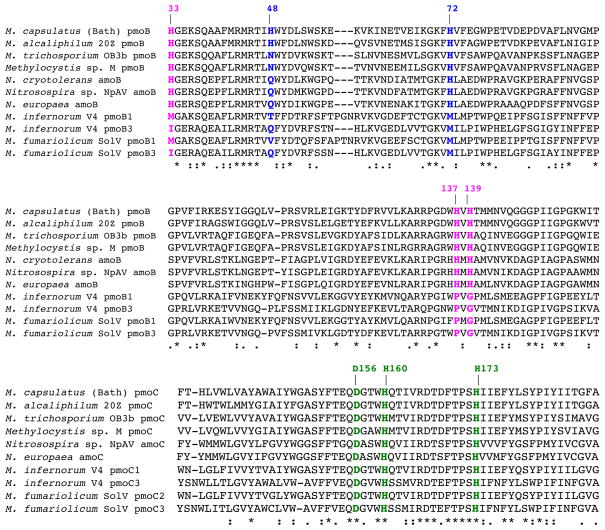
Figure 3.
Multiple sequence alignments of representative pmoB/amoB and pmoC/amoC sequences containing the ligands to the crystallographically observed pMMO metal centers. The ligands to the pmoB dicopper site are shown in magenta, the ligands to the pmoB monocopper site in M. capsulatus (Bath) pMMO are shown in blue, and the ligands to the zinc site are shown in green. The ligands to the dicopper center are not conserved in the bottom four Verrucomicrobia sequences. [A color version of this figure is available online.]
The third metal center is located within the membrane and is occupied by zinc in the M. capsulatus (Bath) and Methylocystis sp. strain M pMMO structures. This zinc is derived from the crystallization buffer and was originally modeled as coordinated by residues Asp 156, His 160, and His 173 from pmoC and Glu 195 from pmoA (Lieberman & Rosenzweig, 2005a). The revised model based on the Methylocystis sp. strain M pMMO structure resulted in the replacement of Glu 195 from pmoA with a solvent molecule and increased exposure of this site in the center of the pMMO trimer (Smith et al., 2011b) (Figure 2). In the case of M. trichosporium OB3b pMMO, zinc was not required for crystallization, and this site is occupied by copper (Hakemian et al., 2008). It is currently unknown whether the copper at this site is biologically relevant or originates from the addition of copper during the protein purification. The three ligands are highly conserved, even in the Verrucomicrobia sequences (Hakemian & Rosenzweig, 2007; Op den Camp et al., 2009) (Figure 3).
The pMMO active site
Metal content and location
A variety of models for the pMMO active site have been proposed in the context of the crystallographic data (Rosenzweig, 2008). One model involves the presence of a trinuclear copper center, first suggested on the basis of electron paramagnetic resonance (EPR) spectroscopic data (Chen et al., 2004; Nguyen et al., 1996). Although not present in the crystal structures, this trinuclear copper center is suggested to reside at a hydrophilic patch of residues within the pmoA and pmoC subunits (Chan et al., 2007) (Figure 1B). This model also includes approximately ten additional copper sites within the C-terminal cupredoxin domain of pmoB (Yu et al., 2007), which are not observed in the crystal structure. A second model invokes iron as the active site metal. On the basis of Mössbauer spectroscopic data, a diiron center similar to that in sMMO was proposed to be present in pMMO at the site of the crystallographic zinc ion (Martinho et al., 2007) (Figure 1). Finally, the consistent observation of copper at the modeled dinuclear site has led to the suggestion that the catalytic site is at this location within the pmoB subunit (Rosenzweig, 2008).
These models were tested experimentally by removing all the metal from membrane-bound M. capsulatus (Bath) pMMO using cyanide and then assessing which metal ions restore both methane oxidation and propylene epoxidation activity (Smith et al., 2011a). Up to 90% of enzyme activity was recovered upon the addition of 2–3 copper ions per 100 kDa pMMO protomer. By contrast, iron addition has no effect on pMMO activity (Balasubramanian et al., 2010). These observations are consistent with a copper active site model, and similar results were obtained with Methylocystis sp. strain M pMMO, although this pMMO has much lower activity upon isolation (Smith et al., 2011b). The addition of more than 2–3 equivalents of copper results in reversible inhibition of activity (Balasubramanian et al., 2010), which can be ameliorated by the addition of catalase, and may result from hydrogen peroxide formation (Miyaji et al., 2009).
To localize the active site within the pMMO structure, constructs were generated corresponding to the two soluble cupredoxin domains of M. capsulatus (Bath) pmoB, both alone (spmoBd1 and spmoBd2) and connected (spmoB) via a synthetic linker that replaces the two transmembrane helices (Balasubramanian et al., 2010; Smith et al., 2011a) (Figure 1B). These proteins, which express in E. coli primarily as inclusion bodies, were refolded in the presence of copper, and their copper binding, spectroscopic, and activity properties were analyzed. Both spmoB and spmoBd1 bind copper, but spmoBd2 does not, contrary to previous reports (Yu et al., 2007). Methane oxidation activity is also observed for both spmoB and spmoBd1, strongly suggesting that the active site is housed within this soluble part of pmoB (Balasubramanian et al., 2010). To pinpoint the active site location, site directed variants designed to disrupt the copper sites were prepared. Activity is observed upon disruption of the mononuclear site (spmoB_H48N), but is abolished in variants that interfere with the dicopper site (spmoB_H137,139A) or both sites (spmoB_H48N_H137,139A) (Balasubramanian et al., 2010). These results suggest that the active site of pMMO is the crystallographically modeled dicopper site (Balasubramanian et al., 2010).
The dicopper active site model
There are several unresolved issues with the assignment of the crystallographic dicopper center as the pMMO active site. First, the three histidine ligands are not conserved in the recently discovered Verrucomicrobia pmoB sequences (Op den Camp et al., 2009) (Figure 3), suggesting that a different type of metal center would have to be present in these pmoB subunits. The Verrucomicrobia sequences are overall quite divergent from the Proteobacterial sequences (although the ligands to the crystallographic zinc site are conserved) and are evolutionarily distant (Op den Camp et al., 2009). It is perhaps not surprising that there is some divergence given that these methanotrophs exist in high temperature environments with pH values ranging from 1–4.5. Although it is not clear what pH the active site is exposed to, it is worth noting that at pH 4.5, methionine has a higher affinity for Cu(I) than histidine (Rubino et al., 2011). Thus, it is conceivable that the extreme environment may necessitate a completely different metal active site.
Another issue is the low activity of the spmoB proteins. The methane oxidation activity is about 10% of that of intact membrane-bound pMMO (Table 1) (Balasubramanian et al., 2010), which is in turn about 10% of whole cell activity (Martinho et al., 2007). This observation could imply that the active site is not within this subunit, but it is unlikely that artifactual methane oxidation would be observed. It may be that the more exposed nature of the site in spmoB, which is also probably only partially loaded with copper, along with inherent problems in refolding proteins from inclusion bodies (Singh & Panda, 2005), account for the low activity. Moreover, the functional roles of the membrane domains have not been elucidated and their absence likely impacts the activity of spmoB. One possibility is that methane enters the active site from the membrane, where it would be more soluble (Miller et al., 1977), and this more favorable and defined substrate entryway is not present in the soluble domain.
Table 1.
| Membrane-bound | Purified | Reference | |||
|---|---|---|---|---|---|
| Coppers per 100 kDa protomer | Activity (nmol propylene oxide min−1mg−1) | Coppers per 100kDa protomer | Activity (nmol propylene oxide min−1mg−1) | ||
| M. capsulatus (Bath) | 20.8 ± 4.5 | 16 | 2.4 ± 0.4 | 17.7 | Leiberman et al., 2003 |
| M. trichosporium OB3b | 4.8 ± 1.1 | 3.0 ± 05 | 1.4 ± 0.6 | 0.11 ± 0.1 | Hakemian et al., 2008 |
| Methylocystis sp. strain M | 2.29 ± 0.22 | 5.3 ± 1.4 | 2.11 ± 0.463 | 1.24 ± 0.213 | Smith et al., 2011b |
|
| |||||
| Coppers per 100 kDa | Activity (nmol CH3OH min−1μmol−1) | Coppers per mole | Activity (nmol CH3OH min −1μmol−1) | ||
|
| |||||
| M. capsulatus (Bath) | 12.4 ± 4 | 2290 ± 60 | Balasubramanian et al., 2010 | ||
| spmoB | 2.84 ± 0.66 | 203.1 ± 20.2 | ” | ||
| spmoBd1 | 1.59 ± 0.84 | 19.3 ± 4.7 | ” | ||
| spmoBd2 | 0.24 ± 0.04 | 0 | ” | ||
| spmoB_H48N | 1.86 ± 0.52 | 14.8 ± 9.2 | ” | ||
| spmoB_H137,139A | 0.75 ± 0.15 | 0 | ” | ||
| spmoB_H48N_H137,139A | 0.82 ± 0.36 | 0 | ” | ||
Data from the Rosenzweig laboratory are summarized. Comprehensive tables can be found in Lieberman & Rosenzweig, 2004 and Semrau et al., 2010.
Activity measured using duroquinol as a reductant.
Copper content and activity values were obtained from detergent solubilized samples.
Finally, the nuclearity of the active site has not been established definitively. EXAFS data for the three pMMOs studied to date indicate the presence of a short Cu-Cu interaction at ~2.5 Å, which increases to ~2.6 Å upon chemical reduction (Hakemian et al., 2008; Lieberman et al., 2006; Lieberman et al., 2003; Smith et al., 2011b). The only location in the crystal structure consistent with a multinuclear copper site is the dicopper center modeled in the pmoB subunit, but the current resolution of the structures (2.7 Å at best) precludes clearly resolving two copper ions separated by < 2.7 Å. This site is strongly occupied with copper in all pMMO structures and can be modeled as dinuclear (Lieberman & Rosenzweig, 2005a) or, in some subunits of the Methylocystis sp. strain M pMMO (Smith et al., 2011b), as mononuclear (Figure 2). The only visible ligands are the three histidine side chains and the amino terminus of pmoB, leaving open the question of whether exogenous bridging or terminal ligands might also be present. The copper binding stoichiometry of M. capsulatus (Bath) pMMO of 2–3 copper ions per protomer (Lieberman & Rosenzweig, 2004) (Table 1) is consistent with the presence of mononuclear and dinuclear sites, assuming that the crystallographic zinc site is not occupied with copper. The copper binding stoichiometries for M. trichosporium OB3b and Methylocystis sp. strain M pMMO are closer to 2 copper ions per protomer (Hakemian et al., 2008; Smith et al., 2011b) (Table 1), which could be interpreted as a dicopper center in pmoB or as a monocopper center in pmoB and a monocopper center in the membrane, as observed in the M. trichosporium OB3b pMMO structure (Hakemian et al., 2008). However, the copper occupancy of this intramembrane site is quite low and may not account for the measured stoichiometry. Moreover, the metal stoichiometry observed in crystal structures is often not consistent with that measured for purified proteins in solution (Sommerhalter et al., 2005). Also relevant is the reduced activity of the M. trichosporium OB3b and Methylocystis sp. strain M pMMOs in comparison to M. capsulatus (Bath) pMMO (Hakemian et al., 2008; Smith et al., 2011b) (Table 1), possibly because of increased lability of the copper active site. Thus, although most of the data, including oxygen binding experiments (vide infra) are compatible with the dicopper active site model, a high resolution structure of the site is essential to ascertain the nuclearity.
Substrate and product binding
Despite the availability of crystallographic data and strong evidence for the active site being located in the pmoB subunit, direct evidence for methane or methanol binding sites and pathways to access the active site has not been obtained. Acetylene has long been known to act as a suicide substrate of pMMO, and studies using 14C-labeled acetylene suggest that the label ends up in the pmoA subunit (Cook & Shiemke, 1996; Zahn & DiSpirito, 1996) and might also bind to the pmoB subunit (Zahn & DiSpirito, 1996). Similar experiments with AMO also resulted in labeling of the pmoA subunit (Hyman & Wood, 1985; Hyman & Arp, 1992), specifically residue His 191, and this residue is assumed to be part of or neighboring the active site (Gilch et al., 2009). The analogous residue in M. capsulatus (Bath) pMMO is Tyr 186, located ~14 Å from the dicopper site in pmoB, ~24 Å from the hydrophilic patch that is proposed to house the tricopper site, and ~26 Å from the zinc site proposed to house the diiron active site. It should also be noted that the retracing of pmoA and pmoC in the Methylocystis sp. strain M pMMO structure places the zinc site within pmoC (Smith et al., 2011b), and if it were to house a diiron active site as suggested (Martinho et al., 2007), pmoC, not pmoA, should be labeled. It thus remains unclear how to interpret the acetylene binding data in the context of the structural and other biochemical data.
Substrate and product binding have also been explored computationally. Using Global Protein Surface Survey (GPSS) analysis, a hydrophobic pocket adjacent to the proposed tricopper center was identified in M. capsulatus (Bath) pMMO. This pocket is lined with Phe 50, Trp 51, and Trp 54 from pmoA, and is suggested to accommodate hydrocarbons up to five carbons in length (Ng et al., 2008). Recent work on M. trichosporium OB3b pMMO using the substrates n-butane and n-pentane indicates that the (R)-2-alcohol product is formed enantioselectively (Miyaji et al., 2011), consistent with previous studies of M. capsulatus (Bath) pMMO (Elliott et al., 1997). Therefore, the binding cavity can orient these substrates. Since pMMO can oxidize pentane, but not hexane, the volume of the substrate binding site can be approximated to ~138 Å3, and the existence of distinct binding sites for methane and larger substrates has been suggested (Miyaji et al., 2011). Little is known about product egress, but cryoelectron microscopic studies suggest that methanol dehydrogenase might interact with the periplasmic soluble domains of pmoB (Myronova et al., 2006), and it is plausible that methanol would exit the enzyme through these regions.
Oxygen binding
The O2 reactivity of the copper sites in pMMO has been addressed computationally (Shiota & Yoshizawa, 2009; Yoshizawa & Shiota, 2006; Chan et al., 2004). According to DFT and QM/MM calculations using the two copper centers modeled in the pmoB subunit of the M. capsulatus (Bath) structure, both a mononuclear CuIII-oxo species and a dinuclear mixed valent bis(μ-oxo)CuIICuIII species are sufficiently reactive to oxidize methane, although formation of the dinuclear species is energetically more favorable than the mononuclear species (Shiota & Yoshizawa, 2009; Yoshizawa & Shiota, 2006). Related to the potential ability of a dicopper species to oxidize methane, the active species in a Cu-ZSM-5 zeolite that converts methane to methanol has been shown to be a mono-μ-oxo CuII2 species that forms from a μ-η2:η2-peroxo CuII2 precursor (Groothaert et al., 2005; Vanelderen et al., 2011; Woertink et al., 2009; Smeets et al., 2010).
The binding of O2 to pMMO has also been investigated experimentally. Recent data show that an optical feature at 345 nm forms upon reaction of reduced, solubilized M. capsulatus (Bath) pMMO with H2O2, and the same feature can also be observed upon reaction of reduced spmoB with either H2O2 or O2 (Culpepper & Rosenzweig, 2012) (Figure 4). The energy and intensity of this band are similar to that of the well characterized μ-η2:η2-peroxo CuII2 species of hemocyanin and tyrosinase (Solomon et al., 1996; Solomon et al., 2011). The feature may alternatively correspond to a met CuII2 species, like that of hemocyanin (Zlateva et al., 1998; Andrew et al., 1993), or to a hydroxo bridged CuII2 species, similar to the type 3 site of multicopper oxidases (Solomon et al., 1996). Sample constraints, including the instability of spmoB and the presence of heme contaminants in pMMO, have precluded further analysis of this species by resonance Raman spectroscopy, however. Importantly, the 345 nm optical feature is also observed for the spmoB_H48N variant, but not for the two variants (spmoB_H137,139A and spmoB_H48N_H137,139A) that disrupt the modeled dicopper center, suggesting that O2 is indeed binding at the proposed active site. Moreover, disappearance of the optical feature is enhanced by the presence of CH4, consistent with it possibly being on the reaction pathway (Culpepper & Rosenzweig, 2012). These data support the assignment of the active site and are consistent with the presence of a dicopper center. The parallel to the Cu-ZSM-5 zeolite is also striking and reinforces the plausibility of the dicopper active site model.
Figure 4.
Optical spectrum of detergent-solubilized pMMO from M. capsulatus (Bath) (top) and difference spectra of recombinant spmoB (bottom) reduced and oxidized with either H2O2 or O2. The feature at 345 nm is similar to that observed for the μ-η2:η2-peroxo CuII2 species formed in hemocyanin and tyrosinase. The peak at 410 nm derives from heme contaminants in the pMMO sample.
Outlook
Since the last Critical Review on pMMO (Lieberman & Rosenzweig, 2004), much progress has been made toward understanding the molecular details of methane oxidation. Three crystal structures of pMMO have been determined, revealing the overall architecture and the locations of several metal centers. Biochemical data indicate that pMMO activity is copper-dependent, and the active site has been localized to the crystallographically modeled dicopper center in the pmoB subunit. Initial spectroscopic evidence for O2 binding at this dicopper site has also been obtained. Taken together, these data are consistent with the active site of pMMO being a dicopper center. Concerns about absence of the ligands in the Verrucomicrobia pmoB sequences and the site nuclearity persist, however. High resolution crystallographic data are absolutely critical to further characterization of the copper active site. The development of the soluble, recombinant spmoB proteins represents the first opportunity to probe pMMO function by site-directed mutagenesis, and suggests that optimized soluble pMMO fragments might be a viable approach toward obtaining additional crystallographic, spectroscopic, and mechanistic data. The function of the transmembrane regions has not been elucidated and will be an important direction for future work. In this regard, the ability to express intact pMMO would be highly desirable. Overall, a multipronged strategy involving pMMO, recombinant models, and studies of related synthetic systems will be required to resolve atomic details of the active site and proceed to the next stages of investigating the catalytic mechanism.
Footnotes
Declaration of Interest
Research in the Rosenzweig laboratory on particulate methane monooxygenase is supported by the National Institutes of Health (grant GM070473 to ACR and grant F32GM097049 to MAC).
References
- Ali H, Murrell JC. Development and validation of promoter-probe vectors for the study of methane monooxygenase gene expression in Methylococcus capsulatus Bath. Microbiol. 2009;155:761–771. doi: 10.1099/mic.0.021816-0. [DOI] [PubMed] [Google Scholar]
- Andrew CR, McKillop KP, Sykes AG. Kinetic studies on the reactions of separated a, b and c subunits of Panulirus interruptus deoxy-hemocyanin with hydrogen peroxide. Biochim Biophys Acta. 1993;1163:17–25. doi: 10.1016/0167-4838(93)90273-t. [DOI] [PubMed] [Google Scholar]
- Arakawa H, Aresta M, Armor JN, Barteau MA, Beckman EJ, Bell AT, Bercaw JE, Creutz C, Dinjus E, Dixon DA, Domen K, DuBois DL, Eckert J, Fujita E, Gibson DH, Goddard WA, Goodman DW, Keller J, Kubas GJ, Kung HH, Lyons JE, Manzer LE, Marks TJ, Morokuma K, Nicholas KM, Periana R, Que L, Rostrup-Nielson J, Sachtler WMH, Schmidt LD, Sen A, Somorjai GA, Stair PC, Stults BR, Tumas W. Catalysis research of relevance to carbon management: progress, challenges, and opportunities. Chem Rev. 2001;101:953–996. doi: 10.1021/cr000018s. [DOI] [PubMed] [Google Scholar]
- Arp DJ, Sayavedra-Soto LA, Hommes NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol. 2002;178:250–255. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Rosenzweig AC. Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc Chem Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R, Smith SM, Rawat S, Stemmler TL, Rosenzweig AC. Oxidation of methane by a biological dicopper centre. Nature. 2010;465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu P, Katterle B, Andersson KK, Dalton H. The membrane-associated form of methane monooxygenase from Methylococcus capsulatus (Bath) is a copper/iron protein. Biochem J. 2003;369:417–427. doi: 10.1042/BJ20020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden R, Cunliffe M, Scanlan J, Moussard H, Kits KD, Klotz MG, Jetten MSM, Vuilleumier S, Han J, Peters L, Mikhailova N, Teshima H, Tapia R, Kyrpides N, Ivanova N, Pagani I, Cheng JF, Goodwin L, Han C, Hauser L, Land ML, Lapidus A, Lucas S, Pitluck S, Woyke T, Stein L, Murrell JC. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J Bacteriol. 2011;193:7001–7002. doi: 10.1128/JB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows KJ, Cornish A, Scott D, Higgins IJ. Substrate specificities of the soluble and particulate methane monooxygenases of Methylosinus trichosporium OB3b. J Gen Microbiol. 1984;130:327–3333. [Google Scholar]
- Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- Chan SI, Chen KH-C, Yu SS-F, Chen C-L, Kuo SS-J. Toward delineating the structure and function of the particulate methane monooxygenase from methanotrophic bacteria. Biochemistry. 2004;43:4421–4430. doi: 10.1021/bi0497603. [DOI] [PubMed] [Google Scholar]
- Chan SI, Wang VCC, Lai JCH, Yu SSF, Chen PPY, Chen KHC, Chen CL, Chan MK. Redox potentiometry studies of particulate methane monooxygenase: support for a trinuclear copper cluster active site. Angew Chem Int Ed. 2007;46:1992–1994. doi: 10.1002/anie.200604647. [DOI] [PubMed] [Google Scholar]
- Chan SI, Yu SSF. Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc Chem Res. 2008;41:969–979. doi: 10.1021/ar700277n. [DOI] [PubMed] [Google Scholar]
- Chen KH-C, Chen C-L, Tseng C-F, Yu SS-F, Ke S-C, Lee J-F, Nguyen HT, Elliott SJ, Alben JO, Chan SI. The copper clusters in the particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath) J Chin Chem Soc. 2004;51:1081–1098. [Google Scholar]
- Chen Y, Crombie A, Rahman MT, Dedysh SN, Liesack W, Stott MB, Alam M, Theisen AR, Murrell JC, Dunfield PF. Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J Bacteriol. 2010;192:3840–3841. doi: 10.1128/JB.00506-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Kunz RC, Boyd ES, Semrau JD, Antholine WE, Han JI, Zahn JA, Boyd JM, de la Mora AM, DiSpirito AA. The membrane-associated methane monooxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J Bacteriol. 2003;185:5755–5764. doi: 10.1128/JB.185.19.5755-5764.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J, Stirling DI, Dalton H. The soluble methane monooxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SA, Shiemke AK. Evidence that copper is a required cofactor for the membrane-bound form of methane monooxygenase. J Inorg Biochem. 1996;63:273–284. [Google Scholar]
- Cook SA, Shiemke AK. Evidence that a type-2 NADH:quinone oxidoreductase mediates electron transfer to particulate methane monooxygenase in Methylococcus capsulatus. Arch Biochem Biophys. 2002;398:32–40. doi: 10.1006/abbi.2001.2628. [DOI] [PubMed] [Google Scholar]
- Csáki R, Bodrossy L, Klem J, Murrell JC, Kovács KL. Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis. Microbiol. 2003;149:1785–1795. doi: 10.1099/mic.0.26061-0. [DOI] [PubMed] [Google Scholar]
- Culpepper MA, Rosenzweig AC. Evidence for oxygen binding at the active site of particulate methane monooxygenase. J Am Chem Soc. 2012;134:7640–7643. doi: 10.1021/ja302195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh SN, Dunfield PF. Facultative and obligate methanotrophs: how to identify and differentiate them. Meth Enzymol. 2011;495B:31–44. doi: 10.1016/B978-0-12-386905-0.00003-6. [DOI] [PubMed] [Google Scholar]
- Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature. 2007;450:879–882. doi: 10.1038/nature06411. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Zhu M, Tso L, Nguyen H-H, Yip JH-K, Chan SI. Regio- and stereoselectivity of particulate methane monooxygenase from Methylococcus capsulatus (Bath) J Am Chem Soc. 1997;119:9949–9955. [Google Scholar]
- Gilch S, Vogel M, Lorenz MW, Meyer O, Schmidt I. Interaction of the mechanism-based inactivator acetylene with ammonia monooxygenase of Nitrosomonas europaea. Microbiol. 2009;155:279–284. doi: 10.1099/mic.0.023721-0. [DOI] [PubMed] [Google Scholar]
- Groothaert MH, Smeets PJ, Sels BF, Jacobs PA, Schoonheydt RA. Selective oxidation of methane by the bis(μ-oxo)dicopper core stabilized on ZSM-5 and mordenite zeolites. J Am Chem Soc. 2005;127:1394–1395. doi: 10.1021/ja047158u. [DOI] [PubMed] [Google Scholar]
- Hakemian AS, Kondapalli KC, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry. 2008;47:6793–6801. doi: 10.1021/bi800598h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Ann Rev Biochem. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–208. doi: 10.1016/0378-1097(95)00311-r. [DOI] [PubMed] [Google Scholar]
- Hou SB, Makarova KS, Saw JHW, Senin P, Ly BV, Zhou ZM, Ren Y, Wang JM, Galperin MY, Omelchenko MV, Wolf YI, Yutin N, Koonin EV, Stott MB, Mountain BW, Crowe MA, Smirnova AV, Dunfield PF, Feng L, Wang L, Alam M. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biology Direct. 2008;3 doi: 10.1186/1745-6150-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman MR, Arp DJ. 14C2H2-Labeling and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J Biol Chem. 1992;267:1534–1545. [PubMed] [Google Scholar]
- Hyman MR, Wood PM. Suicidal inactivation and labelling of ammonia monooxygenase by acetylene. Biochem J. 1985;227:719–725. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T, Jensen S, Reigstad LJ, Larsen O, Birkeland NK. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA. 2008;105:300–304. doi: 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Chen Y, Jiang PX, Zhang C, Smith TJ, Murrell JC, Xing XH. Methanotrophs: Multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J. 2010;49:277–288. [Google Scholar]
- Lieberman RL, Kondapalli KC, Shrestha DB, Hakemian AS, Smith SM, Telser J, Kuzelka J, Gupta R, Borovik AS, Lippard SJ, Hoffman BM, Rosenzweig AC, Stemmler TL. Characterization of the particulate methane monooxygenase metal centers in multiple redox states by X-ray absorption spectroscopy. Inorg Chem. 2006;45:8372–8381. doi: 10.1021/ic060739v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman RL, Rosenzweig AC. Biological methane oxidation: regulation, biochemistry, and active site structure of particulate methane monooxygenase. Crit Rev Biochem Mol Biol. 2004;39:147–164. doi: 10.1080/10409230490475507. [DOI] [PubMed] [Google Scholar]
- Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005a;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- Lieberman RL, Rosenzweig AC. The quest for the particulate methane monooxygenase active site. Dalton Trans. 2005b;21:3390–3396. doi: 10.1039/b506651d. [DOI] [PubMed] [Google Scholar]
- Lieberman RL, Shrestha DB, Doan PE, Hoffman BM, Stemmler TL, Rosenzweig AC. Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc Natl Acad Sci USA. 2003;100:3820–3825. doi: 10.1073/pnas.0536703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho M, Choi DW, DiSpirito AA, Antholine WE, Semrau JD, Münck E. Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J Am Chem Soc. 2007;129:15783–15785. doi: 10.1021/ja077682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew Chem Int Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Miller KW, Hammond L, Porter EG. The solubility of hydrocarbon gases in lipid bilayers. Chem Phys Lipids. 1977;20:229–241. [Google Scholar]
- Miyaji A, Miyoshi T, Motokura K, Baba T. The substrate binding cavity of particulate methane monooxygenase from Methylosinus trichosporium OB3b expresses high enantioselectivity for n-butane and n-pentane oxidation to 2-alcohol. Biotech Lett. 2011;33:2241–2246. doi: 10.1007/s10529-011-0688-3. [DOI] [PubMed] [Google Scholar]
- Miyaji A, Suzuki M, Baba T, Kamachi T, Okura I. Hydrogen peroxide as an effecter on the inactivation of particulate methane monooxygenase under aerobic conditions. J Mol Catal B. 2009;57:211–215. [Google Scholar]
- Myronova N, Kitmitto A, Collins RF, Miyaji A, Dalton H. Three-dimensional structure determination of a protein supercomplex that oxidizes methane to formaldehyde in Methylococcus capsulatus (Bath) Biochemistry. 2006;45:11905–11914. doi: 10.1021/bi061294p. [DOI] [PubMed] [Google Scholar]
- Ng K-Y, Tu L-C, Wang Y-S, Chan SI, Yu SSF. Probing the hydrophobic pocket of the active site in the particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath) by variable stereoselective alkane hydroxylation and olefin epoxidation. ChemBioChem. 2008;9:1116–1123. doi: 10.1002/cbic.200700628. [DOI] [PubMed] [Google Scholar]
- Nguyen H-HT, Nakagawa KH, Hedman B, Elliott SJ, Lidstrom ME, Hodgson KO, Chan SI. X-ray absorption and EPR studies on the copper ions associated with the particulate methane monooxygenase from Methylococcus capsulatus (Bath). Cu(I) ions and their implications. J Am Chem Soc. 1996;118:12766–12776. [Google Scholar]
- Olah GA. Beyond oil and gas: The methanol economy. Angew Chem Int Ed. 2005;44:2636–2639. doi: 10.1002/anie.200462121. [DOI] [PubMed] [Google Scholar]
- Op den Camp HJ, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep. 2009;1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, den Camp H. Methanotrophy below pH1 by a new Verrucomicrobia species. Nature. 2007;450:874–878. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- Prior SD, Dalton H. The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath) J Gen Microbiol. 1985;131:155–163. [Google Scholar]
- Rosenzweig AC. The metal centres of particulate methane monooxygenase. Biochem Soc Trans. 2008;36:1134–1137. doi: 10.1042/BST0361134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig AC, Frederick CA, Lippard SJ, Nordlund P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature. 1993;366:537–543. doi: 10.1038/366537a0. [DOI] [PubMed] [Google Scholar]
- Rubino JT, Chenkin MP, Keller M, Riggs-Gelasco P, Franz KJ. A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics. 2011;3:61–73. [PubMed] [Google Scholar]
- Semrau JD, Dispirito AA, Murrell JC. Life in the extreme: thermoacidophilic methanotrophy. Trends Microbiol. 2008;16:190–193. doi: 10.1016/j.tim.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Semrau JD, Dispirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Lett. 2010;34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- Shiemke AK, Arp DJ, Sayavedra-Soto LA. Inhibition of membrane-bound methane monooxygenase and ammonia monooxygenase by diphenyliodonium: implications for electron transfer. J Bacteriol. 2004;186:928–937. doi: 10.1128/JB.186.4.928-937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota Y, Yoshizawa K. Comparison of the reactivity of bis(μ-oxo)CuIICuIII and CuIIICuIII species to methane. Inorg Chem. 2009;48:838–845. doi: 10.1021/ic8003933. [DOI] [PubMed] [Google Scholar]
- Singh SM, Panda AK. Solubilization and refolding of bacterial inclusion body proteins. J Biosci Bioeng. 2005;99:303–310. doi: 10.1263/jbb.99.303. [DOI] [PubMed] [Google Scholar]
- Smeets PJ, Hadt RG, Woertink JS, Vanelderen P, Schoonheydt RA, Sels BF, Solomon EI. Oxygen precursor to the reactive intermediate in methanol synthesis by Cu-ZSM-5. J Am Chem Soc. 2010;132:14736–14738. doi: 10.1021/ja106283u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DDS, Dalton H. Solubilisation of methane monooxygenase from Methylococcus capsulatus (Bath) Eur J Biochem. 1989;182:667–671. doi: 10.1111/j.1432-1033.1989.tb14877.x. [DOI] [PubMed] [Google Scholar]
- Smith SM, Balasubramanian R, Rosenzweig AC. Metal reconstitution of particulate methane monooxygenase and heterologous expression of the pmoB subunit. Methods Enzymol. 2011a;495:195–210. doi: 10.1016/B978-0-12-386905-0.00013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Rawat S, Telser J, Hoffman BM, Stemmler TL, Rosenzweig AC. Crystal structure and characterization of particulate methane monooxygenase from Methylocystis species strain M. Biochemistry. 2011b;50:10231–10240. doi: 10.1021/bi200801z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon EI, Ginsbach JW, Heppner DE, Kieber-Emmons MT, Kjaergaard CH, Smeets PJ, Tian L, Woertink JS. Copper dioxygen (bio) inorganic chemistry. Faraday Dis. 2011;148:11–39. doi: 10.1039/c005500j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- Sommerhalter M, Lieberman RL, Rosenzweig AC. X-ray crystallography and biological metal centers: is seeing believing? Inorg Chem. 2005;44:770–778. doi: 10.1021/ic0485256. [DOI] [PubMed] [Google Scholar]
- Stafford GP, Scanlan J, McDonald IR, Murrell JC. rpoN, mmoR and mmoG, genes involved in regulating the expression of soluble methane monooxygenase from Methylosinus trichosporium OB3b. Microbiol. 2003;149:1771–1784. doi: 10.1099/mic.0.26060-0. [DOI] [PubMed] [Google Scholar]
- Stanley SH, Prior SD, Leak DJ, Dalton H. Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane oxidizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- Stein LY, Bringel F, DiSpirito AA, Han S, Jetten MSM, Kalyuzhnaya MG, Kits KD, Klotz MG, den Camp H, Semrau JD, Vuilleumier S, Bruce DC, Cheng JF, Davenport KW, Goodwin L, Han SS, Hauser L, Lajus A, Land ML, Lapidus A, Lucas S, Medigue C, Pitluck S, Woyke T. Genome sequence of the methanotrophic alphaproteobacterium Methylocystis sp strain Rockwell (ATCC 49242) J Bacteriol. 2011;193:2668–2669. doi: 10.1128/JB.00278-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LY, Yoon S, Semrau JD, DiSpirito AA, Crombie A, Murrell JC, Vuilleumier S, Kalyuzhnaya MG, Op den Camp HJ, Bringel F, Bruce D, Cheng JF, Copeland A, Goodwin L, Han S, Hauser L, Jetten MS, Lajus A, Land ML, Lapidus A, Lucas S, Médigue C, Pitluck S, Woyke T, Zeytun A, Klotz MG. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J Bacteriol. 2010;192:6497–6498. doi: 10.1128/JB.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecker K, Bendinger B, Schoning B, Nielsen PH, Nielsen JL, Baranyi C, Toenshoff ER, Daims H, Wagner M. Cohn’s Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci USA. 2006;103:2363–2367. doi: 10.1073/pnas.0506361103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenning MM, Hestnes AG, Wartiainen I, Stein LY, Klotz MG, Kalyuzhnaya MG, Spang A, Bringel F, Vuilleumier S, Lajus A, Medigue C, Bruce DC, Cheng JF, Goodwin L, Ivanova N, Han J, Han CS, Hauser L, Held B, Land ML, Lapidus A, Lucas S, Nolan M, Pitluck S, Woyke T. Genome sequence of the arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol. 2011;193:6418–6419. doi: 10.1128/JB.05380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK. Anaerobic oxidation of methane with sulfate: on the reversibility of the reactions that are catalyzed by enzymes also involved in methanogenesis from CO2. Curr Op Microbiol. 2011;14:292–299. doi: 10.1016/j.mib.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Theisen AR, Ali MH, Radajewski S, Dumont MG, Dunfield PF, McDonald IR, Dedysh SN, Miguez CB, Murrell JC. Regulation of methane oxidation in the facultative methanotroph Methylocella silvestris BL2. Mol Microbiol. 2005;58:682–692. doi: 10.1111/j.1365-2958.2005.04861.x. [DOI] [PubMed] [Google Scholar]
- Tinberg CE, Lippard SJ. Dioxygen activation in soluble methane monooxygenase. Acc Chem Res. 2011;44:280–288. doi: 10.1021/ar1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukaegbu UE, Henery S, Rosenzweig AC. Biochemical characterization of MmoS, a sensor protein involved in copper-dependent regulation of soluble methane monooxygenase. Biochemistry. 2006;45:10191–10198. doi: 10.1021/bi060693h. [DOI] [PubMed] [Google Scholar]
- Ukaegbu UE, Rosenzweig AC. Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS. Biochemistry. 2009;48:2207–2215. doi: 10.1021/bi8019614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanelderen P, Hadt RG, Smeets PJ, Solomon EI, Schoonheydt RA, Sels BF. Cu-ZSM-5: A biomimetic inorganic model for methane oxidation. J Catal. 2011;284:157–164. doi: 10.1016/j.jcat.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigliotta G, Nutricati E, Carata E, Tredici SM, De Stefano M, Pontieri P, Massardo DR, Prati MV, De Bellis L, Alifano P. Clonothrix fusca Roze 1896, a filamentous, sheathed, methanotrophic gamma-proteobacterium. Appl Environ Microbiol. 2007;73:3556–3565. doi: 10.1128/AEM.02678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier S, Khmelenina VN, Bringel F, Reshetnikov AS, Lajus A, Mangenot S, Rouy Z, Op den Camp HJM, Jetten MSM, Dispirito AA, Dunfield P, Klotz MG, Semrau JD, Stein LY, Barbe V, Medigue C, Trotsenko YA, Kalyuzhnaya MG. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J Bacteriol. 2012;194:551–552. doi: 10.1128/JB.06392-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward N, Larsen O, Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Metheacute B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonason I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath) PLoS Biol. 2004;2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woertink JS, Smeets PJ, Groothaert MH, Vance MA, Sels BF, Schoonheydt RA, Solomon EI. A [Cu2O]2+ core in Cu-ZSM-5, the active site in the oxidation of methane to methanol. Proc Natl Acad Sci USA. 2009;106:18908–18913. doi: 10.1073/pnas.0910461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa K, Shiota Y. Conversion of methane to methanol at the mononuclear and dinuclear copper sites of particulate methane monooxygenase (pMMO): a DFT and QM/MM study. J Am Chem Soc. 2006;128:9873–9881. doi: 10.1021/ja061604r. [DOI] [PubMed] [Google Scholar]
- Yu SSF, Ji CZ, Wu YP, Lee TL, Lai CH, Lin SC, Yang ZL, Wang VCC, Chen KHC, Chan SI. The C-terminal aqueous-exposed domain of the 45 kDa subunit of the particulate methane monooxygenase in Methylococcus capsulatus (Bath) is a Cu(I) sponge. Biochemistry. 2007;46:13762–13774. doi: 10.1021/bi700883g. [DOI] [PubMed] [Google Scholar]
- Zahn JA, DiSpirito AA. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlateva T, Santagostini L, Bubacco L, Casella L, Salvato B, Beltramini M. Isolation of the met-derivative intermediate in the catalase-like activity of deoxygenated Octopus vulgaris hemocyanin. J Inorg Biochem. 1998;72:211–215. [Google Scholar]