Summary
Background
Penicillins inhibit cell wall synthesis; therefore, H. pylori must be dividing for this class of antibiotics to be effective in eradication therapy. Identifying growth responses to varying medium pH may allow design of more effective treatment regimens.
Aim
To determine the effect of acidity on bacterial growth and the bactericidal efficacy of ampicillin.
Methods
H. pylori were incubated in dialysis chambers suspended in 1.5L of media at various pHs with 5mM urea, with or without ampicillin, for 4, 8 or 16 hours, thus mimicking unbuffered gastric juice. Changes in gene expression, viability and survival were determined.
Results
At pH 3.0, but not at pH 4.5 or 7.4, there was decreased expression of ~400 genes, including many cell envelope biosynthesis, cell division and penicillin-binding protein genes. Ampicillin was bactericidal at pH 4.5 and 7.4 but not at pH 3.0.
Conclusion
Ampicillin is bactericidal at pH 4.5 and 7.4, but not at pH 3.0, due to decreased expression of cell envelope and division genes with loss of cell division at pH 3.0. Therefore, at pH 3.0, the likely pH at the gastric surface, the bacteria are non-dividing and persist with ampicillin treatment. A more effective inhibitor of acid secretion that maintains gastric pH near neutrality for 24 hours/day should enhance the efficacy of amoxicillin, improving triple therapy and likely even allowing dual amoxicillin based therapy for H. pylori eradication.
Keywords: Helicobacter pylori, medium pH, bacterial growth, ampicillin/amoxicillin, gastric acid inhibition, proton pump inhibitor
Introduction
Helicobacter pylori, a Gram-negative neutralophile, is the only organism known to colonize the normal acid-secreting human stomach. About 50% of the world's population is infected1. Colonization is associated with several gastric diseases, including gastritis, peptic and duodenal ulcers, gastric carcinoma, and MALT lymphoma2–5. In 1994, the WHO classified the organism as a type 1 carcinogen6. Despite recent findings that H. pylori infection may have a beneficial effect on gastroesophageal reflux disease (GERD) and other non-gastric manifestations of infection, the consequences of gastric colonization alone argue for its eradication7, 8.
Standard H. pylori eradication therapy involves a complicated regimen of at least two antibiotics (clarithromycin and amoxicillin or metronidazole) and a proton pump inhibitor or antibiotics, acid suppression, and bismuth9,10. It has never been clear as to why two antibiotics, one targeting cell wall biosynthesis and the other targeting protein synthesis, are required for eradication. Acid suppression clearly improves the efficacy of antibiotic treatment11.
Emerging antibiotic resistance to clarithromycin has made successful treatment of infection progressively more difficult, with the success rate of standard triple therapy now at 70%, well below the 80% required for treatment of infectious diseases9. Alternative eradication regimens have been suggested such as sequential therapy (PPI and amoxicillin for 5 days followed by PPI, clarithromycin and metronidazole) or concomitant therapy, but the results of these regimens are not comparable to the initial eradication results with triple therapy (~95%)9. Quadruple therapy (addition of colloidal bismuth subcitrate to standard triple therapy regimens) has shown promise, with cure rates currently between 80 and 90%9, 12. However, since bismuth containing quadruple therapies also rely on antibiotics, this regimen is sensitive to and contributes to increasing antibiotic resistance. Therefore, it is likely that eradication rates will decrease with this regimen as it has for triple therapy.
There have been several hypotheses as to the role of PPIs in eradication therapy. A direct effect of the PPI on the viability of the organism in vitro has been suggested. However, this required the administration of very high doses of the PPI to show direct inhibition13. At these concentrations, there is acid-independent thiol reactivity of the PPI, resulting in non-specific effects14. Another hypothesis is increased gastric bioavailability of amoxicillin after PPI administration, but it is generally agreed that gastric acidity does not influence the pharmacokinetics of this antibiotic15.
H. pylori resistance to amoxicillin is rare, suggesting it would be the antibiotic of choice as long as the bacteria remain susceptible to its action9. Amoxicillin acts by inhibiting the synthesis of the bacterial cell wall. It inhibits cross-linking between the linear peptidoglycan polymer chains that make up a major component of the cell wall. Therefore, bacteria must be actively dividing and synthesizing the cell envelope for this antibiotic to be effective. In the absence of urea, H. pylori enters a non-replicative but viable state when the environmental pH is less than 6 and greater than 316, 17. In vivo gene expression of H. pylori indicates that the average pH at the site of infection in the gerbil stomach is about 3.5, placing the organism in a non-dividing state, decreasing the efficacy of amoxicillin for eradication18. Thus, increasing the intragastric pH to >5.0 should induce the bacteria to enter the replicative state and become more susceptible to both amoxicillin and to the protein synthesis inhibitor, clarithromycin19.
Here, we explore the effects of lengthy exposure of the organisms to pH 7.4, 4.5 and 3.0 in the presence of 5mM urea, taking advantage of a method developed in our laboratory that exposes a small volume of the bacteria in a dialysis chamber to a large volume of pH adjusted medium without added buffer, allowing analysis for 4, 8 or 16 hours. Analysis of the bacterial transcriptome at pH 3.0 reveals down regulation of ~400 genes, including genes encoding envelope synthesis, cell division and protein synthesis. Analysis of viability and growth potential shows that cell viability is decreased at pH 3.0 but not at higher pH, and this pH also decreases the number of colony forming units (CFUs). Also, at pH 3.0, in contrast to the higher pH levels, the bactericidal effect of ampicillin disappears, indicating that the lack of growth at pH 3.0 attenuates penicillin sensitivity.
Methods
Bacterial strains and culture conditions
H. pylori strain ATCC 43504 was used. Bacteria were grown under microaerobic conditions (5% O2, 10% CO2, 85% N2) either on Trypticase Soy Agar (TSA) plates supplemented with 5% sheep blood (Gibco) or in brain heart infusion (BHI) medium (Difco) supplemented with 7% horse serum (Gibco), 0.25% yeast extract (Difco) and Dent selective supplement (Oxoid).
Incubation conditions
To investigate the effect of acid exposure on H. pylori in vitro, it is advantageous to extend the time of acid exposure to allow the organism to fully acclimate to a given environmental pH and to mimic the in vivo intragastric environment during the inter-digestive phase18. It is not possible to maintain constant pH in H. pylori cultures in the presence of urea for this long a time without the use of a buffer, due to urea hydrolysis by urease, leading to an increase in pH in unbuffered solutions. To circumvent this problem and to maintain constant urea levels in unbuffered media, a system was developed using a micro-dialysis chamber (Slide-A-lyzer, MWCO 10kD) containing H. pylori suspended in a large volume of pH-adjusted medium (1.5L) containing 5mM urea. This was capable of maintaining a constant pH for up to 16 hours without depleting urea and allowed for analysis of bacterial status at different medium pHs (Supplement Figure 1).
Bacteria were incubated in BHI containing 7% horse serum, 0.25% yeast extract and 5mM urea with and without 2mg/mL ampicillin for all experiments, and pH was adjusted by the addition of HCl. Bacteria harvested from TSA plates (1 × 108) were suspended in 3mL of BHI and injected into a sterile Slide-A-Lyzer® Dialysis Chamber (Pierce), then placed in 1,500mL of media. Using this technique, it was possible to maintain constant pH and urea levels for up to at least 16 hours without added buffer. H. pylori were incubated at pH 3.0, 4.5 and 7.4 for 4, 8 and 16 hours. The bulk medium pH was recorded at the beginning and end of each experiment and no difference was found.
RNA isolation for transcriptomal analysis
Following incubation at various pH levels for 4 hours, RNA was isolated from H. pylori using a combination of the TRIzol® method and the RNeasy kit (Qiagen) as described previously20. RNA quality was determined using an Agilent 2100 bioanalyzer. Only RNA with an RNA integrity number greater than 8 was used for microarray and qPCR analysis. All experiments were done in triplicate.
Microarray
Microarray analysis was performed using a custom made 4×44 OligoMicroArray from Agilent (Santa Clara, California). This chip contains 10 replicates of three, 60mer oligonucleotide probes to each gene of H pylori strain ATCC 26695. 3 independent microarray experiments were performed on each RNA replicate. All total RNA samples were serially diluted to a concentration of 1000ng in 5.3uL for use with the Quick Amp Labeling kit (Agilent, Santa Clara). In preparation for the microarray, the Agilent One-Color Spike-In Kit was used as a positive control. cRNA labeling and microarray hybridization were performed according to the manufacturers protocol.
Array Quantification
Data were analyzed using Agilent's Feature Extraction Software as well as GeneSpring GX software. Data were filtered by expression (20–80%). All data with at least a two-fold change compared to control were subjected to a T-test. Only those genes whose expression was altered by at least 1.5 fold with a P-value less than 0.05 were considered to represent significantly regulated genes.
Quantitative PCR
RNA was isolated following incubation at pH 3.0, 4.5, and 7.4 as described earlier. cDNA was produced from RNA using Omniscript reverse transcriptase enzyme (Qiagen) and random primers (Invitrogen). The starting quantity of H. pylori RNA for all reverse transcriptase reactions was 6 μg. qPCR primers were designed with the help of Primer3 software21. All primer sequences used are listed in Supplement Table 1 of the supplement. Primers were checked for the presence of a single product between 100–300 base pairs using standard PCR (30 cycles, 92°C denaturing, 55°C annealing, 72°C extension, 1 minute each) with H. pylori gDNA as template.
1μL cDNA was used for all qPCR reactions. H. pylori gDNA (isolated using the CTAB method20) was serially diluted 10 fold × 4 dilutions and, including the original sample, a 5-concentration standard curve was used for all primer pairs. All reported data had standard curve efficiency within acceptable range (between 90–110%). qPCR was completed using the Bio-Rad CFX96 machine and Ssofast EvaGreen Supermix®(Bio-Rad) using hot start (3 minutes × 92°C), followed by 40 cycles of 92°C × 30 seconds, 55°C × 30 seconds, 72°C × 40 seconds. A melt curve was completed at the end of each qPCR run and 1 peak was seen for all primer pairs. Fluorescence data were collected during the extension steps and data were expressed as mean starting quantity. All experiments were done in triplicate both in RNA isolation and qPCR stages.
Quantitative Live/Dead assay
Bacterial viability was assayed using the LIVE/DEAD®BacLight™ Bacterial Viability and Counting Kit (Invitrogen) according to the manufacturer's instructions. Fluorescence intensity was measured on a Fluorolog-3 (Jobin Yvon Horiba) fluorimeter. The ratio of green fluorescence intensity (Syto-9, Ex 488nm, Em 498nm) to red fluorescence intensity (propidium iodide, Ex 488nm, Em 617nm) was calculated to determine changes in viability, where an increase in red fluorescence indicates cell membrane damage and loss of viability.
Measurement of survival at acidic and neutral pH
Following incubation, the bacterial suspension was removed from the Slide-A-Lyzer® and added to 9mL BHI, and serial dilutions were done using 1mL into 9mL BHI. 100μL was plated in duplicate for each condition. Colonies were counted after 3 days.
Results
Transcriptomal Analysis
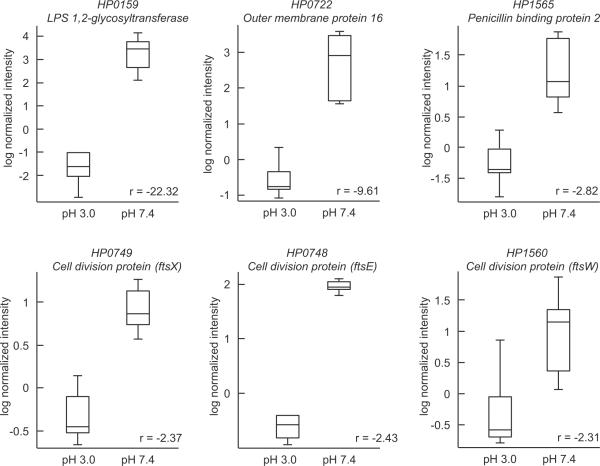
Previous analyses of the gene expression profile of H. pylori have been performed for 30 minutes to 1 hour in the presence or absence of urea at different medium pH levels ranging from 4.5–7.422. Using the constant pH/urea culture system, we analyzed gene expression after a period of 4 hours exposed to pH 7.4 or 3.0 in the presence of 5mM urea. Approximately 463 genes changed expression 2 fold or greater, of which 273 were down regulated and 190 up regulated. Among the down regulated genes there were genes involved in protein synthesis, flagellar activity, cell division and cell wall biosynthesis, including the two penicillin binding proteins expressed by this organism (table 1). Three cell division genes were down regulated by at least four fold. This indicates that, at pH 3.0, the organisms are not dividing, resulting in persistence of the infection due to antibiotic insensitivity. This finding is in contrast to previous studies performed at pH 4.5, where there was no down regulation of cell envelope or cell division genes in the presence of urea22. Other genes that were down regulated, also indicating lack of growth, were several amino acid biosynthesis genes, genes involved in DNA metabolism and several histidine kinases such as the pH sensing HP0165 and HP0244 and its cognate class 2 flagellar regulator HP0703, as well as the HP1364/1365 couple (data not shown). Table 1 shows the microarray data for several cell envelope and cell division genes. The difference in mRNA levels of several genes following 4 hours of incubation at pH 3.0 and 7.4 is represented as box-whisker plots in Figure 1.
Table 1.
Genes involved in cell envelope biosynthesis and cell division down regulated at pH 3.0.
| Accession Number1 | pH3.0/pH7.42 | Description /gene name |
|---|---|---|
| HP1330 | −9.62 | predicted branched-chain amino acid transport protein (azlD) |
| HP0645 | −1.57 | soluble lytic mureintransglycosylase (slt) |
| HP0379 | −3.41 | alpha1,3-fucosyltransferase(futA) |
| HP1063 | −3.97 | glucose-inhibited division protein (gidB) |
| HP1090 | −4.45 | cell division protein (ftsK) |
| HP1556 | −4.15 | cell division protein (ftsI) |
| HP1243 | −5.75 | outer membrane protein (omp28) |
| HP0722 | −5.31 | outer membrane protein (omp16) |
| HP0494 | −3.52 | UDP-N-acetylmuramoylalanine-D-glutamate ligase (murD) |
| HP0597 | −2.40 | penicillin-binding protein 1A (pbp-1A) |
| HP0839 | −5.42 | outer membrane protein P1 (ompP1) |
| HP1565 | −1.60 | penicillin-binding protein 2 (pbp2) |
| HP0896 | −2.59 | outer membrane protein (omp19) |
| HP0421 | −2.43 | type 1 capsular polysaccharide biosynthesis protein J (capJ) |
| HP0648 | −3.86 | UDP-N-acetylglucosamineenolpyruvyltransferase (murZ) |
| HP0867 | −2.28 | predicted lipid A disaccharide synthetase (lpxB) |
| HP1191 | −1.52 | ADP-heptose-lpsheptosyltransferase II (rfaF) |
| HP1372 | −2.60 | rod shape-determining protein (mreC) |
| HP0178 | −2.20 | predicted sialic acid synthase (neuB) |
| HP1155 | −1.81 | transferase, peptidoglycan synthesis (murG) |
| HP1564 | −1.81 | Predictedouter membrane lipoprotein |
| HP1571 | −3.03 | rare lipoprotein A (rlpA) |
| HP0913 | −1.57 | outer membrane protein (omp21) |
Accession number from H. pylori strain 26695 41.
Mean ratio from triplicates of 3 experiments.
Figure 1.
Box-whisker plots representing changes in mRNA from transcriptomal analysis. H. pylori were incubated for 4 hours at pH 3.0 and 7.4 in the presence of 5mM urea. At pH 3.0, there was a decrease in mRNA levels of cell wall biosynthesis (HP0159), cell division (HP0748, HP0749, HP1560), and penicillin binding (HP1565) genes. These data suggest that, at pH 3.0, the organisms are non-replicative, not synthesizing cell wall proteins, and have a reduced metabolism, rendering the bacteria ampicillin insensitive at pH 3.0.
Quantitative polymerase chain reaction
To confirm the microarray results and determine the level of gene expression with increased time of exposure to acid, qPCR was performed on selected cell wall biosynthesis and cell division genes (Supplement Figure 2). The change in gene expression in the qPCR assay corresponded well with the data observed in the microarray. For example, the level of the cell division protein Ftsl (HP1556) mRNA was reduced several fold at pH 3.0 as compared to either pH 4.5 or pH 7.4, as were the penicillin binding protein HP0155, the cell wall gene murein tranglycosylase, and the metabolic NADH oxido-reductase. These data suggest that, at pH 3.0, the organisms are non-dividing and not synthesizing cell wall proteins, rendering the bacteria ampicillin insensitive at pH 3.0.
H. pylori viability in the presence of ampicillin at neutral and acidic pH
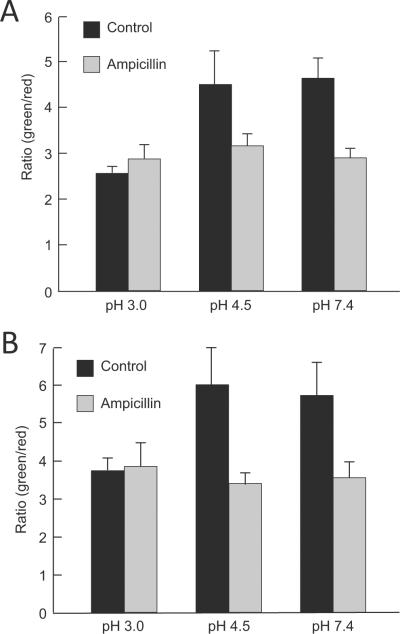
To investigate the bactericidal effects of ampicillin at different pHs, a commercial quantitative viability assay was used. This assay can reveal damage to the bacterial inner membrane by permeation of the fluorescent cation, propidium iodide, and hence demonstrate the deleterious effects of either pH or ampicillin. After 8 hour incubation at pH 7.4, 4.5 or 3.0 supplemented with 5mM urea in the absence of ampicillin, the live/dead ratio was similar at pH 7.4 and pH 4.5. However, incubation at pH 3.0 very slightly decreased the ratio, showing that there was decreased survival at this pH but with still a significant number of viable bacteria. Ampicillin significantly decreased the viability of the bacteria at both pH 4.5 and 7.4, but had no effect at pH 3.0 (Figure 2A). Sixteen hour incubation amplified the data observed after 8 hour incubation. Again, there was a decrease in the ratio of live to dead organisms at pH 3.0 as compared to pH 7.4 or 4.5 in the absence of ampicillin and a large bactericidal effect of ampicillin at pH 4.5 and 7.4, but none at pH 3.0. (Figure 2B).
Figure 2.
H. pylori viability by the live/dead assay method in the absence or presence of ampicillin at neutral and acidic pH. H. pylori incubated for 8 (A) and 16 (B) hours at pH 3.0 in the absence of ampicillin were less viable then at pH 4.5 and 7.4. Ampicillin significantly decreased the viability of the bacteria at both pH 4.5 and 7.4 but had no effect at pH 3.0 at both incubation times.
H. pylori survival in the presence of ampicillin at neutral and acidic pH
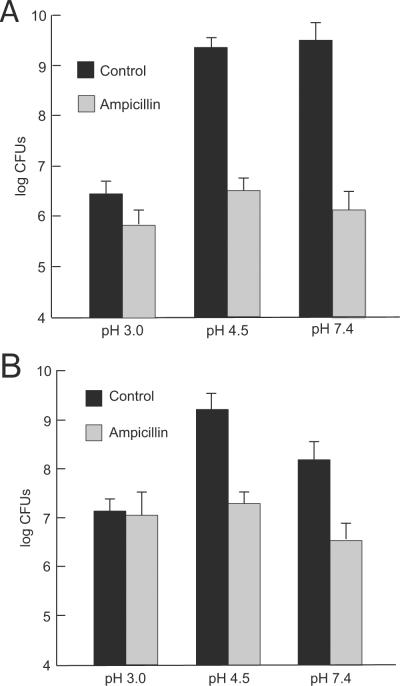
The enumeration of CFUs after exposure to acid or ampicillin is another measure of bacterial survival. After 8 hour incubation, there was a marked decline in survival with exposure to ampicillin at pH 4.5 or 7.4 but no change with ampicillin at pH 3.0 (Figure 3A). Similar data were obtained after 16 hour (Figure 3B) incubation. Incubation at pH 7.4 or 4.5 showed a larger number of CFUs as compared to 16 hour exposure at pH 3.0. Of note, there are still viable bacteria at pH 3.0. The addition of ampicillin significantly reduced the CFUs of organisms incubated at pH 7.4 or 4.5, with only a small, insignificant reduction of CFUs at pH 3.0 (Figure 3A and 3B).
Figure 3.
H. pylori survival in the presence of ampicillin at neutral and acidic pH. After 8 hour exposure to acidity, there was no change in the number of CFUs at pH 4.5 or 7.4, and a marked decline with exposure to ampicillin at these pH levels, but no change with ampicillin at pH 3.0 (A). Similar data were obtained after 16 hour incubation (B).
Discussion
The present study shows that at pH 3.0, H. pylori is viable but in a non-replicative state, diminishing the bactericidal efficacy of penicillin antibiotics and also that of protein synthesis inhibition by clarithromycin. This is supported by the large number of cell envelope and cell division genes down regulated at pH 3.0 and the loss of ampicillin efficacy. Induction of profound acid inhibition using potent H,K-ATPase antagonists, which raise the intragastric pH to between 5.0 and 7.0, should stimulate growth of H. pylori and increase the bactericidal effect of amoxicillin, which should result in eradication.
There has been considerable controversy as to the pH of the gastric habitat of H. pylori. It has been proposed that there is a gastric barrier to proton back diffusion from the gastric lumen to the gastric surface due to both mucus and bicarbonate secretion23. However, recent experiments using fluorescent dyes or microelectrodes in infected mice show that the pH gradient collapses when the luminal pH falls to < 3.024, 25. Also, analysis of the transcriptome of bacteria isolated from the gerbil stomach in comparison to the pH dependent transcriptome in vitro showed that average pH in their gastric habitat was about pH 3.518.
The level and duration of acid suppression affects the success of eradication with PPIs26–28. Nocturnal acid breakthrough is a factor in the failure of triple therapy29. The major location of bacteria resistant to treatment is the more acidic fundus, and hence the lower pH in this region promotes H. pylori growth inhibition and contributes to the failure of triple therapy30.
Improved inhibition of acid secretion increases the rate of eradication. Cure rates of standard triple therapy depend on the efficacy of PPI-dependent inhibition of acid secretion. Although PPIs are covalent inhibitors of the gastric H,K ATPase, their short plasma half-life prevents adequate elevation of pH during the night, when newly synthesized pumps are not exposed to the PPI. This, coupled with the slow growth rate of H. pylori, results in insensitivity of the organisms to growth dependent antibiotics since the pH falls frequently to < 3.017. In a recent study, q.i.d. omeprazole, dosed to maintain intragastric pH above pH 5.5 for 16 hours, and amoxicillin dual therapy eradicated H. pylori infection31, substantiating the idea that improved acid inhibition would improve results of eradication therapy.
Further evidence that increased PPI dwell time, and therefore better acid inhibition, improves H. pylori eradication comes from studies on slow omeprazole metabolizers. PPIs are mainly metabolized by cytochrome 2C19 (CYP2C19), and in patients who are homozygous for the loss of CYP2C19, leading to slower metabolism and increased plasma dwell time of the PPI, inhibition of acid secretion is significantly increased. These patients were successfully treated for H. pylori eradication with omeprazole and amoxicillin alone31, 32. Recently, it was shown that 10 mg rabeprazole q.i.d. maintained plasma PPI levels above the threshold level required for H+,K+-ATPase inhibition for 24 hours, resulting in a median intragastric pH of 6.6 independent of CYP2C19 status33. These results show that increased acid inhibition by PPIs may enable dual therapy, and this can be achieved by increasing the amount of time that this type of drug is in the blood. However, there have been several attempts at dual therapy with lack of success, likely due to inadequate elevation of pH during the night34, 35.
There are several PPIs which would likely raise intragastric pH close to neutrality, tenatoprazole, a slowly metabolized PPI, AGN904, an omeprazole prodrug with a longer dwell time than omeprazole, and the potassium competitive acid blockers TAK-438 and AZD086536–40.
The data presented here demonstrate that an improvement in acid inhibition extending into night time hours, maintaining the intragastric pH close to neutrality without acidic excursions overnight, would greatly improve eradication rates of triple therapy and perhaps allow dual therapy with a potent H,K-ATPase inhibitor and amoxicillin. These results provide a template for new clinical studies on H. pylori eradication.
Supplementary Material
Acknowledgments
Grant support: Supported by NIH and USVA grants P30 DK41301 (EAM), K12 HD034610 (EAM), DK053462 (GS), 1I01BX001006 (GS)
Footnotes
Transcript Profiling: GSE39512 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xhizdyywucikgdg&acc=GSE3951
References
- 1.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002 Oct 10;347(15):1175–86. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 2.Blaser MJ. Helicobacter pylori: its role in disease. Clin Infect Dis. 1992 Sep;15(3):386–91. doi: 10.1093/clind/15.3.386. [DOI] [PubMed] [Google Scholar]
- 3.Nomura A, Stemmermann GN, Chyou PH, Kato I, Perez-Perez GI, Blaser MJ. Helicobacter pylori infection and gastric carcinoma among Japanese Americans in Hawaii. N Engl J Med. 1991 Oct 17;325(16):1132–6. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 4.Parsonnet J. Gastric adenocarcinoma and Helicobacter pylori infection. West J Med. 1994 Jul;161(1):60. [PMC free article] [PubMed] [Google Scholar]
- 5.Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991 Oct 17;325(16):1127–31. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 6.Infection with Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177–240. [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser MJ. Helicobacter pylori and esophageal disease: wake-up call? Gastroenterology. 2010 Dec;139(6):1819–22. doi: 10.1053/j.gastro.2010.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachs G, Scott DR. Helicobacter pylori: Eradication or Preservation. F1000 Med Rep. 2012;4:7. doi: 10.3410/M4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, et al. Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report. Gut. 2012 May;61(5):646–64. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 10.Gisbert JP, Gonzalez L, Calvet X. Systematic review and meta-analysis: proton pump inhibitor vs. ranitidine bismuth citrate plus two antibiotics in Helicobacter pylori eradication. Helicobacter. 2005 Jun;10(3):157–71. doi: 10.1111/j.1523-5378.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 11.Lind T, Megraud F, Unge P, Bayerdorffer E, O'Morain C, Spiller R, et al. The MACH2 study: role of omeprazole in eradication of Helicobacter pylori with 1-week triple therapies. Gastroenterology. 1999 Feb;116(2):248–53. doi: 10.1016/s0016-5085(99)70119-8. [DOI] [PubMed] [Google Scholar]
- 12.Saleem A, Qasim A, O'Connor HJ, O'Morain CA. Pylera for the eradication of Helicobacter pylori infection. Expert Rev Anti Infect Ther. 2009 Sep;7(7):793–9. doi: 10.1586/eri.09.55. [DOI] [PubMed] [Google Scholar]
- 13.Nakao M, Malfertheiner P. Growth inhibitory and bactericidal activities of lansoprazole compared with those of omeprazole and pantoprazole against Helicobacter pylori. Helicobacter. 1998 Mar;3(1):21–7. doi: 10.1046/j.1523-5378.1998.08024.x. [DOI] [PubMed] [Google Scholar]
- 14.Lorentzon P, Eklundh B, Brandstrom A, Wallmark B. The mechanism for inhibition of gastric (H+, K+)-ATPase by omeprazole. Biochim Biophys Acta. 1985 Jul 11;817(1):25–32. doi: 10.1016/0005-2736(85)90064-1. [DOI] [PubMed] [Google Scholar]
- 15.Erah PO, Goddard AF, Barrett DA, Shaw PN, Spiller RC. The stability of amoxycillin, clarithromycin and metronidazole in gastric juice: relevance to the treatment of Helicobacter pylori infection. J Antimicrob Chemother. 1997 Jan;39(1):5–12. doi: 10.1093/jac/39.1.5. [DOI] [PubMed] [Google Scholar]
- 16.Scott D, Weeks D, Melchers K, Sachs G. The life and death of Helicobacter pylori. Gut. 1998 Jul;43(Suppl 1):S56–60. doi: 10.1136/gut.43.2008.s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjostrom JE, Larsson H. Factors affecting growth and antibiotic susceptibility of Helicobacter pylori: effect of pH and urea on the survival of a wild-type strain and a urease-deficient mutant. J Med Microbiol. 1996 Jun;44(6):425–33. doi: 10.1099/00222615-44-6-425. [DOI] [PubMed] [Google Scholar]
- 18.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc Natl Acad Sci U S A. 2007 Apr 24;104(17):7235–40. doi: 10.1073/pnas.0702300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain R, Danziger LH. The macrolide antibiotics: a pharmacokinetic and pharmacodynamic overview. Curr Pharm Des. 2004;10(25):3045–53. doi: 10.2174/1381612043383322. [DOI] [PubMed] [Google Scholar]
- 20.Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J Bacteriol. 2005 Jan;187(2):729–38. doi: 10.1128/JB.187.2.729-738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 22.Wen Y, Marcus EA, Matrubutham U, Gleeson MA, Scott DR, Sachs G. Acid-adaptive genes of Helicobacter pylori. Infect Immun. 2003 Oct;71(10):5921–39. doi: 10.1128/IAI.71.10.5921-5939.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McColl KE. The elegance of the gastric mucosal barrier: designed by nature for nature. Gut. 2012 Jun;61(6):787–8. doi: 10.1136/gutjnl-2011-301612. [DOI] [PubMed] [Google Scholar]
- 24.Baumgartner HK, Montrose MH. Regulated alkali secretion acts in tandem with unstirred layers to regulate mouse gastric surface pH. Gastroenterology. 2004 Mar;126(3):774–83. doi: 10.1053/j.gastro.2003.11.059. [DOI] [PubMed] [Google Scholar]
- 25.Henriksnas J, Phillipson M, Storm M, Engstrand L, Soleimani M, Holm L. Impaired mucus-bicarbonate barrier in Helicobacter pylori-infected mice. Am J Physiol Gastrointest Liver Physiol. 2006 Sep;291(3):G396–403. doi: 10.1152/ajpgi.00017.2006. [DOI] [PubMed] [Google Scholar]
- 26.Hunt RH. Hp and pH--the relevance of gastric acid to the treatment of Helicobacter pylori infection. J Gastroenterol. 1994 Jul;29(Suppl 7):128–33. [PubMed] [Google Scholar]
- 27.Labenz J, Stolte M, Blum AL, Jorias I, Leverkus F, Sollbohmer M, et al. Intragastric acidity as a predictor of the success of Helicobacter pylori eradication: a study in peptic ulcer patients with omeprazole and amoxicillin. Gut. 1995 Jul;37(1):39–43. doi: 10.1136/gut.37.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugimoto M, Furuta T, Shirai N, Kodaira C, Nishino M, Ikuma M, et al. Evidence that the degree and duration of acid suppression are related to Helicobacter pylori eradication by triple therapy. Helicobacter. 2007 Aug;12(4):317–23. doi: 10.1111/j.1523-5378.2007.00508.x. [DOI] [PubMed] [Google Scholar]
- 29.Kim JI, Park SH, Kim JK, Chung IS, Chung KW, Sun HS. The effects of nocturnal acid breakthrough on Helicobacter pylori eradication. Helicobacter. 2002 Dec;7(6):331–6. doi: 10.1046/j.1523-5378.2002.00105.x. [DOI] [PubMed] [Google Scholar]
- 30.Atherton JC, Cockayne A, Balsitis M, Kirk GE, Hawkey CJ, Spiller RC. Detection of the intragastric sites at which Helicobacter pylori evades treatment with amoxycillin and cimetidine. Gut. 1995 May;36(5):670–4. doi: 10.1136/gut.36.5.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang JC, Wang HL, Chern HD, Shun CT, Lin BR, Lin CJ, et al. Role of omeprazole dosage and cytochrome P450 2C19 genotype in patients receiving omeprazole-amoxicillin dual therapy for Helicobacter pylori eradication. Pharmacotherapy. 2011 Mar;31(3):227–38. doi: 10.1592/phco.31.3.227. [DOI] [PubMed] [Google Scholar]
- 32.Furuta T, Sugimoto M, Shirai N, Ishizaki T. CYP2C19 pharmacogenomics associated with therapy of Helicobacter pylori infection and gastro-esophageal reflux diseases with a proton pump inhibitor. Pharmacogenomics. 2007 Sep;8(9):1199–210. doi: 10.2217/14622416.8.9.1199. [DOI] [PubMed] [Google Scholar]
- 33.Sugimoto M, Shirai N, Nishino M, Kodaira C, Uotani T, Yamade M, et al. Rabeprazole 10 mg q.d.s. deceases 24-h intragastric acidity significantly more than rabeprazole 20 mg b.d. or 40 mg o.m., overcoming CYP2C19 genotype. Aliment Pharmacol Ther. 2012 Aug 10; doi: 10.1111/apt.12014. [DOI] [PubMed] [Google Scholar]
- 34.Attumi TA, Graham DY. Increasing the duration of dual amoxicillin plus omeprazole Helicobacter pylori eradication to 6 weeks: a pilot study. Journal of Ggastroenterology and Hepatology. 2012 Jan;27(1):59–61. doi: 10.1111/j.1440-1746.2011.06876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham DY, Javed SU, Keihanian S, Abudayyeh S, Opekun AR. Dual proton pump inhibitor plus amoxicillin as an empiric anti-H. pylori therapy: studies from the United States. J Gastroenterol. 2010 Aug;45(8):816–20. doi: 10.1007/s00535-010-0220-x. [DOI] [PubMed] [Google Scholar]
- 36.Dent J, Kahrilas PJ, Hatlebakk J, Vakil N, Denison H, Franzen S, et al. A randomized, comparative trial of a potassium-competitive acid blocker (AZD0865) and esomeprazole for the treatment of patients with nonerosive reflux disease. Am J Gastroenterol. 2008 Jan;103(1):20–6. doi: 10.1111/j.1572-0241.2007.01544.x. [DOI] [PubMed] [Google Scholar]
- 37.Hori Y, Matsukawa J, Takeuchi T, Nishida H, Kajino M, Inatomi N. A study comparing the antisecretory effect of TAK-438, a novel potassium-competitive acid blocker, with lansoprazole in animals. J Pharmacol Exp Ther. 2011 Jun;337(3):797–804. doi: 10.1124/jpet.111.179556. [DOI] [PubMed] [Google Scholar]
- 38.Hunt RH, Armstrong D, Yaghoobi M, James C. The pharmacodynamics and pharmacokinetics of S-tenatoprazole-Na 30 mg, 60 mg and 90 mg vs. esomeprazole 40 mg in healthy male subjects. Aliment Pharmacol Ther. 2010 Mar;31(6):648–57. doi: 10.1111/j.1365-2036.2009.04219.x. [DOI] [PubMed] [Google Scholar]
- 39.Hunt RH, Armstrong D, Yaghoobi M, James C, Chen Y, Leonard J, et al. Predictable prolonged suppression of gastric acidity with a novel proton pump inhibitor, AGN 201904-Z. Aliment Pharmacol Ther. 2008 Jul;28(2):187–99. doi: 10.1111/j.1365-2036.2008.03725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin JM, Inatomi N, Munson K, Strugatsky D, Tokhtaeva E, Vagin O, et al. Characterization of a novel potassium-competitive acid blocker of the gastric H,K-ATPase, 1-[5-(2-fluorophenyl)-1-(pyridin-3-ylsulfonyl)-1H-pyrrol-3-yl]-N-methylmethanamine monofumarate (TAK-438) J Pharmacol Exp Ther. 2011 Nov;339(2):412–20. doi: 10.1124/jpet.111.185314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997 Aug 7;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.