Abstract
Following genotoxic stress, cells activate a complex signalling network to arrest the cell cycle and initiate DNA repair or apoptosis. The tumour suppressor p53 lies at the heart of this DNA damage response. However, it remains incompletely understood, which signalling molecules dictate the choice between these different cellular outcomes. Here, we identify the transcriptional regulator apoptosis-antagonizing transcription factor (AATF)/Che-1 as a critical regulator of the cellular outcome of the p53 response. Upon genotoxic stress, AATF is phosphorylated by the checkpoint kinase MK2. Phosphorylation results in the release of AATF from cytoplasmic MRLC3 and subsequent nuclear translocation where AATF binds to the PUMA, BAX and BAK promoter regions to repress p53-driven expression of these pro-apoptotic genes. In xenograft experiments, mice exhibit a dramatically enhanced response of AATF-depleted tumours following genotoxic chemotherapy with adriamycin. The exogenous expression of a phospho-mimicking AATF point mutant results in marked adriamycin resistance in vivo. Nuclear AATF enrichment appears to be selected for in p53-proficient endometrial cancers. Furthermore, focal copy number gains at the AATF locus in neuroblastoma, which is known to be almost exclusively p53-proficient, correlate with an adverse prognosis and reduced overall survival. These data identify the p38/MK2/AATF signalling module as a critical repressor of p53-driven apoptosis and commend this pathway as a target for DNA damage-sensitizing therapeutic regimens.
Keywords: AATF, apoptosis, checkpoint, DNA damage, kinase signalling, MK2, p53
Introduction
In response to DNA damage, cells activate a complex signalling network to prevent further cell-cycle progression. Activation of this signalling cascade, which is collectively referred to as the DNA damage response (DDR), provides time for DNA repair, recruits repair machinery to the sites of genotoxic damage, or, if the lesions are beyond repair capacity, leads to the activation of additional pathways mediating apoptosis (Jackson and Bartek, 2009). The DDR has traditionally been divided into two major kinase branches operating through the upstream kinases ATM and ATR together with their respective effector kinases Chk2 and Chk1 (Jackson and Bartek, 2009). Moreover, p38 and its downstream substrate mitogen-activated protein kinase-activated protein kinase-2 (MK2) has recently been identified as a third checkpoint effector kinase complex operating downstream of ATM and ATR and parallel to Chk1 (Manke et al, 2005; Raman et al, 2007; Reinhardt et al, 2007, 2010). p38 and MK2 are components of a general stress kinase pathway that acts in response to a variety of stimuli, including inflammatory signals, reactive oxygen species, heat shock, hyperosmolar stress in addition to DNA damage (Kyriakis and Avruch, 2001).
One of the major downstream targets of the DDR network is the transcription factor p53. DNA damage-induced phosphorylation of p53, through ATM, Chk2, MK2 and others, at amino-terminal sites close to the MDM2-binding region is thought to reduce ubiquitin-dependent degradation allowing accumulation in the nucleus, where p53 contributes to two distinct cellular responses (Toledo and Wahl, 2006): p53 promotes cell death in response to genotoxic stress through the induction of pro-apoptotic target genes such as PUMA, BAX and BAK. In contrast, p53-mediated transactivation of cell-cycle-arresting target genes, such as CDKN1A, GADD45A or RPRM, serves a protective function by allowing time to repair genotoxic lesions. The molecular cues that dictate the cellular decision between a protective p53-mediated cell-cycle arrest or p53-driven apoptosis have remained largely unclear (Reinhardt and Schumacher, 2012). Numerous regulatory mechanisms, involving selective DNA binding of p53 to its target gene promoters, selective transactivation of p53-bound target genes and differential stability of p53-dependent transcripts, have been reported to modulate these p53-dependent cell fate decisions, which are essential to repress the uncontrolled proliferation of incipient cancer cells carrying severely damaged genomic material (Oda et al, 2000b; Seoane et al, 2002; Wei et al, 2006; Das et al, 2007; Espinosa, 2008).
Here, we identify the apoptosis-antagonizing transcription factor (AATF) as a critical regulator of p53-driven apoptosis. AATF had previously been shown to be a substrate of the canonical DDR kinases ATM/ATR and Chk2 (Bruno et al, 2006) and to exert anti-apoptotic activity through promoting expression of XIAP in response to genotoxic stress (Bruno et al, 2008). We now demonstrate that AATF negatively regulates p53-driven apoptosis by preventing efficient DNA damage-induced transactivation of the pro-apoptotic p53 target genes PUMA, BAX and BAK. Moreover, we show that AATF resides in cytoplasmic protein complexes with the cytoskeletal protein myosin-regulatory light chain-3 (MRLC3) in resting cells. Following DNA damage, this interaction is lost through MK2-dependent AATF phosphorylation resulting in nuclear translocation of AATF and the specific engagement of the PUMA, BAX and BAK promoters to repress p53-dependent transcription of these proapoptotic genes. Interestingly, AATF neither binds to the promoters, nor regulates the expression of the cell-cycle-regulating p53 target genes CDKN1A, GADD45α or RPRM. Furthermore, our data show that AATF amplification in tumours results in worse prognosis of neuroblastoma patients. We propose that AATF functions to repress the pro-apoptotic arm of the p53 response to promote a primarily growth-arresting cellular response to genotoxic insults. These data suggest AATF to be an attractive drug target since AATF depletion resulted in substantially increased sensitivity to frontline chemotherapeutic drugs.
Results
A phospho-proteomic screen links MRLC3 to DNA damage kinase signalling
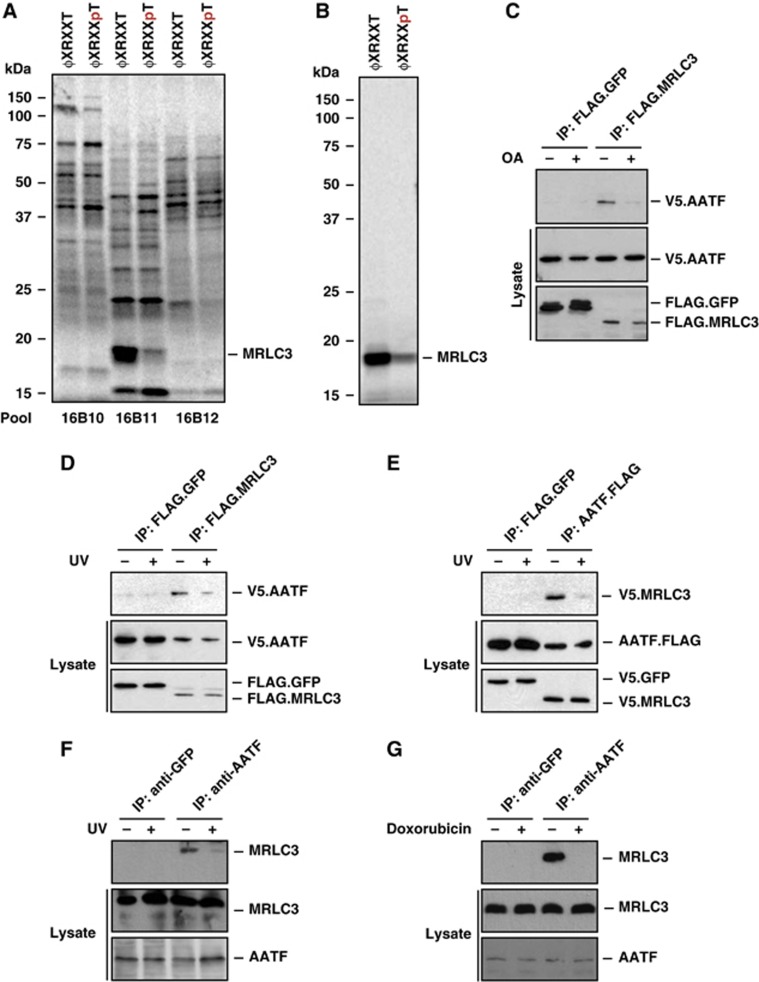
To identify new protein complexes that are regulated through DNA damage kinase signalling, we employed a phosphopeptide library versus protein expression library screening approach. We were interested in protein complexes that were disrupted in a phosphorylation-dependent manner, such as the p53:MDM2 complex, which dissociates upon p53 phosphorylation by numerous DDR kinases (Lavin and Gueven, 2006; Toledo and Wahl, 2006). We used a library of partially degenerate peptides resembling the substrate motif phosphorylated by the DDR kinases Chk1/2 and MK2, corresponding to the sequence [L/I/F]-X-[R/K]-[Q/X]-X-S/T-X-X-X, where X represents any amino acid except cysteine. Both phospho- and non-phospho versions of this library, hereafter denoted ϕ-X-R-X-X-pT and ϕ-X-R-X-X-T, were generated. Streptavidin bead-immobilized peptide libraries were used as bait in an interaction screen against a protein library produced by in vitro transcription/translation (Elia et al, 2003; Manke et al, 2003). We screened a total of ∼200 000 cDNAs arrayed in 2000 pools containing 100 individual, in vitro-translated and 35S-labelled cDNAs. Most in vitro-translated proteins did not bind to either library (Figure 1A). However, we identified 14 distinct pools, in which a 17-kDa protein displayed preferential binding to the ϕ-X-R-X-X-T library (Figure 1A). Progressive subdivision of these pools identified the cytoskeletal protein MRLC3 as the clone that preferentially associated with the ϕ-X-R-X-X-T library (Figure 1A and B). To further validate this interaction, we expressed FLAG-tagged MRLC3 in HEK293T cells and performed in vitro pull down experiments using the streptavidin-immobilized ϕ-X-R-X-X-T and ϕ-X-R-X-X-pT libraries as bait. As shown in Supplementary Figure 1A, MRLC3 displayed robust binding to the ϕ-X-R-X-X-T, but essentially no binding to the ϕ-X-R-X-X-pT library, suggesting that Thr-phosphorylation within the checkpoint kinase motif disrupts the interaction with MRLC3.
Figure 1.
Identification of a phosphorylation-sensitive protein complex consisting of AATF and MRLC3. (A) An oriented (pSer/pThr) phosphopeptide library, biased towards the basophilic phosphorylation motif of Chk1/2 and MK2, was immobilized on streptavidin beads. The phospho ϕXRXXpT and non-phosphorylated ϕXRXXT peptide libraries were screened for interaction against in vitro translated, 35S-Met-labelled proteins. (B) Identification of MRLC3 as a non-phospho binder occurred in pool 16B11 and through progressive subdivision to a single clone. (C) Yeast two-hybrid screening revealed AATF as an interactor of MRLC3. We further characterized this interaction through co-immunoprecipitation (co-IP), performed in the presence or absence of 1 μM okadaic acid (OA). FLAG.MRLC3 was immunoprecipitated from HEK293T cells co-expressing V5.AATF. FLAG.GFP served as a control. Lane 3 shows an interaction of FLAG.MRLC3 with V5.AATF, which was abolished by OA-mediated Ser/Thr phosphatase inhibition 1 h prior to lysis (lane 4). (D) The MRLC3:AATF complex is sensitive to UV-C-induced DNA damage. FLAG.MRLC3 and V5.AATF-expressing HEK293T cells were UV-C irradiated (20 J/m2) 30 min prior to lysis and IP with anti-FLAG beads. FLAG.GFP served as a negative control. While V5.AATF co-precipitated with FLAG.MRLC3 in the absence of UV-C, the interaction was abrogated in the presence of DNA damage. (E) Reversal of the co-IP experiment is shown in (D). Anti-FLAG IP reveals AATF.FLAG:V5.MRLC3 complexes that display strong sensitivity to UV-C-induced DNA damage. FLAG.GFP served as a negative control. (F) Endogenous AATF:MRLC complexes display UV-C sensitivity. AATF was immunoprecipitated from HCT116 cells that were mock-treated or exposed to UV-C (20 J/m2) 30 min prior to lysis and IP. GFP IP served as a negative control (lanes 1 and 2). While substantial amounts of MRLC co-immunoprecipitated with AATF (lane 3), this interaction was abolished by UV-C-induced DNA damage (lane 4). (G) Endogenous AATF:MRLC3 complexes are sensitive to the topoisomerase-II inhibitor doxorubicin. AATF was immunoprecipitated from HCT116 cells that were mock-treated or exposed to doxorubicin (1 μM) 1 h prior to IP. GFP antibody served as a negative control (lanes 1 and 2). Doxorubicin (lane 4) disrupted the interaction between AATF and MRLC (lane 3). Figure source data can be found with the Supplementary data.
We next investigated the interactome of MRLC3 using yeast two-hybrid screening. These experiments identified AATF as a likely MRLC3-interacting protein. To confirm this interaction in mammalian cells, we performed co-immunoprecipitation experiments in HEK293T cells co-expressing V5.AATF and FLAG.MRLC3 or FLAG.GFP, as a control. While AATF could readily be detected in the FLAG.MRLC3 precipitates, it was undetectable in the FLAG.GFP precipitations, thus validating the interaction between AATF and MRLC3 (Supplementary Figure 1B). Since MRLC3 was identified as a protein with strong selective binding to peptides corresponding to the non-phosphorylated forms of checkpoint kinase substrate motifs, but not to these same peptides following phosphorylation, we asked whether the AATF:MRLC3 interaction could be disrupted by phosphatase inhibition. In agreement with the results of the phospho-proteomic screen, treatment of V5.AATF and FLAG.MRLC3-expressing cells with the Ser/Thr phosphatase inhibitor okadaic acid, abrogated the AATF:MRLC3 interaction (Figure 1C).
We then went on to investigate whether the phosphorylation-sensitive interaction between AATF and MRLC3 is regulated by checkpoint kinases in response to genotoxic stress and performed co-immunoprecipitation experiments before and after DNA damage. As we had observed before, V5.AATF co-precipitated with FLAG.MRLC3 in mock-treated cells. In contrast, this interaction was abolished when cells were pre-treated with UV-C, indicating that genotoxic stress negatively regulates MRLC3:AATF complex formation (Figure 1D). Identical co-precipitation behaviour was observed when the FLAG and V5 tags were swapped (Figure 1E). Disruption of the MLRC3:AATF complex was also observed following treatment of cells with doxorubicin, indicating that the complex is sensitive to multiple types of genotoxic stress (Supplementary Figure 1C).
To ask whether endogenous AATF and MRLC3 form similar DNA damage-sensitive complexes, we immunoprecipitated AATF from HCT116 cells and used immunoblotting to detect co-precipitating MRLC3. These experiments confirmed the existence of a physiological interaction between AATF and MRLC3 in resting cells (Figure 1F, lane 3). As expected, application of UV-C or addition of doxorubicin prior to cell lysis abolished this endogenous interaction (Figure 1F and G), recapitulating the effects seen with overexpressed proteins. These data demonstrate that AATF and MRLC3 form a phosphorylation-sensitive protein complex, which is disrupted in response to genotoxic stress, likely mediated through the activity of a basophilic checkpoint kinase.
MRLC3 sequesters AATF in the cytoplasm
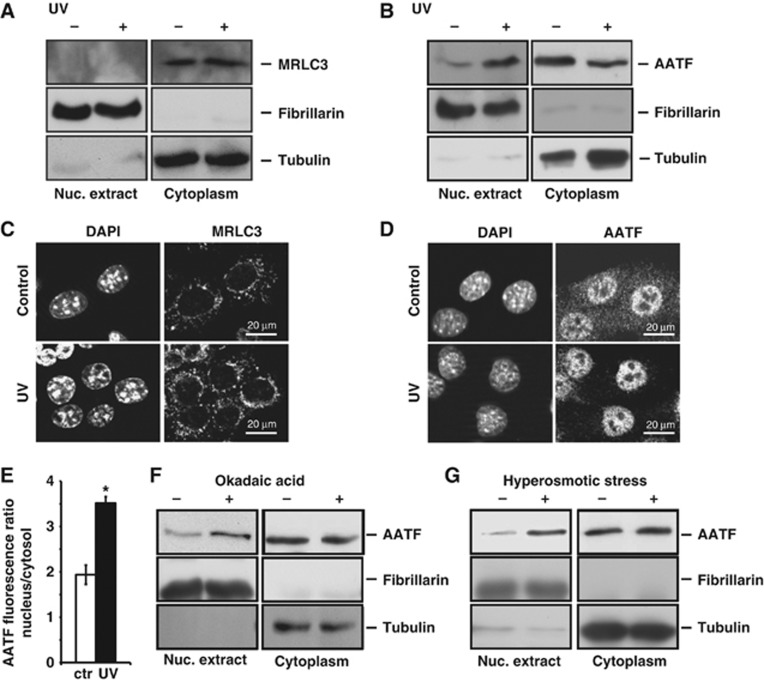
While MRLC3 is believed to reside predominantly in the cytoplasm, the subcellular localization of AATF is less well understood (Watanabe et al, 2007). Furthermore, it remains unclear whether AATF or MRLC3 dynamically shuttle between distinct subcellular compartments upon disruption of the AATF:MRLC3 complex. We directly investigated the spatial dynamics of MRLC3 and AATF in mouse embryonic fibroblasts (MEFs), using biochemical separation of nuclear and cytoplasmic fractions through hypotonic lysis. As shown in Figure 2A, MRLC3 was found exclusively in the cytoplasm and its subcellular distribution was not affected by UV-C-induced genotoxic stress. In marked contrast, AATF showed a DNA damage-dependent dynamic re-localization between cytoplasm and nucleus. While only minuscule amounts of endogenous AATF were detectable in the nuclei of resting cells, we observed a marked enhancement of nuclear AATF after UV-C (Figure 2B). We confirmed these data, using immunofluorescence. As shown in Figure 2C, cells displayed strong cytoplasmic MRLC3 staining before and after UV-C. In contrast, the AATF antibody revealed prominent granular staining in both the cytoplasm and the nucleus of resting cells (Figure 2D). Control experiments performed in AATF-depleted cells revealed that the nuclear staining in resting cells was unspecific (Supplementary Figure 2). Following UV-C irradiation, we observed a statistically significant loss of cytoplasmic AATF staining, consistent with depletion of cytoplasmic AATF pools and subsequent nuclear accumulation (Figure 2D and E). Confirming phosphorylation dependence, treatment of cells with okadaic acid produced a similar nuclear translocation of AATF. As shown in Figure 2F, pre-treatment of MEFs with okadaic acid resulted in a robust nuclear accumulation of AATF, suggesting that AATF phosphorylation, likely by a DNA damage-activated basophilic kinase, leads to disruption of cytoplasmic AATF:MRLC3 complexes with subsequent nuclear AATF accumulation.
Figure 2.
The AATF:MRLC3 cytoplasmic complex is disrupted upon DNA damage. (A) Hypotonic lysis was used to generate nuclear and cytoplasmic fractions. The purity of the fractions was documented with immunoblotting for fibrillarin (nuclear) and tubulin (cytoplasmic). MRLC was exclusively detected in the cytoplasmic compartment. The subcellular distribution of MRLC was not affected by 20 J/m2 UV-C-induced genotoxic stress applied 30 min prior to lysis. (B) AATF resides in the cytoplasm of resting cells and translocates to the nucleus upon genotoxic stress. Nuclear and cytoplasmic fractions of mock- or UV-C-treated (20 J/m2) MEFs were generated as in (A). In the absence of UV, AATF is primarily cytoplasmic. Upon UV-C-induced DNA damage, substantial amounts of AATF are detectable in the nucleus. (C) Indirect immunofluorescence (IF) was performed to verify the subcellular localization of MRLC3 in MEFs 30 min following UV-C (20 J/m2). DAPI staining was used as a counterstain to provide a nuclear reference point. MRLC displayed a granular cytoplasmic staining pattern that was not affected by UV-C irradiation. No MRLC staining could be detected in the nuclei. (D) Indirect IF was performed to verify the subcellular distribution and spatial dynamics of AATF in MEFs 30 min following UV-C (20 J/m2). In resting cells, AATF staining revealed a dominant granular cytoplasmic pattern with some nuclear staining. 30 min following UV-C, only miniscule amounts of AATF remained detectable in the cytoplasm, while the bulk of AATF staining was now nuclear. (E) The DNA damage-induced nuclear re-localization of endogenous AATF was quantified using fluorescence microscopy. Error bars represent s.d. (*P<0.05). (F) Global Ser/Thr phosphatase inhibition promotes the nuclear accumulation of AATF. MEFs were mock-treated or pre-treated with 1 μM OA 60 min prior to lysis. Phosphatase inhibition resulted in nuclear accumulation of AATF. (G) Osmotic stress promotes the nuclear accumulation of AATF. MEFs were mock-treated or exposed to hypertonic Ringer solution 10 min prior to lysis. This non-genotoxic MK2-activating stimulus resulted in nuclear AATF accumulation similar to that seen after DNA damage. Figure source data can be found with the Supplementary data.
Genotoxic stress releases AATF from MRLC3 in an MK2-dependent manner
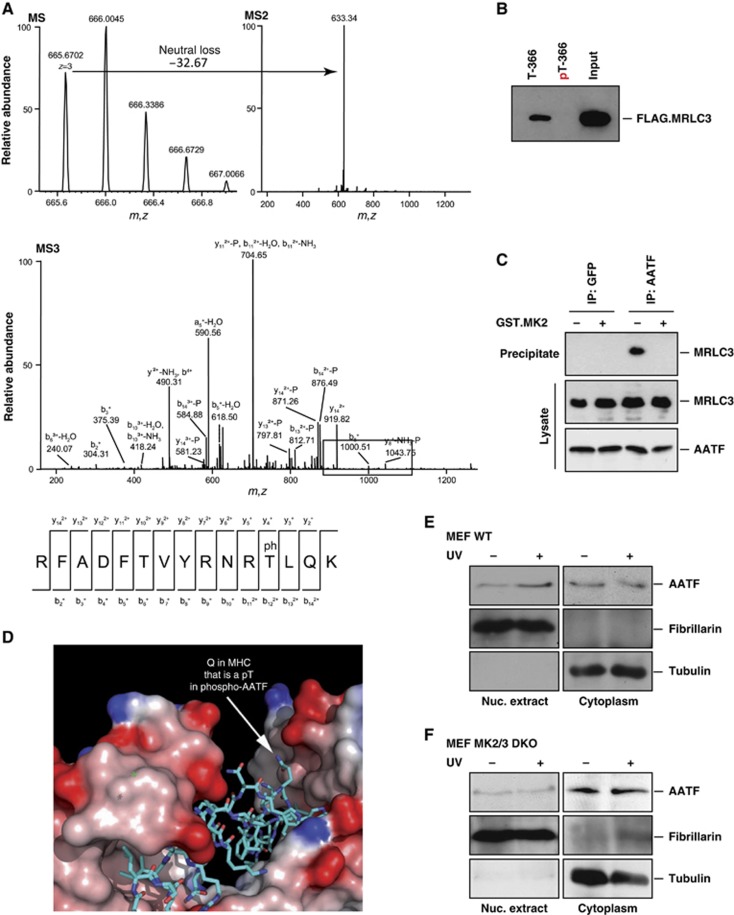
While the DDR is typically initiated within the nucleus, and this is the primary site of action for Chk1 (Supplementary Figure 2), our data strongly suggested that a DNA damage-responsive basophilic kinase(s) was (directly) responsible for the disruption of cytoplasmic AATF:MRLC3 complexes in the cytoplasm. We recently identified MK2 as a checkpoint kinase sharing substrate specificity with Chk1 and Chk2, but whose primary site of action is in the cytoplasm (Reinhardt et al, 2010). Intriguingly, when we repeated our fractionation experiments with cells that had been exposed to osmotic stress, a strong MK2 stimulus (Engel et al, 1998) that has not been reported to induce Chk1/2 activation, we also observed nuclear AATF accumulation (Figure 2G). We next used in vitro kinase assays to test whether MK2 directly phosphorylated AATF. As shown in Supplementary Figure 3, MK2 readily phosphorylated AATF, while no significant 32P incorporation could be detected in control reactions lacking MK2. In silico amino-acid sequence analyses using the Scansite software (Obenauer et al, 2003) revealed that AATF contains a sequence stretch centred around Thr-366 that is an exact match to the kinase motif selected by MK2. Mass spectrometry confirmed phosphorylation of AATF at Thr-366. As shown in Figure 3A, pThr-366-containing peptides were readily detected in Lys-C digests from in vitro kinase assays containing AATF and MK2, suggesting that MK2 directly phosphorylates AATF on Thr-366. To test whether the Thr-366-containing peptide sequence might mediate the phosphorylation-sensitive interaction between AATF and MRLC3, we synthesized Thr-366 and pThr-366-containing biotinylated peptides, spanning amino acid 357–374. Peptides were immobilized on streptavidin beads, incubated with lysates from FLAG.MRLC3 transfected cells and peptide:protein complexes were isolated by affinity purification. While we observed robust binding of MRLC3 to the non-phosphorylated peptide, we were unable to precipitate MRLC3 with the pThr-366 peptide, indicating that the interaction between AATF and MRLC3 is sensitive to AATF-Thr-366 phosphorylation, likely mediated through MK2 (Figure 3B).
Figure 3.
MK2-mediated AATF phosphorylation on Thr-366 disrupts cytoplasmic AATF:MRLC3 complexes to allow nuclear accumulation of AATF. (A) MK2 was subjected to in vitro kinase assays using AATF as a substrate. Reactions were quenched by the addition of SDS-buffer and proteins were resolved on SDS–PAGE before AATF-containing coomassie bands were isolated and proteins subjected to LC–MS/MS analysis after reduction, alkylation and Lys-C digestion. For MS analysis, up to four HCD and CID spectra (MS2) were acquired following each scan. When a neutral loss of phosphoric acid was detected, MS3 spectra were acquired in the linear ion trap to determine the peptide sequence. Mascot 2.2 was used for protein ID. A peptide spanning the phospho-Thr-366 residue was readily detected. (B) A biotinylated phospho-Thr-366-containing AATF peptide fails to interact with FLAG.MRLC3. In vitro pull down assays were performed from FLAG.MRLC3-expressing HEK293T cells using streptavidin-immobilized Thr-366 phosphorylated and non-phosphorylated peptides (AATF amino acid 357–374) as baits. While the non-phosphrylated peptide bound to FLAG.MRLC3, no interaction was observed between the phosphorylated peptide and FLAG.MRLC3. (C) Endogenous AATF:MRLC complexes are disrupted by MK2 in vitro. AATF:MRLC complexes were immunoprecipitated from HCT116 cells using AATF antibodies. GFP-immunoprecipitations served as a negative control. Precipitated material was subjected to a 30-min incubation with MK2 or left untreated before SDS–PAGE and immunoblotting. Incubation with MK2 (lane 4) disrupted the AATF:MRLC complexes immunoprecipitated in the control samples (lane 3). (D) A structural basis for phospho-dependent release of AATF from MRLC. The structure of MRLC bound to MHC is shown (PDB code 2OS8) with MRLC in a surface representation shaded by electrostatic potential (red: acidic, blue: basic). MHC is shown in stick representation with carbons coloured cyan, nitrogens blue, and oxygens red. The position of a Gln residue in MHC corresponding to the position of Thr-366 in AATF based on sequence and secondary structure alignments (Supplementary Figure 6) is indicated. (E) AATF re-localizes to the nucleus following UV-C-induced DNA damage in MK2wt/wt MEFs. Cells were either mock-treated or exposed to UV-C (20 J/m2) 30 min prior to biochemical fractionation. Fibrillarin and tubulin served to control the purity of the fractions. (F) DNA damage-induced nuclear localization of AATF is repressed in MK2/3−/− MEFs. Cells were treated as shown in (E). Figure source data can be found with the Supplementary data.
We next asked whether MK2-mediated AATF phosphorylation is sufficient to disrupt AATF:MRLC3 complexes. To address this question, we used AATF antibodies to precipitate endogenous AATF:MRLC3 complexes from HCT116 cells. Precipitates were incubated with MK2 or mock treated and then resolved on SDS–PAGE followed by immunoblotting to detect MRLC3 that remained bound to AATF throughout the course of the experiment. As depicted in Figure 3C, incubation of endogenous AATF:MRLC3 complexes with active MK2 resulted in a substantial MRLC3 release from bead-immobilized AATF with essentially no MRLC3 being detectable in the precipitated material following the kinase reaction.
Lastly, we examined the known X-ray crystal structure of the interaction interface between MRLC and myosin heavy chain (MHC) (Yang et al, 2007). Using sequence alignment and secondary structure predictions, we found that the peptide surrounding Thr-366 in AATF is likely to assume a helical conformation, like that observed in the MRLC-binding segment of MHC (Figure 3D; Supplementary Figure 4). Mapping of the MRLC-binding segment of AATF onto the known MHC interaction grove in MRLC revealed that AATF Thr-366 is predicted to be located in close proximity to an acidic patch within MRLC (Figure 3D). Phosphorylation of Thr-366 in AATF is thus expected to disrupt the complex between MRLC3 and AATF.
To analyse the role of MK2 in the destabilization of AATF:MRLC3 complexes, we monitored the spatial dynamics of AATF in wild-type or MK2/3−/−MEFs (Supplementary Figure 5) (Ronkina et al, 2007). While a prominent accumulation of nuclear AATF in MK2wt/wt MEFs was detectable (Figure 3E), we failed to monitor any nuclear enrichment of AATF in MK2/3−/− MEFs after UV-C-induced DNA damage (Figure 3F). These data indicate that MK2-mediated AATF phosphorylation on Thr-366 disrupts the interaction between AATF and MRLC3 leading to nuclear translocation of AATF induced by DNA damage.
Chromosomal gains at the AATF locus are associated with reduced survival in human tumours
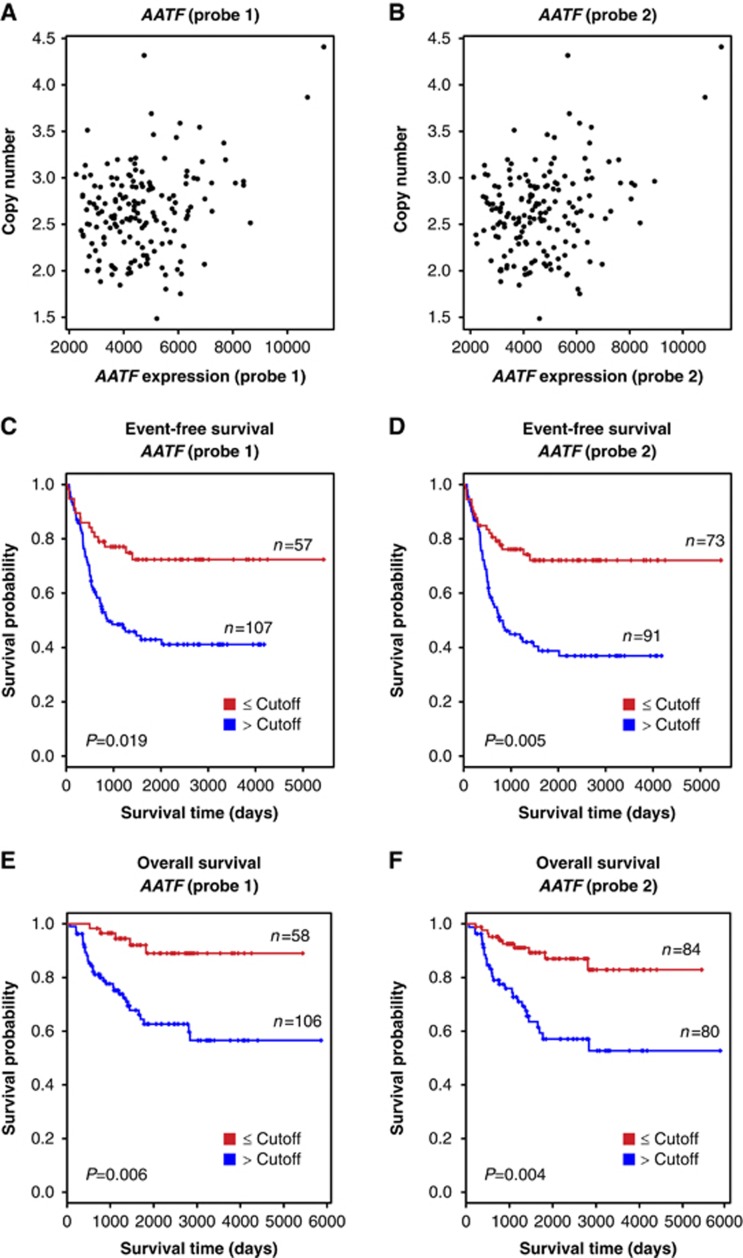
Alterations in genes comprising the DDR network are frequently observed in human tumours and have been shown to promote the acquisition of further cancer-promoting mutations or the evasion of therapy-induced apoptosis (Hanahan and Weinberg, 2000). To test whether the AATF locus might be affected in human tumours, we made use of a cohort of 164 clinically well-characterized neuroblastoma cases. We note that the cancer genes most frequently altered in adult neoplastic disease, such as TP53, CDKN2A or RAS, are rarely aberrant in neuroblastoma. Specifically, inactivating TP53 mutations are extremely rare in primary neuroblastomas (Maris et al, 2007). All tumour samples were obtained at diagnosis prior to any cytotoxic treatment with a poly-chemotherapy regimen and each tumour sample was reviewed by a pathologist to ensure a minimal tumour content of 60%. We used two independent probes to detect AATF mRNA expression. As shown in Figure 4A and B, chromosomal gains on the AATF locus, determined by aCGH, were tightly correlated with increased AATF mRNA expression. We next stratified tumours according to AATF expression to ask whether AATF abundance had an impact on patient survival. As shown in Figure 4C–F, increased AATF expression was associated with reduced event-free and overall survival in these neuroblastoma patients. These data strongly suggest that AATF is affected in human tumours and that increased AATF expression levels are associated with poor prognosis and reduced survival in neuroblastoma patients.
Figure 4.
Association of AATF copy number, expression and clinical outcome in neuroblastoma. In all, 164 neuroblastoma samples were subjected to aCGH-based copy number and mRNA expression analyses to determine whether a correlation between AATF copy number and mRNA expression exists in human tumours. (A, B) Scatter plot showing the correlation between genomic copy number and the expression level of AATF for probe 1 (A, r=0.29, P<0.001) and probe 2 (B, r=0.30, P<0.001). (C–F) Kaplan–Meier survival curves for event-free (C, probe 1; D, probe 2) and overall survival (E, probe 1; F, probe 2) of neuroblastoma patients separated according to AATF expression levels. Red, expression level lower than the cutoff value; blue, expression level higher than the cutoff value.
AATF overexpression promotes the stability of DNA damage-induced cell-cycle checkpoints
Fanciulli and colleagues recently reported that overexpression of AATF in HCT116 cells resulted in an enhanced stability of a doxorubicin-induced G2/M checkpoint. This effect was p53-dependent, since AATF overexpression in p53−/− cells did not result in any detectable difference in checkpoint stability (Bruno et al, 2006). To test whether AATF repression might lead to a destabilization of p53-dependent DNA damage checkpoints, we subjected p53-proficient or -deficient HCT116 cells expressing AATF-specific shRNA to nocodazole trap experiments (Supplementary Figure 5B and C). In brief, addition of nocodazole 2 h following application of genotoxic agents will arrest all cells that prematurely escape DNA damage-induced checkpoints in mitosis. The percentage of mitotically trapped cells that had escaped DNA damage-induced checkpoints was monitored by phospho-histone H3 staining 30 h after infliction of DNA damage. As shown in Supplementary Figure 5D, we observed ∼4% of mitotic cells in asynchronously growing p53-proficient and -deficient cells expressing either control or AATF-specific shRNA. When control shRNA-expressing p53-proficient HCT116 cells were treated with UV-C in the nocodazole trap setting, <1% of these cells had escaped the G2/M checkpoint into mitosis at the end of the time course, indicating a stable UV-C-induced checkpoint in these cells. An almost identical checkpoint stability was observed in p53-deficient HCT116 cells in response to UV-C. To our surprise, RNAi-mediated AATF depletion did not affect the stability of UV-C-induced checkpoints in p53-proficient or -deficient HCT116 cells. We further failed to detect any significant impact of AATF depletion on the stability of doxorubicin- or camptothecin-induced checkpoints in p53-proficient or -deficient settings. However, when we used retroviral overexpression of AATF cDNA, we were able to detect a small, but reproducible and statistically significant increase in the stability of UV-, doxorubicin- and camptothecin-induced DNA damage checkpoints in the p53-proficient cells (Supplementary Figure 5D). These data indicate that AATF repression does not substantially affect p53-dependent cell-cycle checkpoints in HCT116 cells after genotoxic stress, while AATF overexpression has a checkpoint-stabilizing effect in p53-proficient cells.
AATF negatively regulates p53-dependent apoptosis
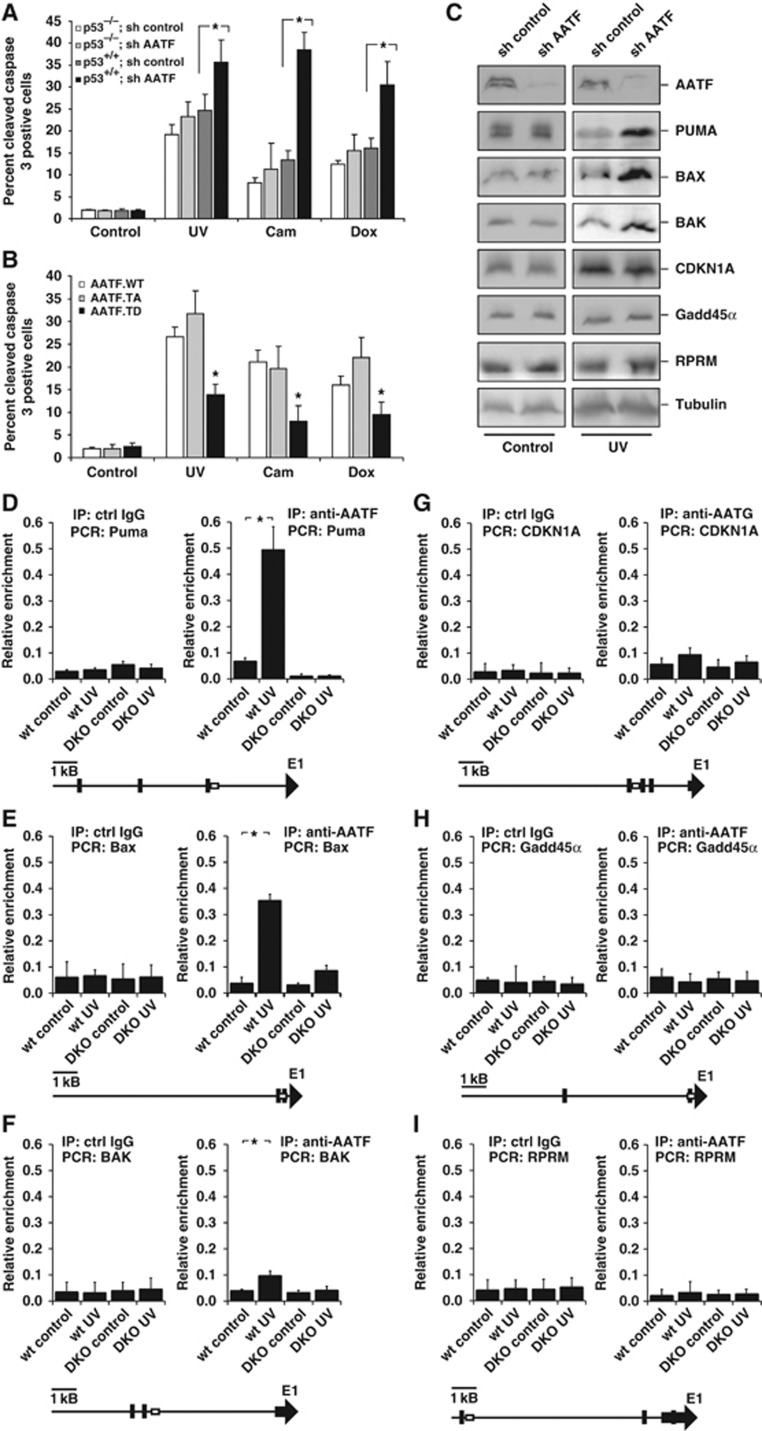
AATF repression has been shown to result in increased apoptosis in response to DNA damage (Di Certo et al, 2007; Bruno et al, 2008). One possible explanation for this observation could be that AATF might be involved in repressing the pro-apoptotic arm of the p53 response to indirectly promote the induction of a p53-dependent cell-cycle arrest following DNA damage. In addition to transactivating cell-cycle-regulating genes, such as CDKN1A or GADD45α, p53 has also been shown to transactivate pro-apoptotic genes, such as PUMA, BAX and BAK (Reinhardt and Schumacher, 2012). Thus, we asked whether AATF affected DNA damage-induced apoptosis in p53+/+ and p53−/− HCT116 cells expressing either control or AATF-specific shRNA. Cells were treated with camptothecin, doxorubicin or UV-C and apoptosis was quantified by FACS 24 h later. The percentage of apoptotic cells following the different treatments did not significantly differ in p53−/− cells that expressed either control or the AATF-specific shRNA (Figure 5A). In striking contrast, AATF depletion in p53+/+ cells resulted in a dramatically increased sensitivity to all three DNA-damaging agents (Figure 5A). Camptothecin treatment of control cells resulted in 13.4±2.1% apoptotic cells 24 h later, which rose to 38.5±4% in AATF-depleted p53+/+ cells (Figure 5A). At 24 h after doxorubicin application, we observed that 16.4±2.2% of control cells were apoptotic, in contrast to 30.5±5.5% of AATF-depleted cells (Figure 5A). Similarly, UV-C irradiation of AATF-depleted p53+/+ cells resulted in 35.7±5.1% of apoptotic cells, compared with only 24.7±3.6% in control cells (experiments in panel A n=16, P<0.05 Student’s t-test). These experiments were then extended to an additional panel of human cancer cell lines containing wt or mutant forms of p53. H23, H2122 (both p53 mutant), SW1573 and H460 (p53 wild type) were transduced with control or AATF-specific shRNA and exposed to UV, camptothecin or doxorubicin, as above. Similar to the HCT116 cells, these cell lines segregated in their phenotypic response based solely on their p53 status. Loss of AATF resulted in significantly enhanced rates of apoptosis in response to all three damaging agents only in cell lines with intact p53, but not in cells carrying mutant p53 (Supplementary Figure 6). Interestingly, MRLC depletion significantly increased resistance to UV-, camptothecin- and doxorubicin-induced apoptosis in p53+/+ cells, consistent with the observation that MRLC3 sequesters anti-apoptotic AATF in the cytoplasm in an inactive state (Supplementary Figures 5C and 6). Thus, loss of AATF appears to enhance the p53-dependent apoptotic response after DNA damage. To finally demonstrate that MK2-mediated AATF phosphorylation on Thr-366 was critical for the release of AATF from MRLC and apoptosis regulation, we repeated the assays in p53+/+ HCT116 cells expressing either wild-type AATF (AATFWT), a non-phosphorylatable AATF Threonine-366 to Alanine mutant (AATFTA) or a phospho-mimicking AATF Threonine-366 to Aspartate mutant (AATFTD). As shown in Figure 5B, camptothecin, doxorubicin and UV-C induced moderate levels of apoptosis in cells overexpressing AATFWT, comparable to those of empty vector-transduced control cells (compare Figure 5A and B). Overexpression of AATFTA slightly increased the number of apoptotic cells when compared with AATFWT-transduced cells, consistent with cytoplasmic trapping of anti-apoptotic AATF, however, this failed to reach statistical significance potentially due to the effects of endogenous AATF (Figure 5B, white and light grey bars). In contrast, overexpression of AATFTD prevented efficient induction of apoptosis in response to camptothecin, doxorubicin and UV-C. While camptothecin treatment resulted in 21.0±2.6% of apoptotic cells in AATFWT-overexpressing cells, this number dropped to 13.8±2.4% in cells transduced with the AATFTD mutant. We observed a similar drop in the rates of apoptosis when we compared AATFWT and AATFTD-expressing cells that were treated with doxorubicin (16.0±2.0% versus 9.4±2.9%, respectively) or UV-C (26.7±2.1% versus 13.8±2.4%, respectively (Figure 5B), experiments in Figure 5A and B n=16, P<0.05, Student’s t-test. As might be expected, we failed to observe such a resistance-mediating effect of AATFTD overexpression in p53−/− cells (Supplementary Figure 7). These data are consistent with a role for AATF in antagonizing p53-driven apoptosis following DNA damage in a manner that depends on MK2-mediated AATF phosphorylation and nuclear translocation in response to genotoxic stress.
Figure 5.
AATF prevents p53-driven apoptosis by repressing DNA damage-dependent induction of pro-apoptotic genes. (A) p53+/+ and p53−/− HCT116 cells expressing control or AATF-specific shRNA were left untreated or exposed to UV (40 J/m2), camptothecin (cam, 10 μM) or doxorubicin (dox, 1 μM) and harvested for quantification of apoptosis 24 h later. AATF depletion in p53+/+ cells resulted in a significant increase in the number of apoptotic cells following all treatment regimens with DNA-damaging agents. No increase in the number of apoptotic cells could be observed in p53−/− cells following DNA damage. Asterisk indicates statistical significance, error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=16. (B) p53+/+ HCT116 cells expressing AATFWT, AATFTA or AATFTD were left untreated or exposed to UV (40 J/m2), camptothecin (10 μM) or doxorubicin (1 μM) and harvested for FACS-based quantification of apoptosis 24 h later. AATFTD expression significantly repressed apoptosis in response to all three genotoxic treatments. Asterisk indicates statistical significance (P<0.05), error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=16. (C) RNAi-mediated AATF depletion in p53+/+ HCT116 cells promotes DNA damage-induced induction of pro-apoptotic p53 target genes, while expression of cell-cycle-regulating p53 target genes remains unaffected. Immunoblotting was used to detect the pro-apoptotic p53-target gene products PUMA, BAX and BAK, and the cell-cycle-arresting p53 target gene products CDKN1A, GADD45α and REPRIMO 12 h after 40 J/m2 UV-C. Tubulin served as a loading control. (D–F) AATF binds to the promoters of the pro-apoptotic p53-target genes PUMA, BAX and BAK in a DNA damage and MK2-dependent manner. ChIP experiments were performed in MK2/3+/+ and MK2/3−/− MEFs that were left untreated or exposed to 40 J/m2 UV-C 60 min prior to cross-linking. DNA was precipitated using AATF antibodies. qPCR was used to quantify PUMA, BAX and BAK promoter-specific DNA in the precipitates. Unspecific IgG served as a control. Asterisk indicates statistical significance, error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=9. (G–I) AATF binding to the CDKN1Ap21, GADD45α and RPRM promoter is not regulated in a DNA damage or MK2-dependent manner. ChIP experiments were performed as in (D–F) and promoter-specific primers were used for qPCR. Unspecific IgG served as a control. Asterisk indicates statistical significance, error bars represent s.d., two-tailed Student’s t-test, P<0.05, n=9. Primers to amplify the genomic DNA of the promoter regions were chosen to cover known p53-binding sites. Schematic drawings (D–I) indicated localization of primer (box) and the known p53-binding sites (black bars). Figure source data can be found with the Supplementary data.
AATF regulates the functional outcome of the p53 response
To elucidate how AATF is involved in determining the outcome of p53 activation, we next monitored the DNA damage-mediated induction of a panel of well-established p53 target genes. We used immunoblotting to analyse the expression of the proapoptotic genes PUMA, BAX and BAK, as well as the growth-arresting genes CDKN1A, GADD45α and RPRM in p53-proficient HCT116 cells expressing control or AATF-specific shRNAs. AATF depletion resulted in a substantially enhanced expression of the proapoptotic genes PUMA, BAX and BAK in response to UV-C, while no difference in the expression of the cell-cycle-regulatory genes CDKN1A and GADD45α could be observed (Figure 5C). Only a miniscule, but consistent, induction of RPRM was seen in AATF-depleted cells upon UV-C (Figure 5C). We next performed chromatin immunoprecipitation (ChIP) to investigate whether AATF selectively binds to the promoter regions of the p53 target genes whose gene products it appears to regulate following genotoxic stress. We designed a set of primers to amplify the genomic DNA of the promoter regions covering known p53-binding sites (indicated by the cartoons accompanying Figure 5D–I). As shown in Figure 5D, <8% of the PUMA promoter DNA subjected to the ChIP experiment could be recovered with AATF-specific antibodies in the absence of genotoxic stress. In contrast, UV irradiation prior to immunoprecipitation allowed the recovery of 49.2±8.9% of the input DNA, indicating enhanced AATF binding to the PUMA promoter in response to genotoxic stress. Deletion of MK2/3 almost completely prevented this interaction of AATF with the PUMA promoter region, indicating a requirement for MK2 to promote AATF binding to the PUMA promoter. Furthermore, we could document a similar MK2/3-dependent interaction of AATF with the promoters of the proapoptotic p53 target genes BAX and BAK in response to UV-C (Figure 5E and F). In contrast, while there was a small and reproducible, yet statistically not significant, increase in the AATF occupancy on the CDKN1A promoter, similar to what has been described by others (Bruno et al, 2006), we failed to detect an increase in AATF occupancy on the GADD45α and RPRM promoters in response to DNA damage despite extensive efforts (Figure 5G–I). Figure 5G–I n=9, P<0.05, Student’s t-test. These data strongly suggest that AATF dampens the p53-dependent expression of a set of selective apoptosis-inducing genes likely by exerting transcriptional repression on the promoter regions, while having little if any effect on a set of canonical p53-regulated genes involved in mediating cell-cycle arrest upon genotoxic stress.
AATF promotes doxorubicin resistance in vivo
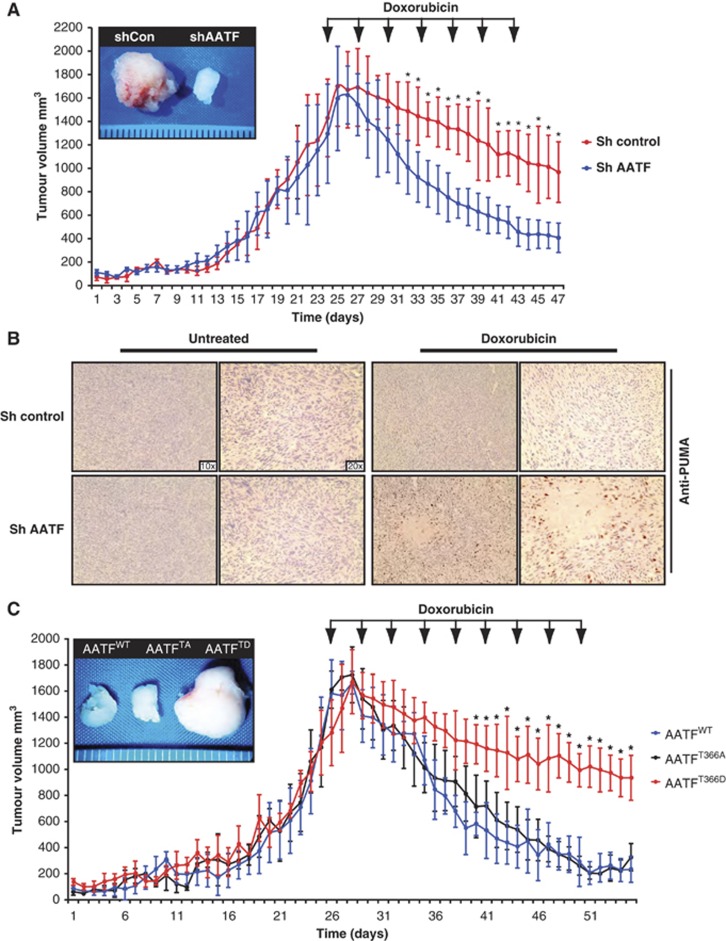
To investigate whether the MK2-dependent apoptosis-antagonizing role of AATF could also be observed in vivo, we performed xenograft experiments. 106 HCT116 cells stably expressing either control, or AATF-specific shRNAs were injected into the flanks of NCRnu/nu mice and doxorubicin treatment was initiated upon tumour formation. Control tumours showed only a mild volume reduction from 1430 (±331) to 967 (±258) mm3, while AATF-depleted tumours showed a remarkable doxorubicin response with tumour volumes decreasing from 1295(±422) to 408(±125) mm3. The differences in mean tumour volumes between control and AATF shRNA-expressing tumours reached statistical significance after only 9 days of treatment (Figure 6A). Using immunohistochemistry, we could show that Puma was specifically induced in doxorubicin-treated AATF-depleted tumours, while little Puma staining was seen in the controls (Figure 6B). These data lend further support to an apoptosis-antagonizing role of AATF through repression of p53-mediated Puma transcription after DNA damage.
Figure 6.
AATF promotes chemo-resistance in vivo. (A) AATF depletion results in a significant increase doxorubicin sensitivity of p53+/+ tumours. In all, 106 HCT116 cells expressing either control or AATF-specific shRNA were xenograftet into the flanks of NCRnu/nu mice (n=6 for each group). (B) Immunohistochemical analysis of control and AATF shRNA-expressing tumours excised at the termination point of the experiments shown in (A). Specimens were stained with PUMA-specific antibodies and counterstained with haematoxylin. (C) AATFTD expression promotes doxorubicin resistance in vivo. p53+/+ HCT116 cells were transduced to express either AATFWT, AATFTA or AATFTD. In all, 106 HCT116 cells of each group were injected into the flanks of NCRnu/nu-mice (n=6 for each group). Arrows indicate doxorubicin injections (5 mg/kg, i.p.). Asterisks indicate significance (Student’s t-test, two-tailed, P<0.05).
To further probe the importance of MK2-mediated AATF phosphorylation on Thr-366, we repeated the xenograft assays with HCT116 cells that were transduced to overexpress AATFWT, AATFTA or AATFTD. Doxorubicin was initiated when AATFWT-overexpressing tumours were 1450(±267) mm3 and AATFTA-overexpressing tumours were 1577(±141) mm3 in volume. AATFWT- and AATFTA-overexpressing tumours subsequently regressed to 212(±98) and 305(±103) mm3 in volume, respectively. In contrast, tumours expressing the phospho-mimicking AATFTD mutant displayed remarkable doxorubicin resistance. AATFTD-expressing tumours measured 1250(±250) mm3 in volume at the initial dose of doxorubicin but showed only a mild volume reduction to 907(±170) mm3 over the treatment cycle (Figure 6C). These data provide in vivo evidence that Thr-366 in AATF serves as a molecular switch that, when phosphorylated, promotes resistance against genotoxic stress, likely through the selective abrogation of transactivation of the proapoptotic p53 target genes PUMA, BAX and BAK.
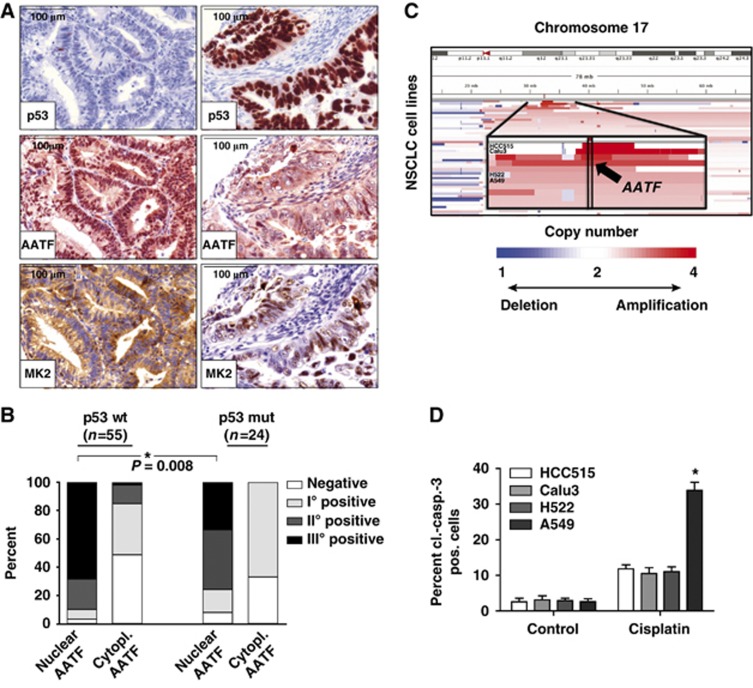
Based on these observations, one might predict that p53-proficient human tumours display an enrichment of nuclear AATF, as this would be critical to counteract p53-driven apoptosis. In contrast, p53-defective tumours might not show such an AATF accumulation, as nuclear AATF localization is likely not to be selected for in the absence of functional p53. To directly test this hypothesis in a relevant human tumour entity, we analysed a panel of 79 individual human endometrial cancer specimens. We used a well-established immunohistochemistry-based algorithm to stratify these tumours based on their p53 status. Staining for p53 was regarded as aberrantly enhanced when >70% of tumour cells showed strong staining. Since the vast majority of p53 mutations stabilize the mutant protein, gross overexpression in cancer cells is suitable to identify the vast majority of the tumour-associated p53 aberrations, with only rare false-negative and probably no false-positive cases (given the internal control of normal protein levels in noncancerous cells on each section) (Jiang et al, 2009). Tumour specimens were further stained with antibodies detecting AATF and MK2 (Figure 7A). As shown in Figure 7B, tumours expressing wild-type levels of p53 showed a robust enrichment of nuclear AATF, with the majority of samples displaying grade II/III nuclear AATF staining. This enrichment of nuclear AATF staining was significantly underrepresented in tumours lacking wild-type p53 (Fisher’s exact test, P< 0.05), strongly suggesting that nuclear localization of AATF is selected for in p53-proficient tumours, potentially to repress p53-driven apoptosis. Intriguingly, the activation and translocation of MK2 could also be observed in most human tumour samples, which also displayed nuclear AATF staining, further substantiating the hypothesis that AATF translocates into the nucleus in an MK2-dependent manner (Figure 7A).
Figure 7.
AATF is amplified in human tumours and displays a nuclear localization pattern in p53-proficient tumours. (A) Nuclear localization of AATF is selected for in p53-proficient tumours. Human endometrial tumours were stained for p53, AATF and MK2. p53-defective tumours show predominantly cytosolic AATF and nuclear MK2 staining (A—right) whereas tumours containing wild-type p53 display prominent nuclear enrichment of AATF accompanied by a prominent cytoplasmic MK2 signal (A—left). (B) Quantification of nuclear and cytoplasmic AATF staining was performed using a four-tier scoring system and subjected to statistical analysis (Fisher’s exact test). (C) AATF amplification is associated with increased resistance of lung cancer cell lines against cisplatin. Copy-number alterations as determined by SNP-arrays (blue=deletion; white=copy number neutral; red=amplification) at chromosome 17 (y axis) across all NSCLC cell lines are depicted (x axis). The chromosomal position of the amplification of AATF is highlighted for the analysed cell lines (zoom-in panel). HCC515 and Calu3 cells with copy number>4, H522 and A549 cells with copy number<4. (D) HCC515 (p53 mutant, AATF amplified), Calu3 (p53 wild type, AATF amplified), H522 (p53 mutant) and A549 (p53 wild type) cells were left untreated or exposed to cisplatin (10 μM) and harvested for FACS-based quantification of apoptosis using a cl.-caspase-3 antibody 24 h later. Asterisk indicates statistical significance, error bars represent s.d., n=12.
We next asked whether analysis of human tumour cells might lend support to our hypothesis that AATF is a p53 co-factor that selectively blunts the pro-apoptotic arm of the p53 response, using a panel of lung cancer cell lines, which were stratified based on p53 status and AATF copy number (Figure 7C). When we analysed cisplatin-induced apoptosis in these cell lines, we found that focal copy number gains at the AATF locus were associated with a significant protection from cisplatin-induced apoptosis in p53-proficient cells (Figure 7D), similar to what we had observed in the neuroblastoma samples described in Figure 4. Thus, AATF amplification emerges as a potential mechanism that is employed by human cancer cells to antagonize the detrimental effects of p53-driven apoptosis.
Discussion
Here, we identified AATF as a key repressor of the pro-apoptotic arm of the p53 response. Moreover, our data provided mechanistic details as well as clinical evidence implicating AATF as a potential drug target for cancer therapy. An important role for AATF in the DDR signalling networks had been suggested before (Bruno et al, 2002, 2006). Previous studies demonstrated that phosphorylation of AATF by the canonical checkpoint kinases ATM/ATR and Chk2 was important for AATF protein stabilization and involved in the control of cell-cycle progression (Bruno et al, 2006). However, much of this understanding of the role of AATF in DDR resulted from genetic gain and loss of function experiments (Bruno et al, 2002). We now report data corroborating the initial observations, showing an enhanced stability of DNA damage-induced cell-cycle checkpoints upon overexpression of exogenous AATF. The effects of AATF overexpression depend at least partially on the activation status of the DDR network with AATF promoting cell-cycle progression in the absence of DNA damage and enforcing a cell-cycle arrest in response to genotoxic stress. These contrasting effects of AATF signalling correlated with distinct promoter binding of AATF in the different scenarios (Bruno et al, 2002, 2006). Expanding on these initial observations with variable control of cell cycle, we now show that AATF serves as a master regulator of the p53-dependent apoptotic programme. Consistent with a clinically important role of apoptosis regulation, AATF expression and localization was indeed regulated in human tumours and gene amplification correlated with worse clinical outcome and reduced overall survival following genotoxic poly-chemotherapy. Intriguingly, chemotherapy-naive neuroblastomas are known to express wild-type copies of TP53 in the vast majority of cases. Thus, these data corroborate the in vitro experiments performed by Fanciulli and colleagues showing that AATF overexpression protected p53-proficient HCT116 cells from chemotherapy-induced apoptosis (Bruno et al, 2006).
Recent studies demonstrated that AATF overexpression results in the induction of antiapoptotic target genes even in the absence of genotoxic stress. Furthermore, AATF depletion prevented the doxorubicin-induced expression of XIAP, one of the identified target genes (Bruno et al, 2008). However, the mechanisms regulating AATF-mediated transcriptional control remained largely unclear. We now provide a molecular rationale for the observations previously reported. We demonstrated that access of endogenous AATF to the genomic DNA is at least in part regulated through cytoplasmic sequestration of substantial pools of functionally inactive AATF in resting cells through a direct interaction with the cytoskeletal protein MRLC3. MK2-mediated AATF phosphorylation on Thr-366 acts as a molecular switch to disrupt cytoplasmic AATF:MRLC complexes. In response to different genotoxic agents, liberated AATF translocates to the nucleus, where it engages the promoters of the pro-apoptotic p53 target genes PUMA, BAX and BAK, repressing their expression. In addition, there was a consistent increase in the binding of AATF to the CDKN1A promoter as demonstrated previously (Bruno et al, 2006). However, in contrast to previous reports, we failed to demonstrate an appreciable increase in cell-cycle regulating p21 protein expression upon AATF knockdown. This might be the result of different doses of genotoxic agents or different time points of analysis.
AATF was previously shown to be stabilized through ATM/ATR- and Chk2-mediated phosphorylation (Bruno et al, 2006). We now report that AATF is also a substrate of the p38/MK2 stress kinase pathway. The p38/MK2 module has been shown to be activated after DNA damage (Kyriakis and Avruch, 2001) and may also involve the canonical DDR kinases ATM and ATR (Raman et al, 2007; Reinhardt et al, 2007). However, in response to UV irradiation, the p38/MK2 pathway is activated independently of ATM and ATR (Reinhardt et al, 2007). These observations suggest that there might be a certain stress specificity to MK2-mediated AATF phosphorylation with maximum AATF effects only when ATM/ATR, Chk2 and MK2 are simultaneously activated. Hyperosmotic stress as a p38/MK2-specific stimulus produced a robust nuclear translocation of AATF, indicating that p38/MK2 activity is sufficient for AATF re-localization. However, further experiments are required to test whether mere nuclear accumulation of AATF is sufficient for its apoptosis-repressing function.
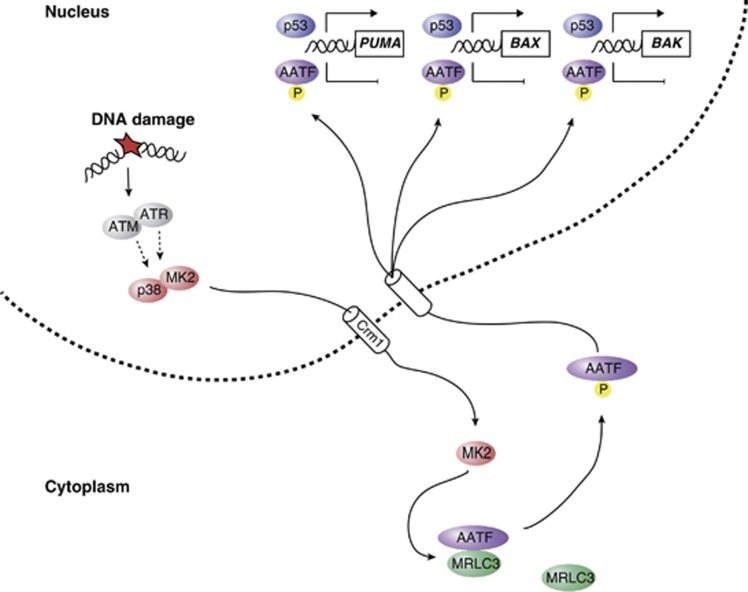
The data presented in this study provide a molecular mechanism that helps explain the regulation of the p53 response through dynamic spatial control of the transcriptional co-factor AATF. The proposed mechanism of AATF-mediated control of p53 signalling (Figure 8) is in line with a model in which the transcriptional activity of p53 is regulated by co-factors engaging a subset of p53-targeted promoters to selectively regulate the transcriptional activity of these promoters, thus shaping the functional outcome of p53 activation. We could show that p53 and AATF engage the PUMA, BAX, BAK, CDKN1A, GADD45α and RPRM promoters independent of each other. Thus, a model is emerging in which DNA damage-mediated nuclear AATF accumulation on the promoters of the pro-apoptotic genes PUMA, BAX and BAK represses the transactivation of these genes, while not restricting the expression of the cell-cycle-regulating genes CDKN1A, GADD45α and RPRM. The net result is a selective repression of p53-driven apoptosis in response to genotoxic stress. These observations are in good agreement with a previously described role for the p38/MK2 pathway in mediating a prolonged cell-cycle checkpoint arrest following DNA damage (Bulavin et al, 2001; Raman et al, 2007; Reinhardt et al, 2010).
Figure 8.
AATF acts as a phosphorylation-dependent molecular switch to dictate the outcome of the p53 response. A simplified model depicting details of the AATF-based molecular switch, controlling the cellular outcome of the p53 response.
However, p53 serves a role as a double-edged sword: Undoubtedly, p53 has enormous potential as a drug target, since p53-mediated transcription of pro-apoptotic genes, has been shown to effectively induce apoptosis (Miyashita et al, 1994; Selvakumaran et al, 1994; Zhan et al, 1994; Oda et al, 2000a; Nakano and Vousden, 2001). However, p53-dependent induction of CDKN1A, 14-3-3σ and other cell-cycle-regulating genes has a protective effect, through prevention of mitotic catastrophe due to premature checkpoint collapse in tumour cells experiencing chemotherapy-induced genotoxic stress (Asada et al, 1999; Abbas and Dutta, 2009; Jiang et al, 2009; Koster et al, 2010). Thus, it is desirable to identify pathways regulating the functional outcome of the p53 response. Here, we identify AATF as a critical modulator selectively repressing p53-driven apoptosis in a phospho-dependent manner by preventing the induction of the pro-apoptotic p53 target genes, PUMA, BAX and BAK while allowing unrestricted CDKN1A, GADD45α and RPRM expression. Thus, the p38/MK2/AATF signalling module emerges as a powerful apoptosis-antagonizing pathway specifically intercepting p53-driven apoptosis (Figure 8). Our data presented here provide a rationale for the development and clinical use of inhibitors of the p38/MK2/AATF pathway to harness the full apoptotic potential of p53, which is non-mutated in ∼50% of all human tumours (Jiang et al, 2009).
Materials and methods
Cell culture and chemicals
HEK293T cells were cultured in DMEM supplemented with 10% FBS and transfected with the calcium phosphate method, p53+/+ and p53−/− HCT116 cells were cultured in McCoy’s 5A plus 10% FBS and transfected with Genejuice (Novagen). WT and MK2/3 DKO MEFs, SW1573, NCI-H460, NCI-H23, NCI-H2122, U2OS cells were grown in DMEM plus 10% FBS. Okadaic acid, camptothecin and doxorubicin were purchased from Sigma. UV-C was applied using a Stratalinker. Hypertonic stress was applied through prewarmed Ringer solution containing 582 mosm/l sucrose 10 min prior to harvesting.
Antibodies and RNAi
Antibodies against FLAG (Sigma), V5 (Chemicon), cl. caspase-3 (BD Biosciences), pHH3 (Upstate), AATF (Sigma, Abnova and homemade), fibrillarin (Abcam), β-tubulin (Dev. St. Hybridoma Bank), Chk1, pSer-345 Chk1, MK2, pThr-334 MK2, Puma, Bax, p53, Gadd45α (Cell Signalling), MRLC (Santa Cruz and homemade), p21 (Santa Cruz), p53 (Dako) were used for immunoprecipitation, immunoblotting, immunofluorescence immunohistochemistry and FACS. AATF and luciferase shRNA sequences were previously described (Bruno et al, 2006; Reinhardt et al, 2007). MRLC shRNA targeting sequence was 5′-TAA CTT CAA ATT CCA GCC AAA-3′.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (BE2212, SFB-829 to TB, RE2246/1-1, RE2246/2-1, SFB-829, SFB-832 to HCR, SFB-566 to MG, SFB-832 to RKT and to BS, TH-1386/3-1 to RKT and MLS), the Volkswagenstifung (Lichtenberg Programme to HCR), the National Institutes of Health (GM68762, CA112967, ES015339 to MBY), the Deutsche Krebshilfe (107954 to RKT), the Ministry of Science, NRW (MIWT, 4000-12 09 to RKT and 313-005-0910-0102 to HCR), the BMBF (0313921 to BS and TB, 01GS08100 to RKT), The Max Planck Society (M.I.F.A.NEUR8061 to RKT), EU Framework Programme CURELUNG (HEALTH F2 2010 258677 to RKT), Stand Up To Cancer American Association for Cancer Research Innovative Research Grant (SU2C AACR IR60109 to RKT), by the Behrens-Weise Foundation (to RKT) and by an anonymous foundation to RKT and the Köln Fortune Programme (KH, SC). MLS is a fellow of the International Association For the Study of Lung Cancer (IASLC). HCT116 cells were from B Vogelstein. AATF shRNA constructs were from M Fanciulli. We thank H Zentgraf for support in generating monoclonal antibodies. We thank the members of our laboratories for helpful discussions.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbas T, Dutta A (2009) p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9: 400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S (1999) Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J 18: 1223–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno T, De Angelis R, De Nicola F, Barbato C, Di Padova M, Corbi N, Libri V, Benassi B, Mattei E, Chersi A, Soddu S, Floridi A, Passananti C, Fanciulli M (2002) Che-1 affects cell growth by interfering with the recruitment of HDAC1 by Rb. Cancer Cell 2: 387–399 [DOI] [PubMed] [Google Scholar]
- Bruno T, De Nicola F, Iezzi S, Lecis D, D'Angelo C, Di Padova M, Corbi N, Dimiziani L, Zannini L, Jekimovs C, Scarsella M, Porrello A, Chersi A, Crescenzi M, Leonetti C, Khanna KK, Soddu S, Floridi A, Passananti C, Delia D et al. (2006) Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell 10: 473–486 [DOI] [PubMed] [Google Scholar]
- Bruno T, Iezzi S, De Nicola F, Di Padova M, Desantis A, Scarsella M, Di Certo MG, Leonetti C, Floridi A, Passananti C, Fanciulli M (2008) Che-1 activates XIAP expression in response to DNA damage. Cell Death Differ 15: 515–520 [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Higashimoto Y, Popoff IJ, Gaarde WA, Basrur V, Potapova O, Appella E, Fornace AJ Jr. (2001) Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature 411: 102–107 [DOI] [PubMed] [Google Scholar]
- Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW (2007) Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 130: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Certo MG, Corbi N, Bruno T, Iezzi S, De Nicola F, Desantis A, Ciotti MT, Mattei E, Floridi A, Fanciulli M, Passananti C (2007) NRAGE associates with the anti-apoptotic factor Che-1 and regulates its degradation to induce cell death. J Cell Sci 120: Pt 111852–1858 [DOI] [PubMed] [Google Scholar]
- Elia AE, Cantley LC, Yaffe MB (2003) Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299: 1228–1231 [DOI] [PubMed] [Google Scholar]
- Engel K, Kotlyarov A, Gaestel M (1998) Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J 17: 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JM (2008) Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 27: 4013–4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT (2009) The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev 23: 1895–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster R, di Pietro A, Timmer-Bosscha H, Gibcus JH, van den Berg A, Suurmeijer AJ, Bischoff R, Gietema JA, de Jong S (2010) Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J Clin Invest 120: 3594–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakis JM, Avruch J (2001) Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev 81: 807–869 [DOI] [PubMed] [Google Scholar]
- Lavin MF, Gueven N (2006) The complexity of p53 stabilization and activation. Cell Death Differ 13: 941–950 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB (2005) MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell 17: 37–48 [DOI] [PubMed] [Google Scholar]
- Maris JM, Hogarty MD, Bagatell R, Cohn SL (2007) Neuroblastoma. Lancet 369: 2106–2120 [DOI] [PubMed] [Google Scholar]
- Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC (1994) Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 9: 1799–1805 [PubMed] [Google Scholar]
- Nakano K, Vousden KH (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694 [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB (2003) Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31: 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N (2000a) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288: 1053–1058 [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y (2000b) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Raman M, Earnest S, Zhang K, Zhao Y, Cobb MH (2007) TAO kinases mediate activation of p38 in response to DNA damage. EMBO J 26: 2005–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB (2007) p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11: 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MATM, Wang X, Linding R, Ong S-E, Weaver D, Carr SA, Yaffe MB (2010) DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol Cell 40: 34–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Schumacher B (2012) The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet 28: 128–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, Milarski K, Askew R, Marusic S, Lin LL, Gaestel M, Telliez JB (2007) The mitogen-activated protein kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol Cell Biol 27: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvakumaran M, Lin HK, Miyashita T, Wang HG, Krajewski S, Reed JC, Hoffman B, Liebermann D (1994) Immediate early up-regulation of bax expression by p53 but not TGF beta 1: a paradigm for distinct apoptotic pathways. Oncogene 9: 1791–1798 [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J (2002) Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Hosoya H, Yonemura S (2007) Regulation of myosin II dynamics by phosphorylation and dephosphorylation of its light chain in epithelial cells. Mol Biol Cell 18: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219 [DOI] [PubMed] [Google Scholar]
- Yang Y, Gourinath S, Kovacs M, Nyitray L, Reutzel R, Himmel DM, O’Neall-Hennessey E, Reshetnikova L, Szent-Gyorgyi AG, Brown JH, Cohen C (2007) Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure 15: 553–564 [DOI] [PubMed] [Google Scholar]
- Zhan Q, Fan S, Bae I, Guillouf C, Liebermann DA, O’Connor PM, Fornace AJ Jr. (1994) Induction of bax by genotoxic stress in human cells correlates with normal p53 status and apoptosis. Oncogene 9: 3743–3751 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.