Abstract
Human single-stranded DNA-binding protein 1 (hSSB1), encoded by OBFC2B, was recently characterized as an essential factor for the initiation of DNA damage checkpoints and the maintenance of genomic stability. Here, we report that loss of Obfc2b in mice results in perinatal lethality characterized by growth delay and skeletal abnormalities. These abnormalities are associated with accumulation of γH2ax, apoptosis and defective pre-cartilage condensation, which is essential for normal bone formation. However, deficiency of Obfc2b does not affect the initiation of DNA damage checkpoints, Atm activation, or the maintenance of genomic stability in B lymphocytes and primary fibroblasts. Loss of Obfc2b results in increased expression of its homologue Obfc2a (hSSB2). In contrast to Obfc2b deficiency, depletion of Obfc2a in fibroblasts results in impaired proliferation, accumulation of γH2ax and increased genomic instability. Thus, the hSSB1 orthologue Obfc2b has a unique function during embryogenesis limited to cell types that contribute to bone formation. While being dispensable in most other cell lineages, its absence leads to a compensatory increase in Obfc2a protein, a homologue required for the maintenance of genomic integrity.
Keywords: apoptosis, DNA damage response, hSSB1, Obfc2b, skeletogenesis
Introduction
Nuclear DNA is normally double-stranded, but single-stranded DNA (ssDNA) is exposed during DNA replication, meiosis, transcription, and DNA double-strand break (DSB) repair. ssDNA, which is an obligate intermediate in these reactions, is more vulnerable to chemical and physical damage than double-stranded DNA (dsDNA). The increased vulnerability of ssDNA is alleviated in part by ssDNA-binding proteins that stabilize, protect and facilitate the repair of damaged ssDNA (reviewed by Mendez and Stillman, 2003). Underlining their importance, loss of function of the ssDNA-binding protein replication protein A (Rpa1) results in embryonic lethality in mice (Wang et al, 2005). Even heterozygous Rpa1 mutant mice show an increase in genomic instability and develop lymphoid tumours (Wang et al, 2005).
Two additional ssDNA-binding proteins, hSSB1 (OBFC2B, NABP2 or SOSS-B1) and hSSB2 (OBFC2A, NABP1 or SOSS-B2), are also thought to be essential for recognition and repair of DNA damage (Richard et al, 2008, 2011a, 2011b; Huang et al, 2009; Li et al, 2009; Zhang et al, 2009). Similarly to RPA1, hSSB1 and hSSB2 form heterotrimeric complexes that are required for their recruitment to DSBs (Huang et al, 2009; Li et al, 2009; Skaar et al, 2009; Zhang et al, 2009). RNA interference (RNAi) experiments indicated that hSSB1 is essential to induce phosphorylation of ataxia telangiectasia mutated (ATM) kinase and its downstream targets in response to DNA damage. Moreover knockdown of hSSB1 is reported to abrogate irradiation-induced G1/S and G2/M cell-cycle arrest and result in genomic instability (Richard et al, 2008; Huang et al, 2009; Li et al, 2009; Zhang et al, 2009). In addition to repair and checkpoint functions, it has been proposed that hSSB1 is also required to produce ssDNA at sites of DSBs and that it does so by recruiting the MRN (MRE11/RAD50/NBS1) complex and the CtBP-interacting protein (CTIP) endonuclease (Richard et al, 2011a, 2011b). However, the role of hSSB1 in DNA repair has only been tested in RNAi knockdown experiments in cell lines.
To study the role of the ssDNA-binding protein hSSB1 in vivo, we produced conditional knockout mice for the hSSB1 orthologue Obfc2b. We find that Obfc2b exhibits an essential, unique and cell type-specific role during embryogenesis. Germline deletion of Obfc2b results in increased replication-associated DNA damage and apoptosis in cell types that are essential for skeletal development and, hence, in severe skeletal defects and perinatal lethality. Furthermore, loss of Obfc2b results in a compensatory increase of its homologue Obfc2a (orthologue to hSSB2). Unexpectedly, these ssDNA-binding proteins are not required to initiate the DNA damage response to irradiation, but play an important tissue-specific role in the suppression of replication-associated DNA damage.
Results
Germline deletion of Obfc2b results in embryonic lethality
Human ssDNA-binding protein 1 (hSSB1 or SOSS-B1) is encoded by the OBFC2B gene (oligonucleotide/oligosaccharide-binding fold containing 2B; Supplementary Figure 1A). To conditionally delete hSSB1, in mice, we introduced loxP sites flanking exons 1 and 2 of Obfc2b in mouse embryonic stem (ES) cells (Obfc2blox; Figure 1A and Supplementary Figure 1B). To generate mice carrying an Obfc2b knockout allele, Obfc2blox mice were bred with mice expressing the EIIACre transgene (Lakso et al, 1996; Supplementary Figure 1B and C). Cre-mediated loss of Obfc2b protein was confirmed by western blotting of B cells from CD19Cre; Obfc2blox/− mice (Supplementary Figure 1D and see below).
Figure 1.
Loss of Obfc2b results in embryonic lethality and developmental abnormalities. (A) Design of the conditional Obfc2b allele. Schematic of the murine Obfc2blox allele with integrated loxP sites before (upper panel), and after Cre-mediated disruption (lower panel) is shown. (B) Obfc2b deficiency results in embryonic lethality. Table of genotypes observed from Obfc2b+/− intercrosses are shown. (C) Obfc2b deficiency results in developmental abnormalities. Appearance of embryos at days E18.5 (left) and P0 (right) is shown. Bar marks size difference and arrows mark developmental abnormalities of the hindlimbs. (D) Obfc2b mRNA analysis in wild-type embryos at day E10.5 by in situ hybridization. Arrows indicate specific staining at the branchial arches (BAs), neural crest (NC), neural tube (NT), forelimbs (FL) and hindlimbs (HL) and somites (So) using the anti-sense probe. An Obfc2b sense probe was used as control (right).
To determine whether Obfc2b is essential for mouse development, Obfc2b+/− mice were interbred. Out of 129 pups analysed at 0–2 weeks of age, we found no viable Obfc2b−/− mice even on the day of delivery (P0) (Figure 1B). However, Obfc2b−/− embryos were present at nearly Mendelian ratios as late as at embryonic day 18.5 (E18.5, Figure 1B). At this time, the homozygous mutant embryos appeared to be viable but exhibited significant growth delay, rudimentary hindlimbs (HL) and an abnormal skull (Figure 1C). We conclude that loss of Obfc2b results in developmental abnormalities during embryogenesis and perinatal death.
To determine whether the developmental abnormalities in Obfc2b−/− mice result from a tissue-specific requirement of Obfc2b function during embryogenesis, we performed in situ hybridization for Obfc2b mRNA expression on wild-type E10.5 embryos. Obfc2b was expressed in several tissues that contribute to the development of skeletal structures (Figure 1D). These include the limb buds that organize the development of fore- and hindlimbs (FL, HL); the somites (So) which form in part the sclerotome and further the vertebrae and part of the skull; the branchial arches (BAs) that contribute to the development of the mandibles and the palate; and the prospective neural crest (NC) that can give rise to craniofacial mesenchyme and further form craniofacial cartilage and bones. In addition, Obfc2b mRNA expression seemed to be specific for the closing neural tube (NT) and different regions of the head (Figure 1D). We conclude that Obfc2b shows a tissue-specific expression pattern during normal embryogenesis.
Obfc2b−/− embryos exhibit severe skeletal defects
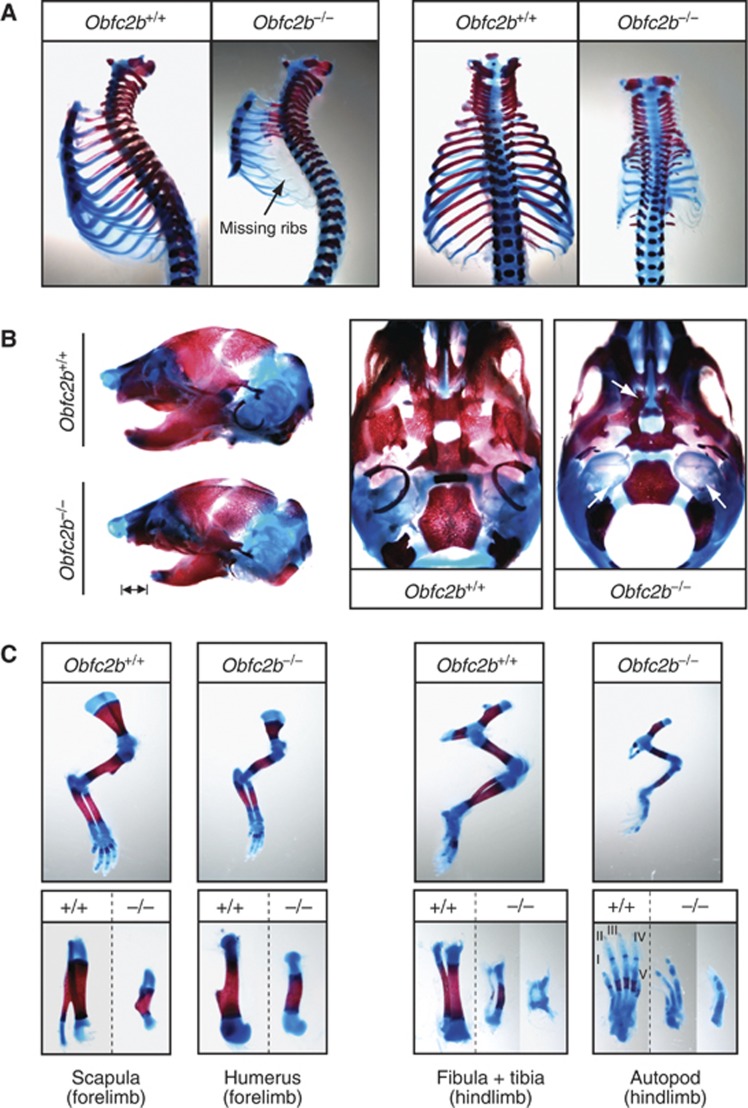
To characterize skeletal defects in more depth, we visualized cartilage and mineralized bone in E18.5 embryos (Figure 2). Obfc2b−/− embryos exhibited severe skeletal abnormalities affecting the rib cage, limbs and skull: the rib cage showed a general decrease in size and ossified rib segments were missing or rudimentary (Figure 2A). Since the lower rib cage serves as an anchor for the thoracic diaphragm, it is likely that the perinatal death of Obfc2b−/− embryos is caused by respiratory failure. Micro-computer tomography (MicroCT) analysis of dead Obfc2b−/− newborns (P0) confirmed the presence of a rudimentary rib cage and revealed that the bones were thinner and showed increased porosity (Supplementary Figure 2). Furthermore, the skull had a hypoplastic lower mandible (Figure 2B, left), the tympanic ring of the inner ear was rudimentary and the palate was cleft (Figure 2B, right). In the region of the forelimbs, the spine of the scapula and the deltoid tuberosity of the humerus were missing or rudimentary, respectively (Figure 2C, left). There was variable penetrance of skeletal abnormalities in the hindlimbs, which included missing digits (Figure 2C, right). MicroCT of the femurs revealed a significant decrease in bone volume (Supplementary Figure 2B–D). This pattern of skeletal defects is consistent with the pattern of Obfc2b expression in wild-type E10.5 embryos described above (Figure 1D). We conclude that Obfc2b deficiency results in multiple skeletal defects during embryogenesis.
Figure 2.
Skeletal abnormalities in Obfc2b−/− embryos. (A) Rib cage preparations of E18.5 embryos visualized by Alcian blue and Alizarin red staining indicating cartilage (blue) and mineralized/ossified tissue (red). (B) Skull preparations: bars mark size differences in the lower mandible (left) and arrows mark the rudimentary tympanic ring and the cleft palate in skulls from Obfc2b−/− embryos (middle and right). (C) Preparations of the forelimbs (left panels) and hindlimbs from wild-type and Obfc2b−/− embryos (right panels).
Osteoblasts, chondrocytes and osteoblasts in Obfc2b −/− mice
To determine whether skeletal defects in Obfc2b−/− mice arise as a consequence of aberrant differentiation or function of bone forming cells, we isolated osteoblasts, chondrocytes and osteoclasts from wild-type and Obfc2b−/− mice. Osteoblasts were isolated from the calvaria of the skull from E18.5 embryos; chondrocytes were isolated from the sternum of the ribcage. Osteoclasts were generated from bone marrow cells by stimulation with RANK-L and M-CSF for 5 days in culture and osteoclast identity was verified by Tartrate-resistant Acidic Phosphatase (TRAP) staining (Supplementary Figure 3A). Isolated cells were then subjected to gene expression analysis using gene arrays. Expression of osteoblast-, chondrocyte- and osteoclast-specific genes confirmed the identity of the isolated cell subsets (Supplementary Figure 3B). Comparison of gene arrays from wild-type and Obfc2b−/− cells showed that Obfc2b was expressed in wild-type but not in Obfc2b−/− cells. However, wild-type and Obfc2b−/− cells were otherwise indistinguishable (Supplementary Figure 3C). We conclude that deficiency of Obfc2b does not lead to significant changes in osteoblast, chondrocyte or osteoclast gene expression.
Skeletal defects are associated with increased apoptosis
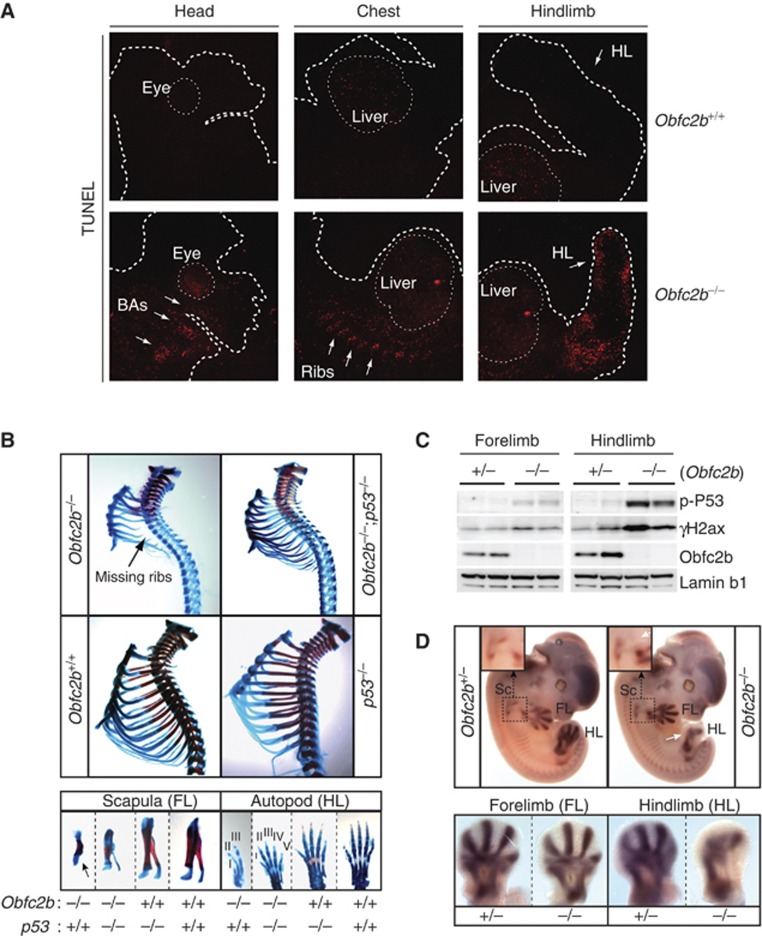
Since the human Obfc2b orthologue hSSB1 has been implicated in DNA repair and DNA repair deficiencies can result in apoptosis during embryogenesis (Gao et al, 1998), we asked whether the skeletal defects in Obfc2b−/− embryos were associated with increased apoptosis. To detect apoptotic cells, we performed terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) on tissue sections from E10.5, E12.5 and E16.5 embryos. Whereas E10.5 and E16.5 Obfc2b−/− embryos displayed no notable abnormalities (Supplementary Figure 4A and B), E12.5 Obfc2b−/− embryos showed significantly increased numbers of apoptotic cells in the developing ribs, hindlimb (HL) bud and branchial arches (BAs) (Figure 3A; Supplementary Figure 4C). This pattern of TUNEL staining is consistent with the majority of the described skeletal defects at E18.5 (see Figure 2).
Figure 3.
Increased skeletal apoptosis in Obfc2b−/− embryos. (A) Obfc2b deficiency is associated with increased apoptosis at E12.5. Terminal deoxynucleotidyl transferase dUTP nick end-labelling (TUNEL) staining of representative tissue sections of E12.5 embryos. Arrows indicate regions of developing branchial arches (BAs), developing ribs and developing hindlimbs (HL). (B) P53 loss partially rescues skeletal abnormalities in Obfc2b−/− embryos. Alcian blue and Alizarin red stained skeletal preparations from P0 Obfc2b−/−;p53+/+, Obfc2b−/−;p53−/−, Obfc2b+/+;p53−/− and Obfc2b+/+;p53+/+ embryos as in Figure 2. Preparations of the rib cage (upper panels), and of the scapula of the forelimb (lower left) and the autopod of the hindlimb (lower right) are shown. (C) Western blot analysis of hind- and forelimbs from E12.5 embryos for phospho-p53 (serine 15), γH2ax, Obfc2b and Lamin b1. (D) In situ hybridization of whole embryos (E12.5) for Sox9 mRNA. Upper panels: images of whole embryos are shown including a magnification of the developing scapula. White arrows indicate aberrant Sox9 staining at the scapula and at the hindlimb. Lower panels: Sox9 staining of forelimbs (left) and hindlimbs (right).
To determine whether the skeletal defects in Obfc2b−/− embryos can be rescued by loss of p53, we interbred the two mutant mouse strains to generate Obfc2b−/−; p53−/− mice. In contrast to Obfc2b−/− newborns that were grossly abnormal and not viable, Obfc2b−/−; p53−/− mice appeared normal, were viable at birth, and survived up to 24 h. Skeletal preparations from Obfc2b−/−; p53−/− mice at P0 showed partial rescue of the thoracic rib cage phenotype (Figure 3B, up), and of the forelimb and hindlimb defects (Figure 3B, low). However, the defects in the skull including the cleft palate were not ameliorated and likely account for the perinatal lethality of the Obfc2b/p53 double knockout pups due to the inability to feed. We conclude that most skeletal defects in Obfc2b−/− embryos are associated with increased apoptosis and, accordingly, can be partially rescued by loss of p53.
Skeletal defects are associated with increased genomic instability at E12.5
To investigate whether increased apoptosis in Obfc2b−/− embryos results from increased genomic instability, we analysed forelimbs and hindlimbs from E12.5 embryos for γH2ax accumulation and p53 phosphorylation by western blotting because both are induced by and serve as markers for DNA damage (Figure 3C). We found that hindlimbs from Obfc2b−/− embryos showed increased γH2ax accumulation and p53 phosphorylation at serine 15 (Figure 3C). In contrast, there was only a minimal increase of γH2ax accumulation and p53 phosphorylation in the forelimbs of Obfc2b−/− embryos, which show only minor skeletal defects. We conclude that the skeletal defects in the limbs of Obfc2b−/− embryos are associated with and likely to result from increased genomic instability and apoptosis at E12.5.
Defective pre-cartilage condensation at E12.5 in Obfc2b −/− embryos
Pre-cartilage mesenchymal condensation precedes the development of chondrocytes and osteoblasts and is essential for the development of skeletal structures (reviewed in Hall and Miyake, 2000 and Kronenberg, 2003). Apoptosis is likely to antagonize pre-cartilage condensation, because defects in skeletogenesis are often associated with increased apoptosis (Akiyama et al, 2002; Cheung et al, 2005; Shim et al, 2010). Conversely, the transcription factor Sox9, which is specifically expressed during condensation in forelimbs and hindlimbs (Wright et al, 1995), is thought to suppress apoptosis (Akiyama et al, 2002). To analyse if increased apoptosis in Obfc2b−/− embryos is associated with defective pre-cartilage condensation we performed in situ hybridization for Sox9 on whole embryos at E12.5 (Figure 3D). In agreement with the presence of a minimal skeletal defect in the forelimb of Obfc2b−/− embryos, pre-cartilage condensation appeared to be mostly normal in the developing forelimbs of Obfc2b−/− embryos (Figure 3D, top and lower left). In contrast, the hindlimbs, which show a high degree of apoptosis and skeletal defects, showed defective pre-cartilage condensation, as did the developing scapula in the forelimb (Figure 3D, up and lower right). We conclude that increased genomic instability and apoptosis in Obfc2b−/− embryos is associated with defective pre-cartilage mesenchymal condensation.
Obfc2b is dispensable for the DNA damage response in B lymphocytes and MEFs
Lymphocytes are especially sensitive to defects in the DNA damage response because they undergo programmed DNA damage during V(D)J recombination, and immunoglobulin class-switch recombination (CSR in B cells). As a result, lymphocytes development and function is abnormal in mice or humans that are mutant in any of a number of different factors that contribute to the recognition and repair of DNA damage (reviewed in Rooney et al, 2004; Dudley et al, 2005 and Jankovic et al, 2007). The Obfc2b orthologue hSSB1 has been postulated to be required for the recognition of DNA damage and its repair by the homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways (Richard et al, 2008, 2011a, 2011b). To circumvent embryonic lethality and analyse Obfc2b function in B cells, we deleted it specifically by combining the conditional allele with a lineage-specific Cre transgene (CD19Cre;Obfc2blox/− mice). Despite undetectable levels of Obfc2b protein in CD19Cre;Obfc2blox/− B lymphocytes, B cell development in the bone marrow was indistinguishable from control mice (Figure 4A). Likewise, T cell development was indistinguishable in lethally irradiated mice reconstituted with fetal liver cells from wild-type or Obfc2b−/− embryos (Figure 4B). Moreover, immunoglobulin CSR in stimulated B cells was unaffected by loss of Obfc2b (Figure 4C). Since CSR and V(D)J recombination require efficient recognition of DNA damage and its repair by the NHEJ pathway we conclude that Obfc2b is not required for DNA damage sensing or its repair by NHEJ in lymphocytes.
Figure 4.
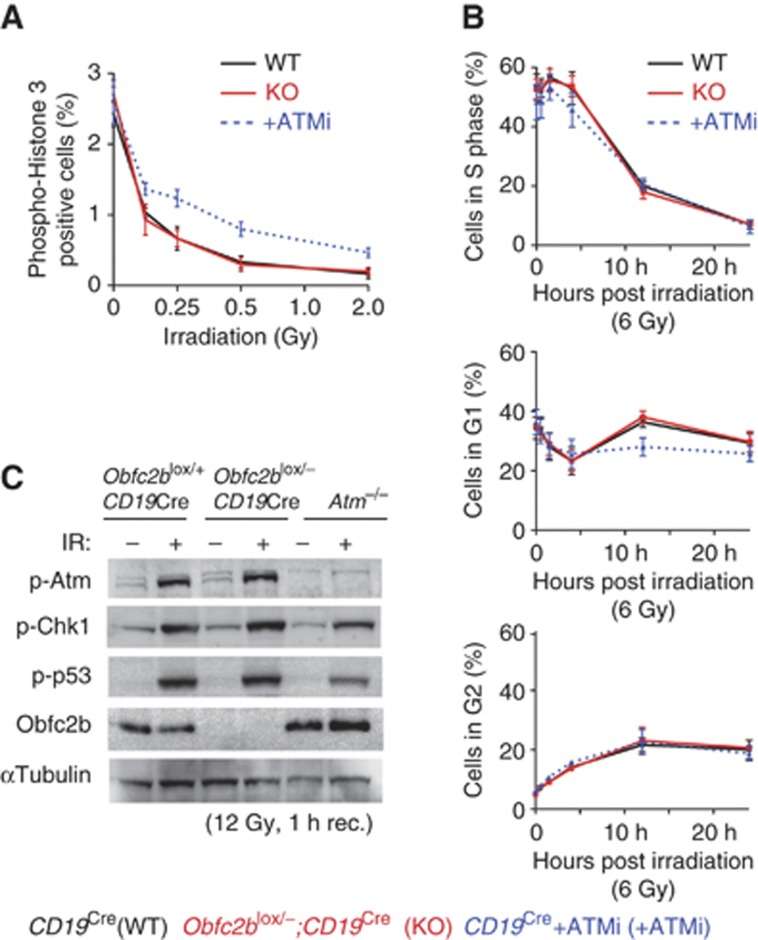
DNA damage response in Obfc2b−/− B cells. (A, B) Normal B and T cell development in Obfc2b−/− mice. (A) Flow cytometry analysis of control (CD19Cre;Obfc2b+/+) and conditional Obfc2b knockout (CD19Cre;Obfc2blox/−) mice. B cell subsets in the bone marrow were identified using the antibodies indicated. (B) Normal T cell development in Obfc2b−/− mice. Thymocytes from lethally irradiated mice reconstituted with fetal liver cells from Obfc2b+/+ or Obfc2b−/− embryos. Analysis was performed >2 months after reconstitution. (C) Normal class switch-recombination (CSR) in Obfc2b−/− B cells. Bar diagram shows mean values of IgG surface expression on conditional Obfc2b knockout (CD19Cre;Obfc2blox/−) and control B cells (CD19Cre/+;Obfc2b+/+ and CD19Cre;Obfc2blox/+) after 4 days of proliferation in vitro, as determined by flow cytometry. IgG1 expression was induced by LPS, IL-4 and RP105 (*) or by LPS and IL-4, IgG3 expression was induced by LPS alone. (D) Normal PARPi sensitivity in Obfc2b−/− B cells. B cells from Obfc2b wild-type mice (CD19Cre;Obfc2b+/+), conditional Obfc2b heterozygous mice (CD19Cre;Obfc2blox/+), conditional Obfc2b knockout mice (CD19Cre;Obfc2blox/−), Atm−/− mice or wild-type B cells treated with 2.5 μM Ku55933 (ATMi) were stained with carboxyfluorescein succinimidyl ester (CFSE) and analysed by flow cytometry after 4 days in culture with LPS and IL-4. Cells were cultured with or without 1 μM Ku58948 (PARPi). Bar diagram shows mean values of five individual experiments. Data observed for Atm−/− B cells and wild-type B cells treated with ATMi have been pooled. For gating of CFSE-positive cells, see Supplementary Figure 5. (E) Normal radiosensivity in Obfc2b−/− B cells. Metaphase analysis of proliferating B cells from control (CD19Cre;Obfc2b+/+ and CD19Cre;Obfc2blox/+) and conditional Obfc2b knockout mice (CD19Cre;Obfc2blox/−) after irradiation with 6 Gy (20 h recovery). Bar diagram shows the mean values of eight pairs of mice analysed in five individual experiments. (F) Obfc2b−/− B cells exhibit a slight increase of c-myc/Igh translocations. PCR analysis of c-myc/Igh translocations in B cells from control (CD19Cre;Obfc2b+/+ and CD19Cre;Obfc2blox/+) and conditional Obfc2b knockout mice (CD19Cre;Obfc2blox/−). B cells were cultured for 4 days with LPS and IL-4 previous to analysis. Bar diagram shows the mean of three and four independent experiments for derivative chromosomes 15 and 12, respectively. Data for both derivative chromosomes has been pooled.
Poly (ADP-ribose) polymerase (PARP) is required for the detection of single-strand DNA breaks. Inhibiting this enzyme with Ku58948 (PARPi) destabilizes the genome by increasing the number of ssDNA breaks, many of which develop into DSBs that are repaired by HR (Bryant et al, 2005; Jackson and Bartek, 2009). Cells defective in HR as well as cells treated with the ATM inhibitor Ku55933 (ATMi) are especially sensitive to PARPi treatment and show reduced proliferation in the presence of PARPi (Bunting et al, 2010). To determine whether Obfc2b is required for HR, we measured B cell proliferation in the presence or absence of PARPi (Figure 4D). Proliferation was measured by cell division-associated decrease of cytoplasmic staining with carboxyfluorescein succinimidyl ester (CFSE). Cells delayed in proliferation remain CFSE positive after 4 days. Atm−/− B cells or wild-type B cells treated with the ATM inhibitor Ku55933 (ATMi) were used as positive controls, and showed a significant increase of cells with delayed proliferation upon PARPi treatment (CFSE-positive cells; Figure 4D and Supplementary Figure 5). However, there was no measurable effect upon loss of Obfc2b. We conclude that Obfc2b is not required for DNA repair by HR in proliferating B lymphocytes.
To further test if Obfc2b is required for the recognition and repair of DNA damage, we irradiated proliferating B cells and analysed metaphases for unrepaired genomic aberrations by fluorescence in situ hybridization (FISH). We found only a small and statistically insignificant increase in genomic aberrations in metaphases from activated Obfc2b−/− B cells after ionizing irradiation (IR; Figure 4E). We next analysed the frequency of c-myc/Igh translocations in proliferating B cells, which are a byproduct of CSR. Defects in recognition and repair of DNA damage typically result in a significant increase in such translocations (Ramiro et al, 2006). The frequency of c-myc/Igh translocations in proliferating Obfc2b deficient B cells was slightly elevated compared to Obfc2b proficient cells (Figure 4F). Nevertheless, the translocation frequencies observed fall within the range of what is typically reported for wild-type B cells (Ramiro et al, 2004, 2006). We conclude that the hSSB1 orthologue Obfc2b is not required to maintain genomic stability in dividing B lymphocytes.
It has been suggested that hSSB1 is required for activation of the G1/S and the G2/M DNA damage checkpoint upon irradiation (Richard et al, 2008). To determine whether the hSSB1 orthologue Obfc2b is required for the initiation of DNA damage checkpoints in vivo, we analysed B cells from CD19Cre;Obfc2blox/− mice for cell-cycle arrest in response to irradiation. Cells entering mitosis undergo histone 3 phosphorylation at serine 10. Accordingly, histone 3 phosphorylation is abrogated in response to G2/M checkpoint activation. As expected, inhibition of Atm kinase activity using Ku55933 (ATMi) resulted in a reduced activation of the G2/M checkpoint in response to irradiation (Fernandez-Capetillo et al, 2002; Figure 5A). However, there was no measurable effect of Obfc2b ablation compared to wild-type cells (Figure 5A; Supplementary Figure 6A). Similarly, loss of Obfc2b did not affect the G1/S checkpoint as measured by 5-bromo-2′-deoxyuridine (BrdU) incorporation (Figure 5B; Supplementary Figure 6B): Upon IR, Obfc2b-deficient cells (KO) were indistinguishable from wild-type controls in terms of cell-cycle distribution (Figure 5B). Consistent with the absence of cell-cycle checkpoint defects in response to IR, Western blot analysis showed no alterations in IR-induced phosphorylation of Atm, Chk1 (a substrate of Atr) or p53 in the absence of Obfc2b (Figure 5C; Supplementary Figure 6C). Similar results were also obtained with Obfc2b−/− primary murine embryonic fibroblasts (MEFs; Supplementary Figure 6D and E). Thus, Obfc2b is dispensable for Atm/Atr activation and the initiation of DNA damage checkpoints in primary B lymphocytes and embryonic fibroblasts.
Figure 5.
DNA damage checkpoint analysis. (A) Normal G2/M checkpoint in Obfc2b−/− B cells. Proliferating B cells from wild-type (WT, CD19Cre;Obfc2b+/+) and conditional knockout (KO, CD19Cre;Obfc2blox/−) mice were analysed for histone 3 (serine 10) phosphorylation before and after irradiation (1 h recovery). As a control, wild-type (CD19Cre/+;Obfc2b+/+) B cells treated with 2.5 μM Ku55933 (ATMi) were analysed. The graph represents the results from three pairs of mice and two independent experiments. (B) Normal G1/S checkpoint in Obfc2b−/− B cells. Proliferating B cells from wild-type (WT, CD19Cre;Obfc2b+/+) and conditional knockout (KO, CD19Cre;Obfc2blox/−) mice were pulsed with BrdU and subjected to cell-cycle analysis. Cells were either mock treated or irradiated with 6 Gy, and allowed to recover for 0.5–24 h. Cells in S phase, G1 and G2 are plotted as individual graphs. Graphs represent the results from three pairs of mice and two independent experiments. (C) Normal IR-induced phosphorylations in Obfc2b−/− B cells. Western blot for phosphorylated Atm (serine 1981), Chk1 (serine 317), p53 (serine 15), and for Obfc2b and αTubulin on lysates from proliferating B cells from CD19Cre;Obfc2blox/+, CD19Cre;Obfc2blox/− and Atm−/− mice. Cells were mock treated or irradiated (12 Gy, 1 h recovery).
Increased Obfc2a expression in Obfc2b-deficient cells
Obfc2b (orthologue to hSSB1) is homologous with Obfc2a (orthologue to hSSB2) and previous studies have suggested that the two may have overlapping functions (Huang et al, 2009; Li et al, 2009). To determine whether Obfc2a might compensate for Obfc2b loss, we analysed Obfc2a protein levels in tissues of Obfc2b−/− embryos (Figure 6A). Western blotting showed that Obfc2b deficiency results in increased Obfc2a protein in all tissues tested (Figure 6A and B). Similar results were obtained by deleting Obfc2b in Obfc2blox/− primary MEFs carrying a Tamoxifen (TAM) inducible Cre transgene (Obfc2blox/−; Cre-ERT2, Figure 6C). In contrast, RNAi-mediated depletion of Obfc2a in primary MEFs resulted in only small changes in Obfc2b levels (Figure 6C). We conclude that Obfc2a protein levels are increased in cells deficient for Obfc2b.
Figure 6.
Loss of Obfc2b increases expression of Obfc2a. (A) Increased Obfc2a protein levels in Obfc2b−/− embryos. Western blot for Obfc2a, Obfc2b and Lamin b1 on tissues isolated from E18.5 embryos genotyped as Obfc2b−/−, Obfc2b+/− and Obfc2b+/+. (B) Bar diagram shows relative protein levels of Obfc2b and Obfc2a detected in five different tissues from wild-type, Obfc2b+/− and Obfc2b−/− mice measured by western blot normalized to Lamin b1. (C) Increased Obfc2a protein levels in Obfc2b-deficient MEFs. Bar diagram shows the relative protein levels of Obfc2b and Obfc2a in primary Obfc2blox/−;Cre-ERT2 MEFs treated with 5 μM Tamoxifen (TAM) for 48 h or infected with Obfc2a-shRNA expressing lentivirus.
Obfc2a is essential for genomic integrity in proliferating MEFs
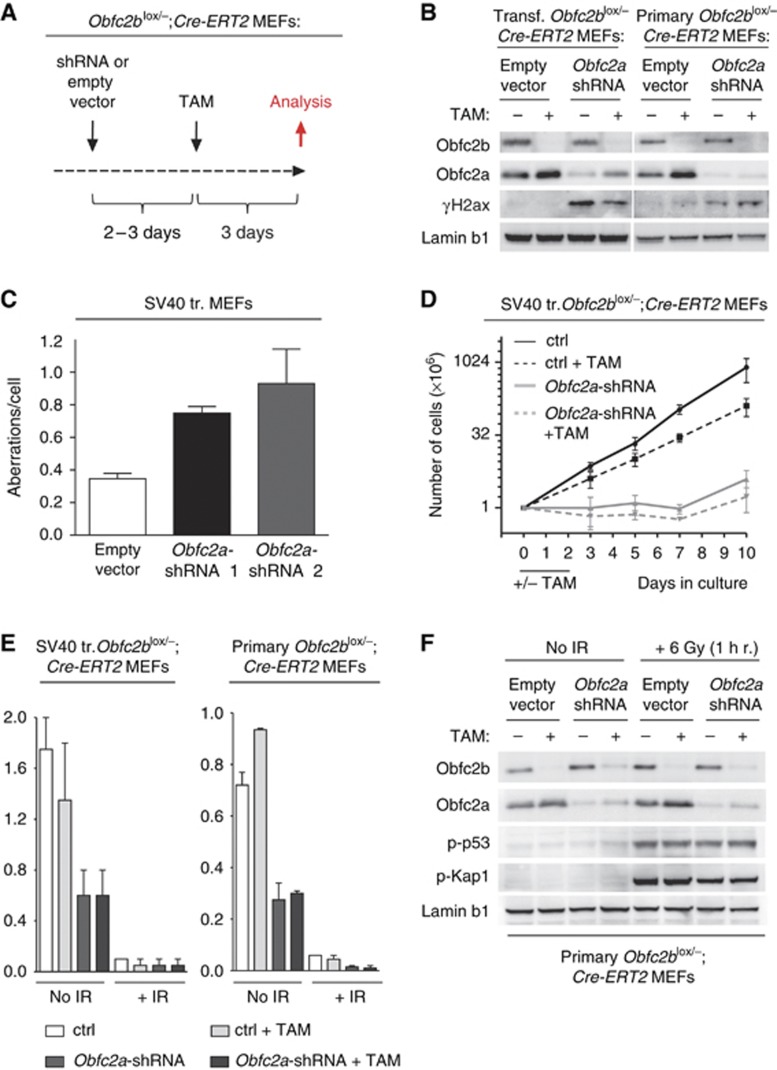
Obfc2b deficiency does not significantly affect the recognition or repair of DNA damage in B cells, or the initiation of DNA damage checkpoints in B cells and MEFs (see Figures 4 and 5). To determine whether the compensatory increase in expression of Obfc2a protein (see Figure 6) masks a defect in these processes, we knocked down Obfc2a by RNAi in Obfc2b-deficient MEFs. To circumvent elevated levels of Obfc2a protein in Obfc2b-deficient cells, we first knocked down Obfc2a by RNAi in Obfc2blox/−; Cre-ERT2 MEFs and subsequently deleted Obfc2b using Tamoxifen (Figure 7A).
Figure 7.
Combined loss of Obfc2b and Obfc2a. (A) Schematic diagram of the sequential knockdown of Obfc2b and Obfc2a in MEFs: Obfc2blox/−;Cre-ERT2 MEFs were double infected with lentivirus harbouring either empty vector (control) or Obfc2a-shRNA expression vectors. Two to three days after first infection, cells were treated with 5 μM Tamoxifen (TAM) for 48 h. Analysis was performed 5–6 days after the first infection. (B) Obfc2a depletion in MEFs results in γH2ax accumulation. Western blot of SV40-transformed or primary Obfc2blox/−;Cre-ERT2 MEFs treated as indicated for Obfc2b, Obfc2a, γH2ax and Lamin b1. (C) Obfc2a depletion in MEFs results in increased genomic instability. Analysis of metaphase spreads from SV40-transformed MEFs for genomic aberrations 3 days post infection with Obfc2a-shRNAs or control vector. 30–50 metaphases have been analysed for each group of infection. The graph represents two independent experiments. (D) Obfc2a depletion in MEFs impairs proliferation. Proliferation of SV40-transformed Obfc2blox/−;Cre-ERT2 MEFs treated as indicated and as described in (A). Diagram represents the mean of two independent experiments. Cell numbers were determined by cell counting. (E) Normal G2/M checkpoint in Obfc2a and Obfc2a/Obfc2b double deficient MEFs. Bar diagrams show the summary of experiments analysing histone 3 phosphorylation in Obfc2blox/−;Cre-ERT2 MEFs by flow cytometry. MEFs were treated as in described in (A). Cells were irradiated with 6 Gy and analysed 1 h post irradiation. Obfc2b and Obfc2a knockdown was verified by western blot in each experiment. (F) Normal IR-induced phosphorylations in Obfc2a and Obfc2a/Obfc2b double deficient MEFs. Western blot of primary Obfc2blox/−;Cre-ERT2 MEFs after sequential knockdown of Obfc2a and Obfc2b as described in (A) for Obfc2b, Obfc2a, phosphorylated p53 (serine 15), phosphorylated Kap1 (serine 824) and Lamin b1. Cells were mock treated or irradiated as indicated.
Knockdown of Obfc2a in primary and SV40 transformed MEFs resulted in increased accumulation of γH2ax as shown by western blotting (Figure 7B; Supplementary Figure 7A) and in an increase in genomic aberrations (Figure 7C). Further, Obfc2a-shRNA expressing MEFs failed to expand in culture (Figure 7D). In contrast, Tamoxifen induced deletion of Obfc2b did not result in γH2ax accumulation (Figure 7B; Supplementary Figure 7A), and had only minor effects on proliferation (Figure 7D). Combined depletion of Obfc2b and Obfc2a did not exacerbate the effects of Obfc2a depletion significantly (Figure 7B and D; Supplementary Figure 7A). We conclude that Obfc2a is essential for normal replication and maintenance of genomic stability.
Obfc2a is dispensable for the DNA damage response to irradiation in MEFs
To examine the effect of Obfc2a deficiency and Obfc2a/Obfc2b double deficiency on the induction of the DNA damage response upon irradiation (IR), we measured IR-induced abrogation of histone 3 phosphorylation by flow cytometry (Figure 7E; Supplementary Figure 7B and C) and IR-induced phosphorylation of p53, Chk1 and Kap1 by western blot (Figure 7F; Supplementary Figure 7A). In contrast to deletion of Obfc2b, depletion of Obfc2a resulted in a strong decrease in histone 3 phosphorylation in unirradiated cells (Figure 7E; Supplementary Figure 7B), which was not exacerbated in double deficient cells. This result is in agreement with the proliferation defect observed for Obfc2a-depleted cells. Following irradiation, Obfc2a-depleted cells showed a similar reduction in phosphorylated histone 3 as Obfc2b-deficient or control cells (Figure 7E; Supplementary Figure 7B and C), and therefore the irradiation-induced G2/M checkpoint appears to be normal in the absence of Obfc2a. Even Obfc2b/Obfc2a double deficient cells efficiently abrogated histone 3 phosphorylation in response to IR. Analysis of irradiation-induced p53, Chk1 and Kap1 phosphorylation by western blot further indicated a normal DNA damage response in the absence of Obfc2a or both, Obfc2a and Obfc2b (Figure 7F; Supplementary Figure 7A). Lower levels of Chk1 phosphorylation in irradiated Obfc2a-depleted cells (Supplementary Figure 7A) correspond to reduced proliferation at the day of analysis (day 5 post infection). This effect is absent at earlier time points after Obfc2a depletion when cells are still proliferating (Supplementary Figure 7D). We conclude that even the combined loss of Obfc2b and Obfc2a does not abrogate the initiation of the DNA damage response in response to irradiation.
Discussion
Previous reports suggested a fundamental role for hSSB1, the protein product of OBFC2B, in the recognition and repair of DNA damage (Richard et al, 2008, 2011a, 2011b; Huang et al, 2009; Li et al, 2009; Skaar et al, 2009; Zhang et al, 2009; Xu et al, 2011). Based on RNAi knockdown experiments in human tumour cell lines and neonatal foreskin fibroblasts, hSSB1 was proposed to regulate DNA damage mediated cell-cycle checkpoints and radiosensitivity by binding to DNA breaks, recruiting MRN and activating ATM.
In experiments with mice that carry a null mutation in Obfc2b we find no evidence to support the idea that this gene is required for the DNA damage response to irradiation, the activation of Atm or the maintenance of genomic integrity in primary B cells or MEFs. Further, the deficiency of its homologue Obfc2a (orthologue to hSSB2) or combined deficiency of Obfc2b and Obfc2a does not affect the DNA damage response to irradiation.
However, Obfc2b seems to play an essential role during embryogenesis. Obfc2b−/− mice exhibit growth delay and severe skeletal defects in the skull, limbs and the rib cage. The latter is likely to result in respiratory failure and cause perinatal death of Obfc2b−/− embryos. These abnormalities correlate with specific expression of Obfc2b in tissues that contribute to bone formation in the embryo and are associated with increased apoptosis at E12.5 in the absence of Obfc2b. Apoptosis is in turn associated with accumulation of γH2ax, suggesting that DNA damage triggers the apoptosis. Consistent with apoptotic cell death as a mechanism for the skeletal abnormalities, we find that many of these defects can be partially rescued by additional loss of p53.
Persistence of some developmental defects in Obfc2b−/−;p53−/− embryos further suggests that Obfc2b deficiency can also interfere with normal cellular function beyond the induction of p53-mediated apoptosis during the development of skeletal structures. Since we show that the Obfc2b homologue Obfc2a is essential for proliferation in MEFs and Obfc2b and Obfc2a appear to have overlapping functions, it is possible that skeletal defects that are not mediated by p53 arise as a consequence of an altered proliferation capacity in cells that specifically require Obfc2b.
Increased DNA damage and apoptosis in Obfc2b−/− embryos is associated with defective pre-cartilage mesenchymal condensation. Apoptosis has been shown to occur before the onset of mesenchymal condensation in many mouse models with skeletal defects (Akiyama et al, 2002; Guha et al, 2002; Cheung et al, 2005; Li et al, 2005; Shim et al, 2010), and it is likely to prevent condensation. Conversely, expression of the transcription factor Sox9 has been suggested to favour bone formation by suppressing apoptosis during mesenchymal condensation (Akiyama et al, 2002). We propose that Obfc2b supports the development of normal skeletal structures by preventing apoptosis arising from replication-associated DNA damage during embryogenesis (Figure 8). Obfc2b may do so by protecting ssDNA during replication, which is particularly prone to DNA damage.
Figure 8.
Obfc2b supports normal skeletogenesis by suppressing replication-associated DNA damage. Model of Obfc2b function during embryogenesis. During the development of pre-cartilage mesenchymal condensation at E12.5 the single-stranded DNA-binding protein Obfc2b prevents replication-associated DNA damage. In the absence of Obfc2b such DNA damage results in increased apoptosis, which interferes with pre-cartilage condensations. As a consequence, Obfc2b−/− embryos develop skeletal defects.
Skeletal defects that arise as a consequence of increased genomic instability are not uncommon among human genetic disorders. For example, the Cockayne Syndrome (CS) results from defects in transcription-coupled repair during repair of DNA damage by the nucleotide-excision repair pathway (Venema et al, 1990; de Boer and Hoeijmakers, 2000). Besides increased genomic instability, patients suffering from CS exhibit defects in their extremities, the spine and the skull (de Boer and Hoeijmakers, 2000). Similarly, patients suffering from Fanconi Anaemia (FA) or Seckel Syndrome exhibit increased genomic instability and skeletal abnormalities (McKusick et al, 1967; Chaganti and Houldsworth, 1991) and reviewed in Kerzendorfer and O’Driscoll (2009). In mice, only the Seckel Syndrome-related mutation leads to skeletal abnormalities (Murga et al, 2009) while CS and FA mutations do not. Seckel mice exhibit improper alternative splicing of the Atr gene and, as a consequence, Seckel mice express less Atr protein, leading to increased replicative stress and increased apoptosis (Murga et al, 2009). Similarly to the human disorder, Seckel mice exhibit skeletal defects in the skull. Together, the Seckel and Obfc2b−/− mouse models suggest that replication-associated DNA damage and apoptosis are a significant cause for skeletal malformations.
The best-characterized ssDNA-binding protein in eukaryotes is replication protein A (RPA). RPA is primarily recruited to ssDNA lesions that are exposed in collapsed replication forks or resected dsDNA breaks during the S phase of the cell-cycle. RPA recruits ATR and the HR machinery to these lesions (Zou and Elledge, 2003). As might be expected, Rpa1 mutation in mice predominantly affects proliferation and the maintenance of genome integrity (Wang et al, 2005). In contrast, we show that the ssDNA binding protein Obfc2b is not required for proliferation of B lymphocytes or MEFs. However, this analysis is complicated by the finding that Obfc2b (hSSB1) depletion leads to a compensatory increase in protein of its homologue Obfc2a (hSSB2), which forms similar heterotrimeric ssDNA-binding complexes as Obfc2b (Huang et al, 2009; Li et al, 2009; Skaar et al, 2009; Zhang et al, 2009). Furthermore, depletion of Obfc2a resulted in a severe impairment in proliferation, γH2ax accumulation and an increase in genomic aberrations. Therefore, Obfc2b/Obfc2a proteins (hSSB1/hSSB2) may share common functional characteristics with Rpa1 including a requirement for these factors in cell division. However, their requirement appears to be cell type specific.
In agreement with previous studies we find that deficiency of Obfc2b (hSSB1) or Obfc2a (hSSB2) results in genomic instability and in a reduction in phosphorylated histone 3 (Richard et al, 2008; Zhang et al, 2009). Moreover deletion of Obfc2b results in a compensatory increase in Obfc2a protein (Huang et al, 2009). However, hSSB1 has also been reported to control the G1/S and G2/M DNA damage checkpoints, ATM activation, and the repair of DNA damage by NHEJ and HR (Richard et al, 2008; Huang et al, 2009). None of these is altered by Obfc2b deletion in mice, or by Obfc2a knockdown in our experiments. However, our work differs from previous experiments in that the latter were performed using continuously growing cell lines, which may have acquired defective DNA damage responses while undergoing transformation (e.g., Broceno et al, 2002). We speculate that such additional abnormalities made the cultured cells more prone to show defects in cell-cycle checkpoints or the DNA damage response after depletion of hSSB1.
In summary, we report an essential, unique and cell type-specific role of the hSSB1 orthologue Obfc2b during embryogenesis. Loss of Obfc2b results in apoptosis of cell lineages that are essential for skeletal development and depend on Obfc2b. Moreover, Obfc2b and its homologue Obfc2a (orthologue to hSSB2) appear to be co-regulated in that loss of Obfc2b leads to a compensatory increase in Obfc2a protein. These ssDNA-binding proteins are not required to initiate the DNA damage response to irradiation, but together play an important role in normal cellular proliferation and the suppression of replication-associated DNA damage. Proliferation defects and replication-associated DNA damage are likely to result in apoptosis in rapidly dividing cells as observed during early embryogenesis in select domains of Obfc2b−/− embryos.
Materials and methods
Mice
Obfc2blox/+ mice were produced by HR in C57BL/6 albino ES cells. Details on the targeting vector, screening by Southern blot, and genotyping PCR are provided in the legends to the Supplementary Figures. FLPe (a gift by Dr Susan Dymecki; Rodriguez et al, 2000), EIIACre (Lakso et al, 1996), CD19Cre (Rickert et al, 1997), Cre-ERT2 (de Luca et al, 2005), Atm−/− (Barlow et al, 1996), and p53−/− mice (Jacks et al, 1994) were used for breeding with Obfc2bNEO-lox/+, Obfc2blox/+ or Obfc2b+/− mice as indicated. All experiments were in agreement with protocols approved by the Rockefeller University and National Institutes of Health (NIH) Institutional Animal Care and Use Committee.
Fetal liver transfer
Fetal liver transfers were performed with SJL mice as hosts (The Jackson Laboratory) as described in Horwitz et al (1997), with the exception that mice were irradiated with a dose of 2 × 500 rad with a 3-h recovery between each irradiation and before injection.
MEF culture and shRNA infection
Primary MEFs were isolated from E12.5 embryos and frozen as aliquots at passage 2 (1 week). Immortalized MEFs were generated by infection with SV40 large T retrovirus at 2 weeks of cell culture. Viral supernatant was generated as described in Robbiani et al (2008). Lentiviral supernatant was produced by transfection of 293T cells with Δ8.9 and VSV encoding helper plasmids (Naldini et al, 1996) and either empty vector (pLKO-IRES-GFP; Open Biosystems) or Obfc2a-shRNA encoding expression vectors (TRCN0000177652 [sh1], TRCN0000178020 [sh2]; Open Biosystems). 4-Hydroxytamoxifen (TAM) was from Sigma.
Tissue sections and TUNEL analysis
Frozen tissue sections were prepared from E10.5 and E12.5 embryos as described (Lindquist et al, 2004). Tissue sections from E16.5 embryos were prepared by paraffin embedding and further sectioning by the Laboratory of Comparative Pathology (Rockefeller University). TUNEL of tissue sections was performed using the In Situ Cell Death Detection Kit, TMR red (Roche).
B cell cultures
Primary B cells were isolated from spleen and treated as described (Bothmer et al, 2010). B cells were kept in culture with 25 μg/ml lipopolysaccharide (LPS) (Sigma) and 5 ng/ml of mouse recombinant interleukin 4 (IL-4) (Sigma) for 4 days and percentage of immunoglobulin (Ig) switching was measured by flow cytometry. If indicated, 0.5 μg/ml RP105 (RP/14, BD) was added to the culture medium. Irradiation was performed after 48 h of culture using the dosages and recovery times indicated. CFSE (Molecular Probes) labelling for proliferation analysis was performed as described in Bothmer et al (2010). PARP inhibitor (PARPi, Ku58948) and ATM inhibitor (ATMi, Ku55933) were from KuDOS and used at a concentration of 1 and 2.5 μM, respectively.
DNA damage checkpoint analysis
For the analysis of G2/M arrest, irradiated or mock-treated cells were stained using an anti-phosphorylated histone 3 (serine 10, clone D2C8) antibody (Cell Signaling) according to manufacturer’s instructions. For the analysis of G1/S arrest, the APC-BrdU flow kit was used (BD). B cells were irradiated at 48 h of culture with LPS and IL-4 and harvested at time points indicated.
Western blot
Western blot was performed as described in Feldhahn et al (2005). The following antibodies were used: Anti-phospho-Histone H2a.x (serine 139) clone JBW301 (Millipore), anti-phospho-Atm (serine 1981) (Rockland), anti-phospho-Chk1 (serine 317) (R&D), anti-phospho-p53 (serine 15) (Cell Signaling), anti-αTubulin (Abcam), anti-Lamin b1 (Abcam), anti-hSSB1 (Bethyl Laboratories), anti-Obfc2a (Proteintech Group) and anti-phospho-Kap1 (serine 824) (Bethyl Laboratories).
Flow cytometry
Single cell suspensions from bone marrow, spleen or thymus were obtained as described (Robbiani et al, 2008) and stained using the following antibodies: B220-PE, B220-APC, B220-PerCPcy5, Cd19-APC, IgM-PE, IgD-FITC, Cd3-PE, Cd8-PE, Cd4-FITC, Cd42.2-PerCPcy5, TCRγ/δ-PE, TCRα/β-FITC (BD). For analysis of CSR, an anti-IgG1-APC antibody (BD) or an anti-IgG3-biotin antibody (Southern Biotech) in combination with Steptavidin-APC (BD) was used.
Metaphase spreads analysis
Metaphase spreads were prepared and imaged as described (Callen et al, 2007). B cells were incubated for 1 h with 0.1 μg/ml Colcemid (Roche) before metaphase preparation; MEFs were incubated for 3 h with Colcemid.
c-myc/Igh translocation analysis
PCR analysis of c-myc/Igh translocations was performed as described in Robbiani et al (2008). In brief, genomic DNA (gDNA) from B cells stimulated with 25 μg/ml LPS (Sigma) and 5 ng/ml mouse recombinant IL-4 (Sigma) for 4 days was extracted, and 500 ng gDNA (equivalent of 100 000 cells) was used in each PCR. After gel electrophoresis of PCR products, agarose gels were subjected to Southern blotting. Bands detected by both a c-myc and an Igh probe were scored as translocation.
Skeletal preparations
Skeletal preparations and staining with Alcian blue (Sigma) and Alizarin red (Sigma) to visualize cartilage and bone/mineralized tissue was performed on day E18.5 p.c. or P0 embryos as indicated according to McLeod (1980), Selleri et al (2001) and Ferretti et al (2011).
Micro-computer tomography
P0 embryos were formalin embedded and subjected to MicroCT. In all, 3.5 μm voxel size, 45KVp, 0.36 degrees rotation step, 180 degrees angular range, 400 ms exposure and 1 averaged frame per view were used for the scans, which were performed in air. The Scanco μCT software (HP, DECwindows Motif 1.6) was used for 3D reconstruction, evaluation and viewing of images. After 3D reconstruction, the volumes of interest were segmented and analysed using a global threshold of 0.3 g/ccm. Directly measured bone volume fraction (BV/TV) and tissue mineral density (TMD) were calculated for mid-diaphysis for estimating the differences between Obfc2b+/+ and Obfc2b−/− embryos.
In situ hybridization
Whole-mount in situ hybridization on E10.5 and E12.5 embryos and probe purification was performed as described (Ferretti et al, 2011, Selleri et al, 2001). Murine Obfc2b mRNA-specific sense and anti-sense probes were produced by PCR amplification of the last exon and 3′UTR of Obfc2b using 5′-TTCCGAGAACCAGAACGG-3′ as forward and 5′-AAGGAGGGCAGGCAGAGG-3′ as reverse primer on cDNA from wild-type B cells. The PCR product was ligated into the pCR4-TOPO vector (Invitrogen) and the plasmid sequenced and linearized using PmeI (NEB) or NotI (NEB). Sox9 probes were produced using a Sox9 cDNA encoding plasmid (Wright et al, 1995). Digoxigenin (DIG)-labelled sense and anti-sense probes were produced by in vitro transcription using T3 and T7 primers and the DIG RNA labeling kit (Roche).
Isolation of chondrocytes, osteoblasts and osteoclasts and gene array analysis
Primary chondrocytes were isolated from the sternum of E18.5 p.c. old mice by pronase and collagenase treatment as described in Lefebvre et al (1994) and Retting et al (2009). After expanding for 7 days in cell culture, chondrocytes were harvested by trypsination. Primary osteoblasts were isolated from E18.5 p.c. old mice from the part of the calvaria of the skull that has been identified in the skeletal preparations to be already mineralized (Alizarin Red staining, see Figure 2B). Calvaria plates were then pre-digested twice by 0.1 mg/ml collagenase in aMEM medium at 37°C for 15 min. Bone plates were then cultured in aMEM medium (Invitrogen) +10% fetal calf serum +1% Penicillin/Streptomycin (Invitrogen) for 13 days and outgrowing osteoblasts were harvested by trypsination. Osteoclasts were isolated by culturing erythrocyte-depleted bone marrow cells from the femur in αMEM medium +10% fetal calf serum +1% Penicillin/Streptomycin including 20 ng/ml murine M-CSF (PeproTech) for 24 h. Non-adherent cells were transferred to new media containing 30 ng/ml murine M-CSF and 40 ng/ml RANK-L (PeproTech) and cultured for 5 more days. Media was replaced by fresh media every 2 days. Multinucleated giant cells were stained by TRAP staining (Sigma-Aldrich) to verify osteoclast identity. RNA was extracted from 2.5 × 106 chondrocytes and osteoblasts, and from one 10 cm dish for osteoclasts using Trizol (Invitrogen). RNA was subjected to whole genome gene expression analysis using the MouseRef-8 v2.0 Expression BeadChip (Illumina). Data was analysed using GeneSpring GX software.
Supplementary Material
Acknowledgments
All members of the Nussenzweig laboratory for discussions. Klara Velinzon for cell sorting. The Rockefeller University Gene Targeting Facility for the generation of mutant mice. The work was supported by an NIH grant to MCN (R01 AI037526-18) and an NIH grant to LS (RO1 HD043997). NF was a Fellow of the Leukemia and Lymphoma Society. MCN is a Howard Hughes Medical Institute Investigator.
Author contributions: NF designed and performed most experiments. EF performed skeletal preparations on E18.5 embryos and assisted in in situ hybridizations. DFR assisted in the design of the Obfc2bflox targeting vector. SD helped on metaphase preparations. EC performed the analysis of all prepared metaphase spreads. LS supervised experiments performed on embryos. AN and MCN supervised all experiments. NF and MCN wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B (2002) The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16: 2813–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris A (1996) Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86: 159–171 [DOI] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC (2010) 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 207: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broceno C, Wilkie S, Mittnacht S (2002) RB activation defect in tumor cell lines. Proc Natl Acad Sci USA 99: 14200–14205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917 [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, Sleckman BP, Ried T, Nussenzweig M, Nussenzweig A (2007) ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell 130: 63–75 [DOI] [PubMed] [Google Scholar]
- Chaganti RS, Houldsworth J (1991) Fanconi anemia: a pleotropic mutation with multiple cellular and developmental abnormalities. Ann Genet 34: 206–211 [PubMed] [Google Scholar]
- Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J (2005) The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell 8: 179–192 [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH (2000) Nucleotide excision repair and human syndromes. Carcinogenesis 21: 453–460 [DOI] [PubMed] [Google Scholar]
- de Luca C, Kowalski TJ, Zhang Y, Elmquist JK, Lee C, Kilimann MW, Ludwig T, Liu SM, Chua SC Jr (2005) Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J Clin Invest 115: 3484–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Chaudhuri J, Bassing CH, Alt FW (2005) Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv Immunol 86: 43–112 [DOI] [PubMed] [Google Scholar]
- Feldhahn N, Klein F, Mooster JL, Hadweh P, Sprangers M, Wartenberg M, Bekhite MM, Hofmann WK, Herzog S, Jumaa H, Rowley JD, Muschen M (2005) Mimicry of a constitutively active pre-B cell receptor in acute lymphoblastic leukemia cells. J Exp Med 201: 1837–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, Carpenter PB, Bonner WM, Chen J, Nussenzweig A (2002) DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol 4: 993–997 [DOI] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, Wells V, Hebert JM, Karner C, Anderson MJ, Williams T, Dixon J, Dixon MJ, Selleri L (2011) A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell 21: 627–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME et al. (1998) A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95: 891–902 [DOI] [PubMed] [Google Scholar]
- Guha U, Gomes WA, Kobayashi T, Pestell RG, Kessler JA (2002) In vivo evidence that BMP signaling is necessary for apoptosis in the mouse limb. Dev Biol 249: 108–120 [DOI] [PubMed] [Google Scholar]
- Hall BK, Miyake T (2000) All for one and one for all: condensations and the initiation of skeletal development. Bioessays 22: 138–147 [DOI] [PubMed] [Google Scholar]
- Horwitz BH, Scott ML, Cherry SR, Bronson RT, Baltimore D (1997) Failure of lymphopoiesis after adoptive transfer of NF-kappaB-deficient fetal liver cells. Immunity 6: 765–772 [DOI] [PubMed] [Google Scholar]
- Huang J, Gong Z, Ghosal G, Chen J (2009) SOSS complexes participate in the maintenance of genomic stability. Mol Cell 35: 384–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA (1994) Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic M, Nussenzweig A, Nussenzweig MC (2007) Antigen receptor diversification and chromosome translocations. Nat Immunol 8: 801–808 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C, O’Driscoll M (2009) Human DNA damage response and repair deficiency syndromes: linking genomic instability and cell cycle checkpoint proficiency. DNA Repair (Amst) 8: 1139–1152 [DOI] [PubMed] [Google Scholar]
- Kronenberg HM (2003) Developmental regulation of the growth plate. Nature 423: 332–336 [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA 93: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Garofalo S, Zhou G, Metsaranta M, Vuorio E, De Crombrugghe B (1994) Characterization of primary cultures of chondrocytes from type II collagen/beta-galactosidase transgenic mice. Matrix Biol 14: 329–335 [DOI] [PubMed] [Google Scholar]
- Li C, Xu X, Nelson DK, Williams T, Kuehn MR, Deng CX (2005) FGFR1 function at the earliest stages of mouse limb development plays an indispensable role in subsequent autopod morphogenesis. Development 132: 4755–4764 [DOI] [PubMed] [Google Scholar]
- Li Y, Bolderson E, Kumar R, Muniandy PA, Xue Y, Richard DJ, Seidman M, Pandita TK, Khanna KK, Wang W (2009) HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem 284: 23525–23531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist RL, Shakhar G, Dudziak D, Wardemann H, Eisenreich T, Dustin ML, Nussenzweig MC (2004) Visualizing dendritic cell networks in vivo. Nat Immunol 5: 1243–1250 [DOI] [PubMed] [Google Scholar]
- McKusick VA, Mahloudji M, Abbott MH, Lindenberg R, Kepas D (1967) Seckel’s bird-headed dwarfism. N Engl J Med 277: 279–286 [DOI] [PubMed] [Google Scholar]
- McLeod MJ (1980) Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 22: 299–301 [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2003) Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 25: 1158–1167 [DOI] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montana MF, Soria R, Mulero F, Canamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O (2009) A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet 41: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272: 263–267 [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC (2006) Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 440: 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC (2004) AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118: 431–438 [DOI] [PubMed] [Google Scholar]
- Retting KN, Song B, Yoon BS, Lyons KM (2009) BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136: 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, Takeda S, Hay RT, Gautier J, West SC, Paull TT, Pandita TK, White MF, Khanna KK (2008) Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 453: 677–681 [DOI] [PubMed] [Google Scholar]
- Richard DJ, Cubeddu L, Urquhart AJ, Bain A, Bolderson E, Menon D, White MF, Khanna KK (2011a) hSSB1 interacts directly with the MRN complex stimulating its recruitment to DNA double-strand breaks and its endo-nuclease activity. Nucleic Acids Res 39: 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DJ, Savage K, Bolderson E, Cubeddu L, So S, Ghita M, Chen DJ, White MF, Richard K, Prise KM, Schettino G, Khanna KK (2011b) hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res 39: 1692–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K (1997) B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25: 1317–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC (2008) AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135: 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25: 139–140 [DOI] [PubMed] [Google Scholar]
- Rooney S, Chaudhuri J, Alt FW (2004) The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev 200: 115–131 [DOI] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O’Gorman S, Cleary ML (2001) Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128: 3543–3557 [DOI] [PubMed] [Google Scholar]
- Shim M, Foley J, Anna C, Mishina Y, Eling T (2010) Embryonic expression of cyclooxygenase-2 causes malformations in axial skeleton. J Biol Chem 285: 16206–16217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Richard DJ, Saraf A, Toschi A, Bolderson E, Florens L, Washburn MP, Khanna KK, Pagano M (2009) INTS3 controls the hSSB1-mediated DNA damage response. J Cell Biol 187: 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA 87: 4707–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, Edelmann W (2005) Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 37: 750–755 [DOI] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P (1995) The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet 9: 15–20 [DOI] [PubMed] [Google Scholar]
- Xu S, Feng Z, Zhang M, Wu Y, Sang Y, Xu H, Lv X, Hu K, Cao J, Zhang R, Chen L, Liu M, Yun JP, Zeng YX, Kang T (2011) hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene 30: 2219–2229 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wu J, Yu X (2009) Integrator3, a partner of single-stranded DNA-binding protein 1, participates in the DNA damage response. J Biol Chem 284: 30408–30415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.