Abstract
Biallelic mutations in the untranslated regions (UTRs) of mRNAs are rare causes for monogenetic diseases whose mechanisms remain poorly understood. We investigated a 3′UTR mutation resulting in a complex immunodeficiency syndrome caused by decreased mRNA levels of p14/robld3 by a previously unknown mechanism. Here, we show that the mutation creates a functional 5′ splice site (SS) and that its recognition by the spliceosomal component U1 snRNP causes p14 mRNA suppression in the absence of splicing. Histone processing signals are able to rescue p14 expression. Therefore, the mutation interferes only with canonical poly(A)-site 3′ end processing. Our data suggest that U1 snRNP inhibits cleavage or poly(A) site recognition. This is the first description of a 3′UTR mutation that creates a functional 5′SS causative of a monogenetic disease. Moreover, our data endorse the recently described role of U1 snRNP in suppression of intronic poly(A) sites, which is here deleterious for p14 mRNA biogenesis.
Keywords: polyadenylation, U1 site, 3′ UTR mutation
Introduction
The 3′untranslated region (UTR) harbours the signals for proper mRNA 3′ end formation (Proudfoot, 2011) and additional elements with regulatory potential (Mayr and Bartel, 2009). Nascent mRNAs mature by cleavage and poly(A) tail addition in close proximity to the transcription site (Gilmartin, 2005; West and Proudfoot, 2009). Poly(A) tails are synthesized by poly(A) polymerase (Barabino and Keller, 1999) and they influence downstream events including RNA export, translation, and stability (Grey et al, 2000; Fuke and Ohno, 2008; Qu et al, 2009).
Disease-related mutations in 3′UTRs mainly affect the polyadenylation signal (PAS) and surrounding sequences (Conne et al, 2000; Chen et al, 2006; Danckwardt et al, 2008). The remaining unclassified sequence variations may reveal new aspects of gene regulation as they could target miRNA binding sites (Fabian et al, 2010) or elements that influence mRNA localization and stability (Shyu et al, 2008; Andreassi and Riccio, 2009). A point mutation in the 3′UTR of the gene encoding p14/robld3 results in a complex immunodeficiency syndrome (Bohn et al, 2007). Patients are characterized by a congential neutropenia resulting in recurrent bacterial lung infections. P14/ROBLD3 is an endosomal scaffold protein involved in signalling via endocytosed receptors (Teis et al, 2006; Bohn et al, 2007). The mutation (C-A, +23, 3′UTR) strongly decreases p14 mRNA and protein levels. Previously, Bohn et al (2007) fused the p14 3′UTR to a heterologous mRNA and observed a half-life-independent mRNA reduction implying that the mutation causes a defect in RNA biogenesis; however, the mechanism remained elusive. We now demonstrate that the mutation creates a 5′ splice site (SS) and that its recognition by U1 snRNP interferes with 3′ end formation in the absence of splicing. Interestingly, the wild-type sequence also functions as a 5′SS when paired with a strong 3′SS raising the possibility that p14 expression may also be endogenously regulated by U1 snRNP-mediated poly(A) site suppression (Kaida et al, 2010; Vorlova et al, 2011).
Results
Increasing the complementarity of the mutated region to U1 snRNA enhances p14 mRNA suppression
To study the mechanisms of defective RNA biogenesis in P14/ROBLD3 deficiency, the mutant p14 cDNA was cloned with the 3′UTR, the PAS, the cleavage site as well as 314 nt of downstream genomic sequences (Figure 1A). In our system, the point mutation caused a 60% reduction in p14 mRNA in HeLa cells (Figures 1B and 2D), indicating that the transient p14/ROBLD3 expression system partially recapitulated the results from patients’ cells, where a 95% reduction was observed (Bohn et al, 2007). The ∼3.6-kb band in the northern blot represents a read-through transcript, which terminates at a downstream PAS close to the SV40 promoter in the circular plasmid backbone. Interestingly, the amount of read-through is increased by the p14 mutation (Figure 1B).
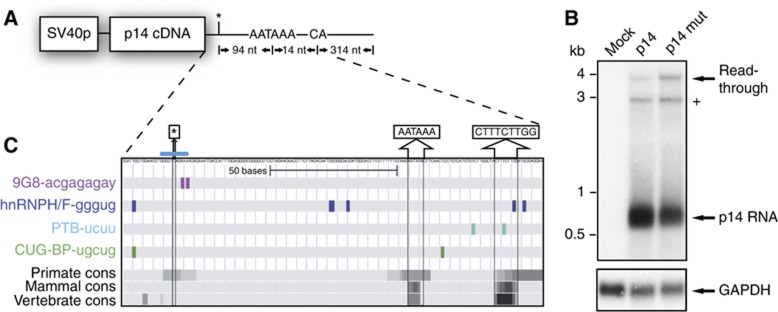
Figure 1.
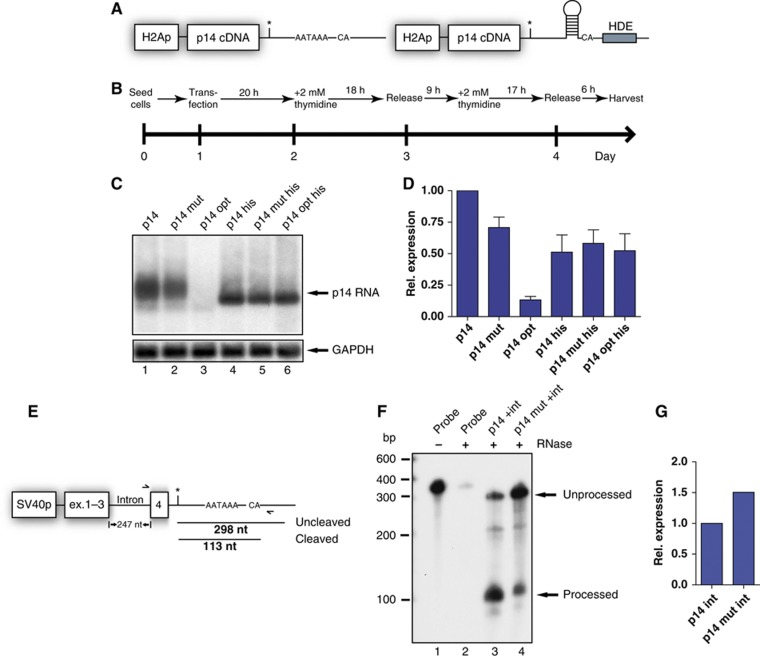
Establishment of a p14 minigene and bioinformatic analysis of the 3′UTR. (A) Depiction of the p14 expression plasmid driven by the SV40 promoter. The asterisk marks the position of the point mutation. 3′ of the mutation the PAS and the cleavage site are drawn. The 3′UTR was extended into the genomic locus of p14. (B) Northern blot using total RNA harvested after 36 h from transiently transfected HeLa cells using the indicated plasmids. Equal transfection efficiency was monitored by co-transfection of an eGFP-encoding plasmid and analysis by flow cytometry in all experiments (data not shown). The blot was hybridized with a 32P-radiolabelled probe corresponding to the p14 cDNA. A marker in kb is shown on the left and the transcripts are indicated on the right. The (+) marks a DNA contamination originating from the transfected plasmid as verified by DNase digestion (data not shown). The blot was re-hybridized with a probe corresponding to the GAPDH cDNA as a loading control. The endogenous HeLa p14 expression was only detected after prolonged exposure (data not shown). (C) Bioinformatic analysis of the p14 3′UTR. The conservation values calculated for the 3′UTR region are presented in grey scale, from white to black, representing phastcons values from 0 (no conservation) to 1 (high conservation), respectively. The locations of predicted binding sites of four splicing factors CUG-BP, PTB, hnRNPH/F, and 9G8 are shown in coloured rectangles. The position of the C-A, +23 mutation is indicated by an asterisk as well as the sequence of the PAS and the downstream conserved element. The putative 5′SS is marked by a blue bar.
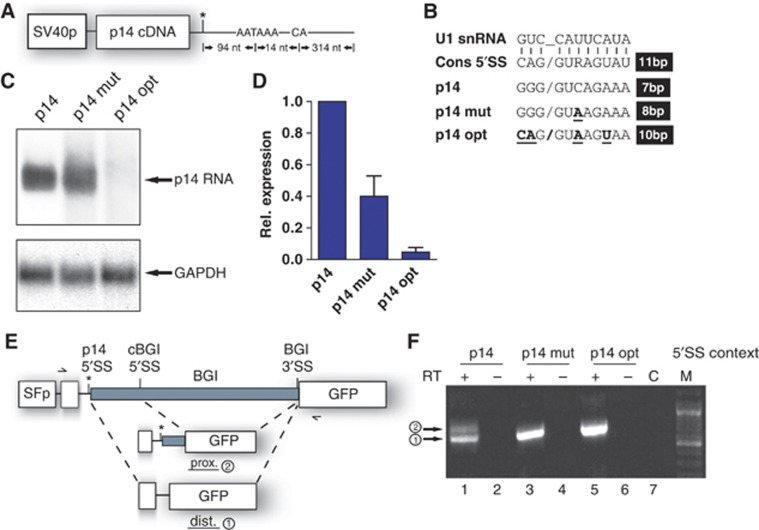
Figure 2.
Increasing the complementarity of the mutated region to U1 snRNA enhances p14 mRNA downregulation. (A) Scheme of the p14 expression plasmid driven by the SV40 promoter. (B) The sequence surrounding the mutation is drawn like a putative 5′SS. The consensus sequence pairing to U1 snRNA is depicted on the top. Vertical lines represent base pairing by hydrogen bonds. Below, the p14 wild-type, mutated, and optimized sequences are displayed. Changes to the wild-type p14 sequence are indicated in bold. The slash indicates the exon/intron border as in an authentic 5′SS. On the right, the number of possible base pairs to U1 snRNA are counted including G:U base pairs. (C) Northern blot using total RNA performed as in Figure 1B. (D) Quantitation of the northern blot shown in (C) by phosphoimager analysis. The p14-specific signal was corrected for the loading control GAPDH. The p14 wild type was set to 1. The relative expression values represent the average and standard deviation from four independent experiments. (E) The mutation in the p14 3′UTR creates a functional 5′SS. For the splicing reporter plasmid, the second intron from the rabbit β-globin gene (grey box; BGI) was fused to the GFP ORF. The endogenous globin 5′SS was exchanged to the p14-derived sequence (black line and asterisk) plus part of the p14 ORF (white box). The reporter is driven by the spleen focus forming promoter (SFp). The cryptic 5′SS in the BGI is named as cBGI. The arrows mark the primers used for the RT–PCR in (F). The distal product is obtained from the p14-derived 5′SS and the proximal when the cryptic BGI 5′SS is used. (F) RT–PCR using total RNA from 293T cells transfected with the splicing reporters. Each RT reaction was performed without enzyme and a control without template. The numbering on the left is according to (E). The correct exon junctions were verified by nucleotide sequencing (data not shown).
We analysed the sequence conservation of the p14 3′UTR using the phastcons table from the UCSC genome browser. The overall conservation was very high among placental mammals, including the nucleotide that is mutated (C-A, +23) in the p14 immunodeficiency (Supplementary Figure S1A; Figure 1C). Only rodents, but not the closely related order of lagomorphs displayed a gap at this position (Supplementary Figure S1A). Comparison of available vertebrate species showed conservation of the PAS and a downstream GU-rich region, most likely representing the downstream sequence enhancer (Proudfoot, 2011; Figure 1C). Strikingly, the mutation site is embedded in a putative 5′SS, and the mutation increases its match to the 5′SS consensus sequence (Figures 1C and 2B). In addition, we performed a bioinformatic analysis, using the SFmap algorithm (Akerman et al, 2009), to identify conserved sequence motifs that may be involved in splicing regulation. This analysis identified an SR protein-binding site (9G8) just downstream of the mutation (Figure 1C; Supplementary Figure S1B). A cluster of conserved motifs matching the binding sites for hnRNPH/F and CUGBP-1 are located upstream of the mutation (Figure 1C). Interestingly, both hnRNPH/F and CUGBP-1 are known to be involved in the regulation of weak 5′SS (Caputi and Zahler, 2002; Goraczniak and Gunderson, 2008). The binding sites for hnRNPH/F downstream of the PAS may represent positive elements for 3′ end formation (Arhin et al, 2002).
Given the direct overlap with the mutation site, we focused on the putative 5′SS. 5′SS are recognized by the RNA component of U1 snRNP, thereby committing pre-mRNAs to the splicing pathway (Wahl et al, 2009). The Analyzer Splice Tool revealed that the putative p14 5′SS can anneal to 8 base pairs to U1 snRNA, 7 of which are uninterrupted giving a score of 82.6 and a free energy of −8 kcal/mol (G:U pairs included; Figure 2B, compare second and fourth lines). Whereas the wild-type p14 sequence anneals to 7 base pairs to U1 snRNA, interrupted at position +3, resulting in a score of 72.4 and a free energy of −5.2 kcal/mol (Figure 2B, third line). Thus, the mutation may convert the wild-type sequence into a functional 5′SS. If the 5′SS created by the mutation (C-A, +23) splices to a downstream 3′SS, then the p14 PAS could be omitted, leading to both the mRNA reduction and the enhanced read-through (Figure 1B). Such a splicing event would not be predicted to elicit non-sense-mediated RNA decay, because the 5′SS is in too close proximity to the termination codon (Rebbapragada and Lykke-Andersen, 2009). However, no splicing was detected by RT–PCR assay with the p14 minigene (Supplementary Figure S2). Furthermore, splicing of the endogenous p14 3′UTR was not detected using 3′ RACE on total RNA prepared from immortalized patient’s B cells (data not shown). Thus, aberrant splicing is not the cause of the P14/ROBLD3 deficiency.
Previous studies showed that recruitment of U1 snRNP to 3′UTRs can diminish mRNA abundance (Gunderson et al, 1998; Fortes et al, 2003; Goraczniak et al, 2009). Thus, we predicted that creating an optimal U1 snRNA recognition site should abrogate p14 RNA expression (p14 opt; Figure 2B, bottom line). Indeed, p14 mRNA expression was barely detectable when an optimal 5′SS was introduced into the p14 minigene (Figure 2C and D), emphasizing that increased complementarity to U1 snRNA enhanced p14 suppression. Therefore, creation of a 5′SS by the C-A, +23 mutation is a probable explanation for the observed p14 mRNA downregulation.
The mutation in the p14 3′UTR creates a functional 5′SS
Since we were not able to detect any splicing originating from the putative p14 5′SS both in the minigene (Supplementary Figure S2) and in the genomic context, we wondered whether the mutant p14 sequence represents a bona fide 5′SS. We constructed a splicing reporter containing the second β-globin intron (BGI; Figure 2E). The main BGI 5′SS was replaced with the putative p14 5′SS. Note that the BGI harbours an additional cryptic 5′SS (cBGI). In this way, processing can lead to the formation of a distal (p14-derived 5′SS; #1) and a proximal (cBGI 5′SS-derived; #2) splice product (Figure 2E). RT–PCR analysis showed that both the p14 mut and opt sequences function as efficient 5′SS when combined with a strong 3′SS (Figure 2F, lanes 3 and 5). The wild-type p14 sequence can also serve as 5′SS, consistent with a report that 7 bp complementarity to U1 snRNA can function as active SS (Guan et al, 2007), albeit less efficiently (Figure 2F, lane 1). We also observed competition of the weak p14 wild-type 5′SS with the cBGI 5′SS, thereby producing the proximal splice product (Figure 2F, lane 1). This prompted us to search in the available deep sequencing databases for a splice event originating from the wild-type sequence into the 3′ genomic region. However, no reads covering such a splice event were found (J Castle, personal communication) further supporting the hypothesis that U1 snRNP represses p14 expression in the absence of splicing. The GFP expression of the splicing reporter was used to monitor the efficiency of splicing. Due to upstream ORFs, the GFP cannot be efficiently translated from the proximal splice product. Consequently, FACS analysis showed an increase in GFP expression in case of the p14 mut and opt sequences (Supplementary Figure S3).
In summary, our splicing reporter assay demonstrated that the point mutation in the p14 3′UTR creates a functional and efficient 5′SS when combined with a strong 3′SS. Interestingly, the wild-type sequence can inefficiently function as a 5′SS, raising the possibility that p14 is normally regulated by an U1 snRNP-dependent mechanism. To test this, we destroyed the invariant GU dinucleotide, which further decreases complementarity to U1 snRNA (Supplementary Figure S4A). Consistent with our prediction, the U2C mutation increased the p14 mRNA levels by 1.7-fold (Supplementary Figure S4B and C). Thus, p14 is the second human gene whose expression is regulated via a suboptimal 5′SS in the 3′UTR (Guan et al, 2007).
Terminal intron splicing increases p14 mRNA expression only in the wild-type context
Several groups reported coupling of terminal intron splicing and 3′ end processing (Kyburz et al, 2006; Danckwardt et al, 2007). For instance, splicing of the second β-globin intron drastically increases mRNA levels by enhancing 3′ end processing (Lu and Cullen, 2003; Rigo and Martinson, 2008).
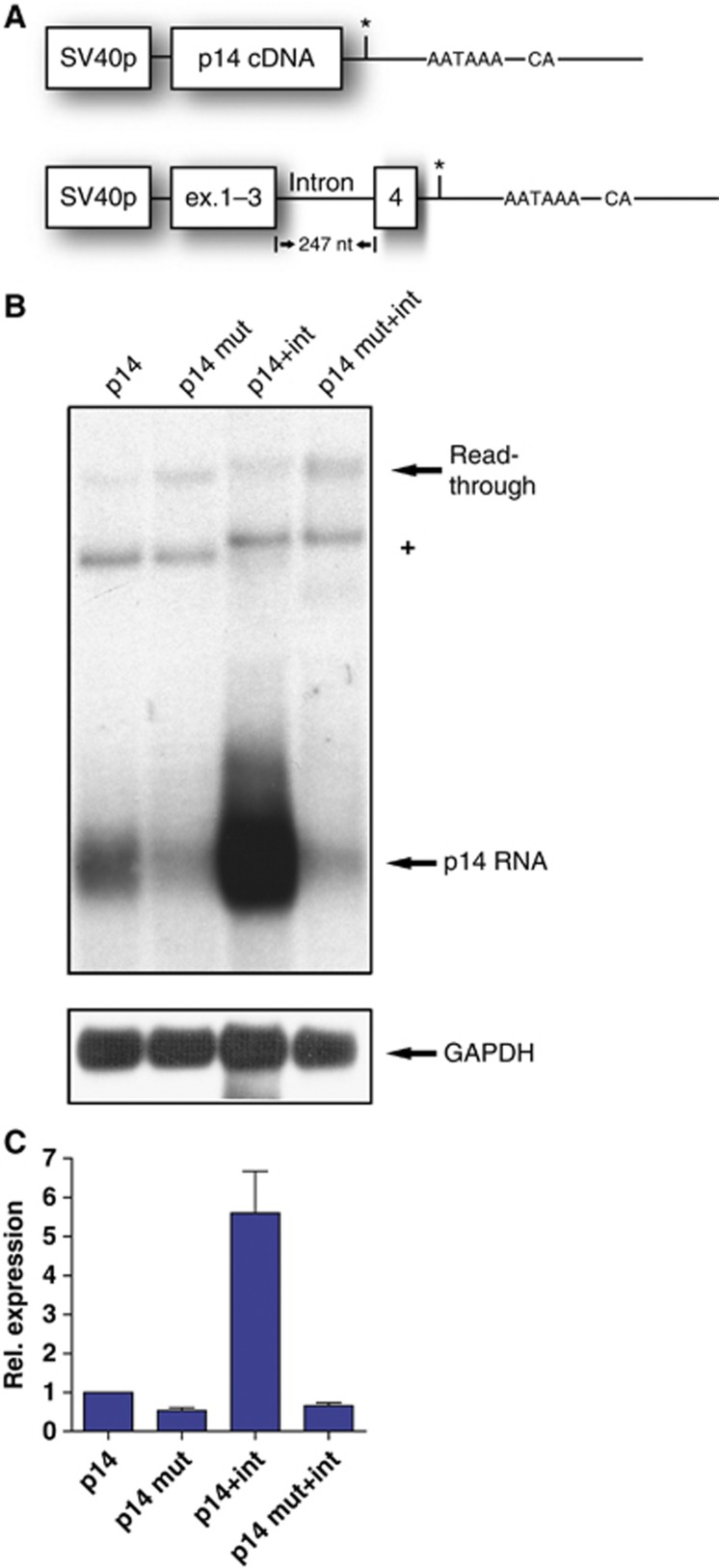
To determine the influence of terminal intron splicing on p14 mRNA, we included the terminal p14 intron in our plasmid (Figure 3A). This led to strongly elevated p14 mRNA levels by six-fold in the wild-type context (Figure 3B and C). Thus, p14 RNA expression depends on terminal intron splicing to the same extent as β-globin (Lu and Cullen, 2003; Nott et al, 2003). Remarkably, there is no increase in mutant p14 mRNA when the terminal intron is present (Figure 3B and C). This indicates that the positive effect on 3′ end processing is counteracted by the mutation. In comparison to the minigene harbouring the p14 cDNA, inclusion of the terminal intron now recapitulates the substantial p14 downregulation that was observed in patients’ cells (Figure 3C; Bohn et al, 2007).
Figure 3.
Splicing of the terminal intron enhances p14 RNA expression of the wild-type but not of the mutant transcript. (A) Schematic drawing of the p14 cDNA minigene and the plasmid containing the terminal p14 intron between exons 3 and 4. The 3′UTR including the mutation remained unaltered. (B) Northern blot performed from total RNA as in Figure 1B. Note that the DNA contamination (+) and the read-through transcript migrate higher due to the inclusion of the intron. The intron is not removed in the read-through transcript arguing for a fixation on the proximal p14 PAS if splicing occurs. (C) Quantitation of the northern blot (B) as performed in Figure 2D. The wild-type p14 expression was set to 1. The relative expression values represent the average and standard deviation from seven independent experiments.
U1 snRNP causes p14 mRNA suppression
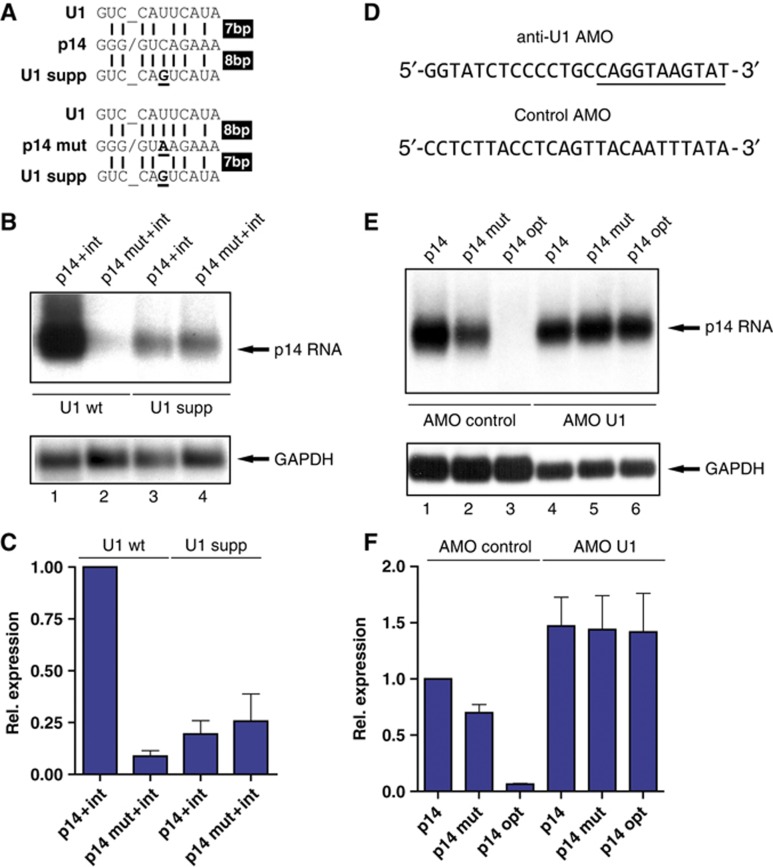
To provide additional evidence that U1 snRNA binding causes p14 downregulation, we used a genetic U1 snRNA suppressor system. U1 snRNA suppressor mutants are incorporated into U1 snRNP particles capable of recognizing a different set of 5′SS depending on the introduced mutations (Zhuang and Weiner, 1986; Furth et al, 1994). Our U1 suppressor mutant is able to anneal to 8 bp of the p14 wild-type sequence or 7 bp with the p14 mutant (Figure 4A). Co-transfection of the p14 plasmid with suppressor U1 snRNA yielded a strong downregulation of p14 wild-type mRNA levels, whereas expression was unaffected by co-transfection of wild-type U1 snRNA (Figure 4B, compare lanes 1 and 3). The suppressor U1 snRNA downregulated p14 wild-type mRNA almost to the same extent as the C-A, +23 mutation, demonstrating that the suppressor U1 snRNP elicited p14 mRNA suppression (Figure 4B, compare lanes 1 and 3; Figure 4C). On the other hand, suppressor U1 snRNPs should enhance expression of the mutated p14 mRNA, because the recognition of the mutated sequence as a 5′SS should be reduced. As shown in Figure 4B, co-transfection of the U1 suppressor is able to increase p14 mut expression (compare lanes 2 and 4), but not to the same extent as the wild-type mRNA is downregulated (Figure 4C). The amount of p14 mutant mRNA is increased by three-fold compared to a five-fold downregulation in case of the p14 wild-type mRNA (Figure 4C). This is likely due to the fact that U1 suppressors cannot outcompete the high levels of endogenous U1 snRNPs, which execute the downregulation (p14 mut) compared to the opposite situation, where the suppressor U1 snRNPs can mediate the negative effect (p14 wild type).
Figure 4.
Suppression of mutated p14 mRNA is mediated by U1 snRNP. (A) Depiction of the U1 snRNA suppressor system. Top, the p14 wild-type sequence is shown in the middle. Vertical lines display the possible base pairs to U1 snRNA (top) and the suppressor U1 snRNA (bottom). The introduced mutation into U1 snRNA is depicted in bold and underlined. A slash (underlined space in U1 snRNA) marks the putative exon/intron border in the p14 sequence. Bottom, the same scheme is drawn for the p14 mutation. Note, that the number of base pairs between p14 and the mutation to wild-type U1 snRNA are inverted by the suppressor snRNAs as indicated on the right. (B) Northern blot performed as in Figure 1B. The p14 wild-type and mutated plasmids were co-transfected with expression plasmids for U1 wild-type and suppressor snRNAs in a ratio of 1:3. (C) Quantitation by phosphoimager analysis as in Figure 2D. The transfections in the presence of U1 wild type and suppressor are indicated above the graph. The relative expression values represent the average and standard deviations from five independent experiments. (D) The sequence of the antisense morpholinos (AMOs) is given (Kaida et al, 2010). In U1 AMO nucleotides complementary to the free 5′ end of U1 are underlined. (E) The indicated p14 cDNA minigenes were transfected and AMOs were microporated 4 h later. Total RNA was harvested 16 h later and subjected to northern blot analysis as in Figure 1C. (F) Quantitation by phosphoimager analysis as in Figure 2D. Control and U1 AMOs are indicated above the graph. The relative expression values represent the average and standard deviations from three independent experiments.
To avoid the competition between suppressor and endogenous U1 snRNPs, we sought to target U1 snRNA directly. We chose anti U1 morpholinos (AMO U1) directed against the 5′ end of U1 snRNA (Kaida et al, 2010; Figure 4D). Morpholinos elicit neither siRNA responses nor cleavage of the duplex by RNaseH (Kaida et al, 2010). As an independent positive control we used our previously described HIV splicing reporter (Bohne et al, 2005). The reporter was first transfected and the AMOs subsequently microporated. Application of AMO U1 but not of a control AMO led to a 2.5-fold increase in ratio of unspliced versus spliced RNA (Supplementary Figure S5), showing that splicing is partially inhibited in the HIV reporter under these conditions. To test the effect of AMO U1 on p14 expression, we used the intron-less minigene (Figure 1A) to avoid negative effects of splicing inhibition on terminal intron splicing (Figure 3). As shown in Figure 4E blocking U1 snRNP by microporation of AMO U1 resulted in a complete rescue of p14 expression for the wild type, mutant, and even the optimized 5′SS. p14 expression was slightly increased compared to the control AMOs (Figure 4F).
To directly assess binding of U1 snRNP to the p14 5′SS, we performed electromobility shift assays (EMSAs) using purified U1 snRNP and radiolabelled 22mer RNA oligos encompassing the wild-type or mutant sequences (Supplementary Figure S6). Both wild-type and mutant p14 5′SS RNA oligos formed a U1 complex, but not the control oligo (Supplementary Figure S6). We observed a similar phenomenon in vivo using our splicing reporter where both wild-type and mutant represent bona fide 5′SS (Figure 2E and F).
Taken together, our data demonstrate that recognition of the mutated p14 3′UTR by U1 snRNP is responsible for the observed downregulation of p14 mRNA, which therefore represents the molecular trigger of the P14/ROBLD3 immunodeficiency.
The p14 mutation does not affect expression in the context of histone 3′ end processing signals
To gain insights into how binding of U1 snRNP to the mutated p14 3′UTR decreases expression, we tested the effects of knockdown or chemical inactivation of the RNA exosome either by applying siRNAs directed against Rrp6 (PM/Scl 100; Staals and Pruijn, 2010) or by using 5′ fluorouracil (Kammler et al, 2008). Neither approach rescued p14 expression although an unstable endogenous transcript was upregulated (Supplementary Figure S7). Next, we asked if the mutation could affect p14 mRNA created by an alternative 3′ end processing pathway. The p14 PAS and the cleavage site were replaced by histone 3′ end formation signals (Figure 5A). Replication-dependent histone genes are intron-less and non-polyadenylated. Instead, histone 3′ end processing depends on an RNA stem loop structure and a histone downstream element (HDE; Figure 5A; Dominski and Marzluff, 2007). We exchanged the promoter (Figure 5A, left) and in addition replaced the sequences downstream of the mutation by histone 3′ end formation signals (Figure 5A, right). Since histone expression is restricted to S phase of the cell cycle (Dominski and Marzluff, 2007), we used a double thymidine block protocol as outlined in Figure 5B to release a synchronized culture into S phase. In the context of the polyadenylated mRNA, the p14 mutation and the optimized 5′SS decreased mRNA levels (Figure 5C, lanes 2 and 3). The downregulation was not as pronounced as in the non-synchronized cells, where expression was directed by the SV40 promoter (Figure 2C and D). However, the trend that mutations which increased complementarity to U1 snRNA led to stronger downregulation of p14 mRNA persisted (Figure 5D).
Figure 5.
The p14 mutation does not affect expression in the context of histone 3′ end processing signals. (A) Depiction of the modified p14 expression plasmids. The SV40 promoter was exchanged to the histone H2A promoter to confer cell cycle-dependent transcriptional activity. The right side displays the p14 his plasmids, where in addition to the promoter part of the 3′UTR was replaced by the H2A 3′ end consisting of stem loop, the CA dinucleotide and histone downstream element (HDE; grey box). (B) Experimental design of the double thymidine block to synchronize the cells and to release the culture into S phase. (C) Northern blot performed as in Figure 1B. A probe corresponding to the p14 cDNA was used. (D) Quantitation by phosphoimager analysis as in Figure 2D. Relative expression values represent the average and standard deviation from five independent experiments. (E) Schematic representation of the p14 intron-containing minigene (Figure 3). The length of the probe used for RNase protection assay (RPA) is drawn as black line (uncleaved). Cleavage at the p14 PAS would lead to a shorter protected fragment (black line below; cleaved). The exact length of the fragments in nt is given. In addition, the primers used for the RT–qPCR are indicated by arrows. (F) RPA using total RNA from HeLa cells transfected with intron-containing minigenes. A size marker in base pairs is given on the left. As digestion control, the probe was loaded with or without RNase treatment. The p14-specific products are indicated by arrows on the right. (G) RT–qPCR data are presented as relative values setting the wild type (p14 int) to 1. Only one representative experiment out of two is shown.
In comparison expression from the histone constructs is lower (Figure 5D) due to relatively inefficient histone processing in a transient transfection system (EJ Wagner, personal communication). However, the p14 mutation and the optimized 5′SS sequence did not affect 3′ end formation in the histone context (Figure 5C, lanes 5 and 6). All p14 variants now displayed equal levels of RNA. Note that since histone mRNAs are not polyadenylated, the mRNA band is shorter and more discrete compared to other Pol II transcripts (Figure 5C). Hence, the p14 mutation affects only PAS-dependent 3′ end formation.
To identify which step during p14 3′ end processing is suppressed by U1 snRNP, we used RNase protection assay (RPA) and RT–qPCR to analyse mRNAs produced by the intron-containing minigenes (Figure 5E). The longer protected RPA probe indicates unprocessed or read-through transcripts (Figure 5E). The shorter probe denotes cleaved mRNAs at the p14 PAS. RPA using total mRNA from cells transfected with both versions of the intron-containing minigenes revealed more processed versus unprocessed transcripts for the wild-type p14 mRNA (Figure 5F, lane 3). This ratio is reversed in the mutant (Figure 5F, lane 4). This proves that the mutation led to an inhibition of 3′ end formation at the p14 PAS. The RPA detected both unspliced and spliced RNAs. For the RT–qPCR, we used an intronic forward primer to sense only unspliced pre-mRNA (Figure 5E). Thus, the assay should measure unspliced and unprocessed p14 pre-mRNA or read-through transcripts, where the intron has not been removed. As shown in Figure 5G, the mutation leads to a 1.5-fold, increase indicating that the initial transcript levels are similar and, more importantly, that unspliced/unprocessed pre-mRNA accumulates in case of the p14 mutation.
Discussion
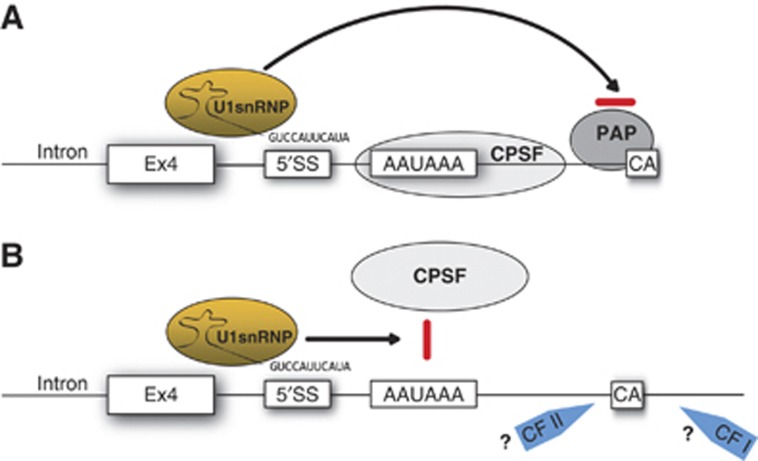
The p14 C-A, +23 mutation creates a 5′SS capable of recruiting an inhibitory U1 snRNP complex leading to a failure of proper 3′ end formation and rapid degradation of p14 mRNA. This U1 nsRNP-mediated suppression is the ultimate cause of the p14 congenital immunodeficiency described by Bohn et al (2007). The question remains how U1 snRNP interferes with 3′ end processing? Gunderson et al (1998) proposed that U1 snRNP binding in proximity to a PAS may specifically inhibit poly(A) tail addition (Figure 6). This implies that a fraction of the transcribed mutant p14 mRNA is not polyadenylated leading to rapid decay close to the site of transcription (Custodio et al, 1999; Milligan et al, 2005; Conrad et al, 2006; Kazerouninia et al, 2010). Furthermore, it was recently proposed that degradation of SS and PAS mutants is initiated before complete termination of transcription (Davidson et al, 2012). This model is consistent with two of our observations. First, mutated p14 mRNA that escapes U1-mediated suppression displays average-length poly(A) tails (Figure 1B; data not shown; Bohn et al, 2007). Second, after transcriptional induction, p14 mRNA levels are already decreased, presumably due to rapid decay (Bohn et al, 2007; data not shown). Again, mutant p14 mRNA, which did not bind a U1-inhibitory complex, decays with the same half-life as wild-type mRNA (Bohn et al, 2007).
Figure 6.
A model of U1 snRNP-mediated p14 suppression. (A) In the classical view recognition of a 5′SS in close proximity to a PAS by U1 snRNP inhibits poly (A) polymerase (PAP). A failure of poly(A) tail addition then leads to RNA degradation. (B) U1 snRNP is also implicated in cellular surveillance mechanism called PCPA (details see text). Here, U1 snRNP may inhibit already the recognition of the PAS by CPSF and/or the activity of cleavage stimulating factors I and II.
Recently, a surveillance function was ascribed to U1 snRNP, referred to as inhibition of premature cleavage and polyadenylation (PCPA) whereby U1 suppresses 3′ end processing at non-canonical, intronic PAS (Kaida et al, 2010). During normal 3′ end formation cleavage precedes polyadenylation (Proudfoot, 2011). Thus, if U1 solely inhibits poly(A) polymerase (see above), cleavage would occur at intronic PAS multiple times during transcription and pre-mRNA integrity is lost. This integrity is not absolutely required for splicing, but is important for efficient RNA synthesis and processing (Pastor et al, 2011). Our data instead suggest that U1 snRNP interferes with cleavage or recognition of the PAS (Figure 5).
Theoretically, anti-U1 AMOs may lead to the liberation of U1-70K and thus to secondary effects beside blockage of U1 snRNA. However, recent work by the Cartegni laboratory showed PCPA of receptor tyrosine kinases using three different approaches: morpholinos directed against 5′SS upstream of intronic PAS, anti-U1-70K siRNAs, and SS mimicry (Vorlova et al, 2011). In addition, the splicing inhibition observed in the U1 AMO-treated splicing reporter cannot be explained by displacement of U1-70K (Supplementary Figure S5).
Replacing the p14 PAS by histone processing signal rescued p14 expression (Figure 5). Thus, U1 snRNP interferes with cleavage/polyA site selection but not with the histone 3′ end formation. Histone and poly(A) processing complexes share many factors including the putative endonuclease, which cleaves the transcript (Kolev and Steitz, 2005). In addition, the U2 snRNP component SF3b was shown to enhance both histone and PAS processing (Kyburz et al, 2006; Friend et al, 2007). Thus, U1 snRNP binding to 3′UTRs targets a factor or activity unique to PAS 3′ end processing. Possible candidates are cleavage factors I and II as they are unique to PAS processing (Danckwardt et al, 2008; Figure 6). As an alternative, PAS selection itself makes the process vulnerable to U1 binding (Figure 6). The AAUAAA hexamer is recognized by CPSF160 (Di Giammartino et al, 2011). In contrast, site selection in histone genes functions via a conserved stem-loop structure and its cognate binding protein (Dominski and Marzluff, 2007). Very recently, the nuclear version of the poly-A binding protein (PABP-N1) was shown to be a repressor of 3′ end formation at non-canoncial PAS (Jenal et al, 2012). It will be interesting to examine PAS selection and the interplay between U1 snRNP, CPSF160 and PABP-N1 in the future.
The wild-type p14 sequence can bind U1 snRNP in vitro and constitutes a weak 5′SS in vivo, suggesting an U1 snRNP-dependent regulation of endogenous P14 expression (Supplementary Figures S4 and S6). This is supported by our observation that blocking U1 snRNA by morpholinos enhanced p14 expression (Figure 4). A similar mechanism is exploited both by bovine (Furth et al, 1994) and by human papillomavirus, where four weak 5′SS are followed by binding sites for CUGBP-1 (Goraczniak and Gunderson, 2008). Interestingly, the p14 5′SS is preceded by a CUGBP-1 binding site (Figure 1C). It will be of high interest to address CUGBP’s role in the regulation of p14 3′ end processing. In addition, the cellular U1A gene is also regulated via U1 snRNP-mediated suppression (Guan et al, 2007). Recently, a study of soluble isoforms of receptor tyrosine kinases revealed that PCPA is also functional in this group of genes and may be important for their regulation (Vorlova et al, 2011).
In conclusion, we established a novel pathogenic mechanism for a 3′UTR mutation. Intriguingly, the factor IX (F9) 3′UTR mutation A-G, +1.156 leading to severe haemophilia B (Vielhaber et al, 1993) may also create a 5′SS in a similar distance to the PAS compared to p14. Thus, 5′SS created by point mutations within 3′UTRs illustrate not only a novel mechanism for a primary immunodeficiency, but may also be at the base for other disorders characterized by defective mRNA biogenesis.
Materials and methods
Plasmids
The reporter plasmids p14 and p14 mut were generated from a SV40 promoter-driven pGL3 plasmid (Promega) by replacing firefly luciferase with the p14 cDNA amplified from human neutrophils. The SV40 late poly(A) signal was replaced by the authentic p14 3′UTR including 314 bp of downstream genomic sequences. The p14 opt and p14 U2C mutants were generated by PCR mutagenesis. The terminal p14 intron was amplified from human genomic DNA and cloned into the pre-existing Van91I/BsmI sites. The H2A promoter was amplified using the EWP22 plasmid as a template (kind gift of E Wagner, Houston, USA) and inserted into the p14 plasmid. The histone 3′end was fused to p14 by an overlap PCR using EWP22 and the p14 plasmids as templates. The U1 snRNA mutant was cloned by PCR mutagenesis using a forward primer containing the U1 mutation. The PCR product was cloned as a BamHI/BglII fragment into the pUC19 U1wt plasmid (kind gift from A Weiner, Seattle, USA). The splicing reporter harbouring the second BGI from rabbit was constructed by shuffling the eGFP ORF from pEGFP-N1 into pcDNA3. The spleen focus forming virus (SF) promoter was inserted and the BGI was fused to the SF promoter by overlap PCR. Finally, the BGI 5′SS was replaced with the p14, p14 mut, and p14 opt sites.
Cells and transfections
HeLa, 293T, and 293 cells were grown in DMEM supplemented with 10% fetal calf serum, 1 mM sodium pyruvate, and 1% penicillin/streptomycin. The day before transfection, 6 × 105 HeLa cells were seeded into a 6-cm dish. Transfections were performed using Icafectin 441 (Eurogentec, Brussels) and 3 μg p14 plasmid plus 0.2 μg of a GFP plasmid as a transfection control. For the U1 suppressor experiments, 0.8 μg of the p14 int plasmid was co-transfected with 2.4 μg of the respective U1 plasmids. Medium was changed 16 h post transfection and cells were harvested after 36 h. The morpholino experiments were performed as follows. 1.5 × 106 cells were seeded into 6 cm dishes. Twenty-four hours later, cells were transfected as described above. Four hours later cells were trypsinized and counted. In all, 9 × 105 cells were incubated with 10 μM morpholinos. The morpholino/cell solution was microporated using the Neon™ microporator (Invitrogen, Darmstadt, Germany). In all, 1 × 105 cells were microporated in a 10-μl tip. This procedure was repeated nine times to reach 9 × 105 cells per six well. Cells were harvested 16 h later. In case of 293T cells, 5 × 106 cells were seeded into a 10-cm plate the day before transfection. Transfections were performed using the calcium phosphate precipitation method. For the splicing reporter, 5 μg of the pSF BGI and 0.5 μg of a DsRed Express encoding plasmid to assess transfection efficiency were used. Medium was changed 8 h post transfection and cells were harvested after 48 h. 293 cells were cultivated and transfected under the same conditions as the 293T cells with 5 μg of the H2A p14 plasmids. The transfected cells were first synchronized using a double thymidine block as described (Whitfield et al, 2000).
RNA preparation and analysis
RNA methods were performed as described previously (Zychlinski et al, 2009). For detection of p14 RNA, a specific probe corresponding to the p14 cDNA was generated from the p14 plasmid by NcoI/BamHI digestion. The GAPDH-specific probe was prepared by an EcoRI digestion of a GAPDH plasmid (gift of K Habers, HPI Hamburg). For reverse transcription, 5 μg of total RNA was DNase digested using the Ambion (Austin, TX, USA) TURBO™ DNase protocol. In all, 900 ng of this RNA was reverse transcribed using the MoMLV-RT (Fermentas, St Leon-Rot, Germany). The RT reaction was performed using a mixture of an oligo dT primer and a GFP-specific primer (rv: 5′-GGACTGGGTGCTCAGGTAGTGG-3′). The final PCR amplification utilized a forward primer (fw: 5′-GTCCTCCGATTGATCTAGAGCGGCATTGG-3′) and the reverse GFP primer. The resulting PCR fragments were gel-purified, subcloned into pCR2.1 vector (Invitrogen, Karlsruhe, Germany) and sequenced.
For the RPA analysis, a PCR fragment of the p14 3′UTR encompassing nucleotides 23 (site of the mutation) to 321 was PCR amplified using primers riboprobe fw (5′-AAGAAAAGAGAAATGACCATTTGGAGGGGC-3′) and SP6 rv 5′-GATTTAGGTGACACTATAGGTCATGCCATTGGTGAGGAC-3′). The SP6 promoter is highlighted in bold face letters. The fragment was cloned into the pCR2.1 vector (Invitrogen). The 358-bp template DNA for in vitro transcription was generated by PCR using the primers pCR2.1 p14 as (5′-GGATCCACTAGTAACGGCCGCCAGTGT-3′) and pCR2.1 p14 Sp6 as (5′-ATTTAGGTGACACTATAGAGGTCATGCCATTGGTGAGGAC-3′). The amplicon was purified with PCR-purification columns (Sigma-Aldrich). RNA probes were synthesized using 20U SP6 RNA polymerase (Fermentas), 0.74 MBq of 32P-labelled α[32P]-UTP (Hartmann Analytic), 1 × transcription buffer (Fermentas), 60 ng template DNA, 0.5 mM AGC mix, and 100 μM UTP. The RNA probe was subsequently purified by Urea (8%)-PAGE. The RPAs were performed using the RPAIII kit (Ambion) according to manufacturer’s manual. RPA products were separated by Urea-PAGE and visualized by autoradiography.
For the RT–qPCR, 900 ng of total RNA from transfected HeLa cells was digested with TurboDNase (Ambion) and purified with RNeasy (Qiagen). The reverse transcription (RT) of p14 RNA and U1 snRNA was performed using Quantitect (Qiagen) and RevertAid (Thermo Scientific) Reverse Transcriptase, respectively. Detection and quantification were performed as previously described (Zychlinski et al, 2009). As standards, serial dilutions of pGL3 SV40 p14 wt lint and pUC19 U1 wt constructs were used.
Bioinformatic analysis
The p14 3′UTR was analysed for putative SS using the SS prediction program neural network (NNSPLICE 0.9) embedded in the fruit fly website (http://www.fruitfly.org/seq_tools/splice.html). The identified 5′SS was further scored using the Analyzer Splice Tool accessible at http://ibis.tau.ac.il/ssat/SpliceSiteFrame.htm, based on the Shaprio score (Shapiro and Senapathy, 1987). Alignments were extracted from UCSC Genome Browser hg18 (Kent et al, 2002). Conservation of the p14 3′UTR (chr1:154294789–15429495) was calculated for 9 primates, 32 mammalian, and 44 vertebrates separately using phastcons (Siepel et al, 2005). Binding site predictions for the RNA-binding proteins were calculated using the SFmap algorithm (Akerman et al, 2009), applying the default parameters (Paz et al, 2010).
Statistics
The mean average and the standard deviation of the individual experiments were calculated and a standard two sample Student’s t-test was performed. The results were compared to a t-table (Skylab) to determine the P-value.
Supplementary Material
Acknowledgments
We would like to thank Sven Danckwardt, Zbigniew Dominski, and John Castle for helpful discussions and especially Aaron Goldstrohm for critical reading of the manuscript. We are grateful to Eric Wagner for providing reagents; Vanessa Melhorn, Natalia Bulanõs, and Darya Haas for technical help; and Thomas Schulz for ongoing aid. This work was funded by DFG grant BO 2512/2-1 to J Bohne and Israeli-Lower Saxony cooperation grant VWZN2628 to J Bohne and Y Mandel-Gutfreund. E-M Schrom and J Bodem were supported by DFG BO 3006/2-1. Christoph Klein is supported by the DFG Gottfried-Wilhelm-Leibniz program and the BMBF E-RARE program.
Author contributions: JL, EMS, MR, JB and JB designed and performed the experiments and analysed the data. DZ provided constructs and cloning strategies. AR and YM-G performed the bioinformatic analysis. AS and GB contributed constructs and performed initial experiments. JL, CK, and JB supervised the study and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akerman M, David-Eden H, Pinter RY, Mandel-Gutfreund Y (2009) A computational approach for genome-wide mapping of splicing factor binding sites. Genome Biol 10: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Riccio A (2009) To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474 [DOI] [PubMed] [Google Scholar]
- Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J (2002) Downstream sequence elements with different affinities for the hnRNP H/H’ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res 30: 1842–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Keller W (1999) Last but not least: regulated poly(A) tail formation. Cell 99: 9–11 [DOI] [PubMed] [Google Scholar]
- Bohn G, Allroth A, Brandes G, Thiel J, Glocker E, Schaffer AA, Rathinam C, Taub N, Teis D, Zeidler C, Dewey RA, Geffers R, Buer J, Huber LA, Welte K, Grimbacher B, Klein C (2007) A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med 13: 38–45 [DOI] [PubMed] [Google Scholar]
- Bohne J, Wodrich H, Krausslich HG (2005) Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5′ end. Nucleic Acids Res 33: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Zahler AM (2002) SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J 21: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN (2006) A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet 120: 1–21 [DOI] [PubMed] [Google Scholar]
- Conne B, Stutz A, Vassalli JD (2000) The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med 6: 637–641 [DOI] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA (2006) Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell 24: 943–953 [DOI] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M (1999) Inefficient processing impairs release of RNA from the site of transcription. EMBO J 18: 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE (2008) 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J 27: 482–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE (2007) Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J 26: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Kerr A, West S (2012) Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J 31: 2566–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL (2011) Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF (2007) Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 396: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI (2003) Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci USA 100: 8264–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Lovejoy AF, Steitz JA (2007) U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation. Mol Cell 28: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke H, Ohno M (2008) Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res 36: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC (1994) Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol 14: 5278–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM (2005) Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev 19: 2517–2521 [DOI] [PubMed] [Google Scholar]
- Goraczniak R, Behlke MA, Gunderson SI (2009) Gene silencing by synthetic U1 adaptors. Nat Biotechnol 27: 257–263 [DOI] [PubMed] [Google Scholar]
- Goraczniak R, Gunderson SI (2008) The regulatory element in the 3′-untranslated region of human papillomavirus 16 inhibits expression by binding CUG-binding protein 1. J Biol Chem 283: 2286–2296 [DOI] [PubMed] [Google Scholar]
- Gray NK, Coller JM, Dickson KS, Wickens M (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J 19: 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan F, Caratozzolo RM, Goraczniak R, Ho ES, Gunderson SI (2007) A bipartite U1 site represses U1A expression by synergizing with PIE to inhibit nuclear polyadenylation. RNA 13: 2129–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW (1998) U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264 [DOI] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kuhn U, Menzies FM, Vrielink JA, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, Agami R (2012) The poly(a)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 538–553 [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G (2010) U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468: 664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammler S, Lykke-Andersen S, Jensen TH (2008) The RNA exosome component hRrp6 is a target for 5-fluorouracil in human cells. Mol Cancer Res 6: 990–995 [DOI] [PubMed] [Google Scholar]
- Kazerouninia A, Ngo B, Martinson HG (2010) Poly(A) signal-dependent degradation of unprocessed nascent transcripts accompanies poly(A) signal-dependent transcriptional pausing in vitro. RNA 16: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA (2005) Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev 19: 2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W (2006) Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Mol Cell 23: 195–205 [DOI] [PubMed] [Google Scholar]
- Lu S, Cullen BR (2003) Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan L, Torchet C, Allmang C, Shipman T, Tollervey D (2005) A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol Cell Biol 25: 9996–10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Meislin SH, Moore MJ (2003) A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor T, Dal Mas A, Talotti G, Bussani E, Pagani F (2011) Intron cleavage affects processing of alternatively spliced transcripts. RNA 17: 1604–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz I, Akerman M, Dror I, Kosti I, Mandel-Gutfreund Y (2010) SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res 38: W281–W285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2011) Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C (2009) Assembly of an export-competent mRNP is needed for efficient release of the 3′-end processing complex after polyadenylation. Mol Cell Biol 29: 5327–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J (2009) Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol 21: 394–402 [DOI] [PubMed] [Google Scholar]
- Rigo F, Martinson HG (2008) Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol 28: 849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15: 7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A (2008) Messenger RNA regulation: to translate or to degrade. EMBO J 27: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Pruijn GJ (2010) The human exosome and disease. In Advances in Experimental Medicine and Biology Jensen TH (ed.)Vol. 702: RNA Exosome, Springer, pp132–142 [PubMed] [Google Scholar]
- Teis D, Taub N, Kurzbauer R, Hilber D, de Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, Klein C, Hess MW, Huber LA (2006) p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J Cell Biol 175: 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber E, Jacobson DP, Ketterling RP, Liu JZ, Sommer SS (1993) A mutation in the 3′ untranslated region of the factor IX gene in four families with hemophilia B. Hum Mol Genet 2: 1309–1310 [DOI] [PubMed] [Google Scholar]
- Vorlova S, Rocco G, Lefave CV, Jodelka FM, Hess K, Hastings ML, Henke E, Cartegni L (2011) Induction of antagonistic soluble decoy receptor tyrosine kinases by intronic PolyA activation. Mol Cell 43: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- West S, Proudfoot NJ (2009) Transcriptional termination enhances protein expression in human cells. Mol Cell 33: 354–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF (2000) Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol 20: 4188–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM (1986) A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835 [DOI] [PubMed] [Google Scholar]
- Zychlinski D, Erkelenz S, Melhorn V, Baum C, Schaal H, Bohne J (2009) Limited complementarity between U1 snRNA and a retroviral 5′ splice site permits its attenuation via RNA secondary structure. Nucleic Acids Res 37: 7429–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.