Abstract
Nuclear pore complexes (NPCs) fuse the two membranes of the nuclear envelope (NE) to a pore, connecting cytoplasm and nucleoplasm and allowing exchange of macromolecules between these compartments. Most NPC proteins do not contain integral membrane domains and thus it is largely unclear how NPCs are embedded and anchored in the NE. Here, we show that the evolutionary conserved nuclear pore protein Nup53 binds independently of other proteins to membranes, a property that is crucial for NPC assembly and conserved between yeast and vertebrates. The vertebrate protein comprises two membrane binding sites, of which the C-terminal domain has membrane deforming capabilities, and is specifically required for de novo NPC assembly and insertion into the intact NE during interphase. Dimerization of Nup53 contributes to its membrane interaction and is crucial for its function in NPC assembly.
Keywords: nuclear envelope formation, nuclear pore complex assembly, nuclear membrane, Nup35, Nup53
Introduction
The defining feature of the eukaryotic cell is the compartmentalization of genetic material inside the nucleus. The spatial and temporal separation of transcription and translation has enabled eukaryotes to achieve a level of regulatory complexity that is unprecedented in prokaryotes. This is accomplished by the nuclear envelope (NE), which serves as the physical barrier between the cytoplasm and the nucleoplasm. Nuclear pore complexes (NPCs) are the exclusive gateways in the NE allowing diffusion of small substances and regulated trafficking of macromolecules up to a size of 15 nm for ribosomal subunits or even 50 nm for Balbiani ring particles (for review, see Wente and Rout, 2010; Hoelz et al, 2011; Bilokapic and Schwartz, 2012). NPCs form large pores in the NE, having a diameter of ∼130 nm. Unlike other transport channels, NPCs span two lipid bilayers, at sites where the outer and inner membranes of the NE are fused. Therefore, NPCs are assumed to play an important role in deforming these membranes to form a pore as well as in stabilizing this highly curved pore membrane (Antonin et al, 2008).
Given the structural complexity and extraordinary transport capabilities of NPCs, it is quite surprising that these huge macromolecular assemblies of 40–60 MDa are only composed of ∼30 different proteins. These nucleoporins (Nups) can be roughly categorized into those forming the structure of the pore and those mediating transport through the NPC. The latter class is characterized by a high number of FG repeats. Two evolutionary conserved subcomplexes form the major part of the structural scaffold. The Nup107–160 complex in metazoa, or the corresponding Nup84 complex in yeast, is the best characterized of these owing to an extensive set of genetic, biochemical and structural data. Computational and structural analyses suggest that this complex is related to vesicle coats (Devos et al, 2004; Mans et al, 2004; Brohawn et al, 2008). It is possible that these proteins form a coat-like assembly stabilizing the curved pore membrane of the NPC, which is analogous to clathrin or COPI and II during vesicle formation (Field et al, 2011; Onischenko and Weis, 2011). Notably, neither clathrin nor COP coats interact directly with the lipid bilayers. They are rather linked to the deformed membrane via adaptor and integral proteins (McMahon and Mills, 2004). Although three integral membrane proteins are known in the vertebrate NPC, it is unclear how a possible link between the Nup107–160 complex and the membrane is established.
The second major structural and evolutionarily conserved subcomplex of the NPC is the metazoan Nup93 complex, Nic96 in yeast, which might serve as a link to the pore membrane. In vertebrates, it is composed of five nucleoporins: Nup205, Nup188, Nup155, Nup93 and Nup53. Nup53, also referred to as Nup35 (Cronshaw et al, 2002), interacts with Nup93 and Nup155 (Hawryluk-Gara et al, 2005), corresponding to Nup170 and Nic96 in yeast (Marelli et al, 1998; Fahrenkrog et al, 2000). Nup155, Nup93 and Nup53 are each indispensable for NPC formation in vertebrates (Franz et al, 2005; Hawryluk-Gara et al, 2008; Mitchell et al, 2010; Sachdev et al, 2012). Interestingly, Nup53 and its corresponding yeast homologues Nup53p and Nup59p interact with the transmembrane nucleoporin NDC1, thereby potentially linking the NPC to the pore membrane (Mansfeld et al, 2006; Onischenko et al, 2009). Although NDC1 is essential in both vertebrates and yeast (Winey et al, 1993; West et al, 1998; Mansfeld et al, 2006; Stavru et al, 2006) it is not found in all eukaryotes (Mans et al, 2004; DeGrasse et al, 2009; Neumann et al, 2010), suggesting that in some organisms NPCs can form in the absence of a Nup53–NDC1 interaction. Indeed, Aspergillus nidulans is viable in the absence of all three known transmembrane nucleoporins (Liu et al, 2009). This suggests that there are alternative modes of interaction between the nucleoporins and the pore membrane.
Here, we show that Nup53 binds membranes directly and independently of other proteins. It possesses two membrane interaction regions, which are important for NPC assembly. Although either site is sufficient for NPC assembly at the end of mitosis, the C-terminal membrane binding site of Nup53 is specifically required for NPC assembly during interphase, probably because of its membrane deforming capabilities.
Results
Nup53 is a membrane binding protein
Overexpression of Nup53 in yeast causes expansion of the NE (Marelli et al, 2001). Similar membrane proliferation phenotypes have been observed upon overexpression of nuclear membrane binding proteins, such as lamin B (Prufert et al, 2004). Yeast Nup53 contains a C-terminal region predicted to form an amphipathic helix (Marelli et al, 2001; Patel and Rexach, 2008), which could serve as a membrane binding module. However, Nup53 interacts with the integral pore membrane protein NDC1 in both yeast and metazoa (Mansfeld et al, 2006; Onischenko et al, 2009) and thus might be linked to the membrane via this interaction. We therefore tested whether Nup53 is able to interact with membranes independently of other proteins.
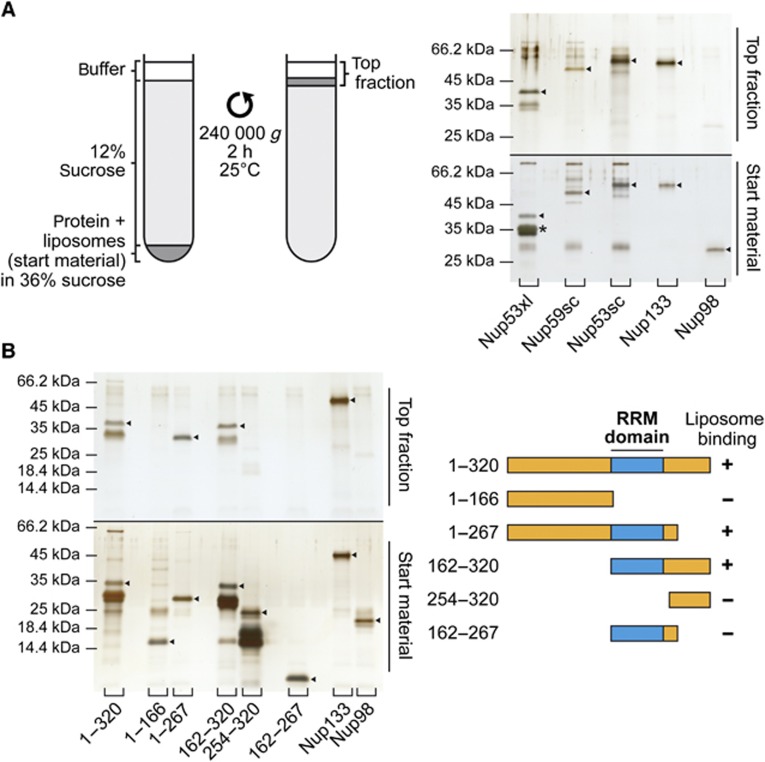
To assay for membrane binding, we generated liposomes with an average radius of ∼150 nm. These liposomes were incubated with different, recombinantly expressed nucleoporins and floated through sucrose cushions. Liposome binding proteins were recovered after centrifugation from the top fraction (Figure 1A). A Nup133 membrane binding fragment (Drin et al, 2007) was used as a positive control and found in the liposome containing top fraction (Figure 1A). Similarly, Xenopus Nup53 was found in the top fraction, indicating membrane interaction. In contrast, a fragment of the FG repeat-containing nucleoporin Nup98 did not bind liposomes. Thus, Xenopus Nup53 binds directly to membranes independently of other interacting proteins. To determine whether this feature is conserved during evolution, we tested the two yeast homologues Nup53p and Nup59p, which both bound liposomes (Figure 1A).
Figure 1.
Nup53 directly binds membranes. (A) 3 μM recombinant Xenopus Nup53 (Nup53xl), the two yeast orthologues Nup59sc and Nup53sc as well as fragments of Nup133 and Nup98 as positive and negative controls, respectively, were incubated with 6 mg/ml fluorescently labelled liposomes prepared from E. coli polar lipids and floated through a sucrose gradient as indicated on the left. Top fractions of the gradient, as well as 3% of the starting material, were analysed by SDS–PAGE and silver staining. Please note that Nup53 is sensitive to C-terminal degradation (*) and that the full-length protein significantly enriched in the liposome bound fraction. (B) Full-length (1–320) and different fragments of Xenopus Nup53 as well as fragments of Nup133 and Nup98 were analysed for liposome binding as in (A). Only fragments comprising the RRM domain (indicated in blue in the schematic representation) bound liposomes.
We next sought to define the regions of Xenopus Nup53 important for its membrane interaction. Nup53 can be roughly divided into three parts: the N-terminus (amino acid (aa) 1–166), a middle region (aa 166–267) comprising a conserved RNA recognition motif (RRM) domain and the C-terminus (aa 267–320) (Figure 1B). We generated different N- and C-terminal truncations of Nup53 and tested them for liposome binding (Figure 1B). While full-length Nup53 (aa 1–320) bound to liposomes, the N-terminal region of the protein (aa 1–166) showed no binding. Extending this fragment by 100 aa to include the RRM domain rendered the protein capable of membrane binding (aa 1–267). The C-terminal half of Nup53 (aa 162–320), which included the RRM domain, also interacted with liposomes. However, a fragment consisting of only the C-terminal region of Nup53 but lacking the RRM domain (aa 254–320) could not bind liposomes. Surprisingly, a fragment comprising only the RRM domain (aa 162–267) did not bind liposomes showing that the RRM domain is necessary but not sufficient for Nup53 membrane binding.
Nup53 dimerization is necessary for membrane binding and NPC formation
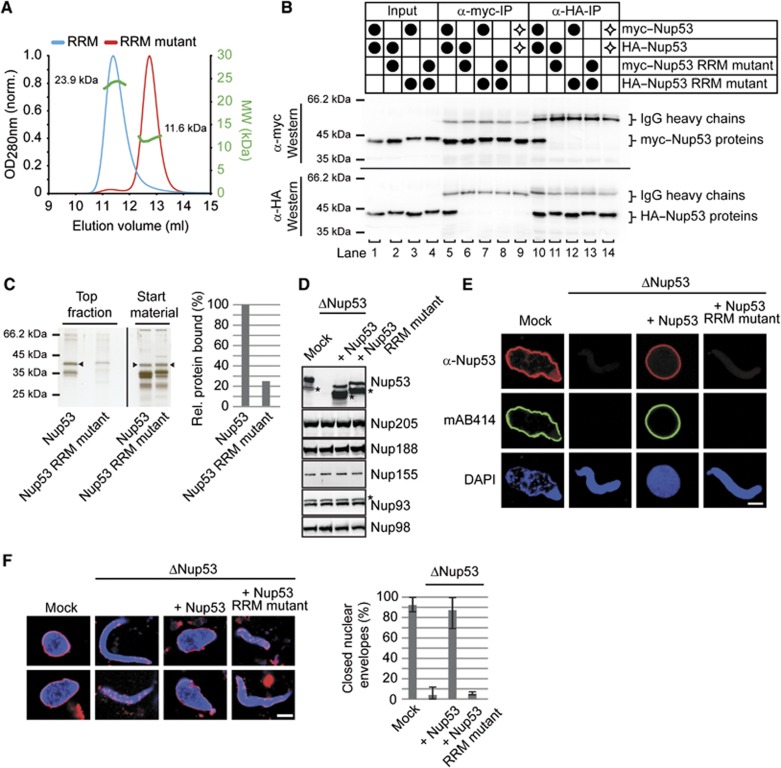
As the RRM domain is crucial for Nup53 membrane interaction we investigated the function of this domain. The crystal structure of the mouse domain suggests that it acts as a dimerization rather than an RNA binding module (Handa et al, 2006). We designed a mutant of this domain by exchanging two amino acids (F172E/W203E) in the dimerization surface. Size exclusion chromatography in combination with multi-angle laser light scattering revealed that the resulting fragment was monomeric (Figure 2A).
Figure 2.
Dimerization of the RRM domain is essential for Nup53 membrane binding and nuclear pore complex formation. (A) Size exclusion chromatography on a Superdex75 10/300 GL column followed by multi-angle static laser light scattering of the Xenopus Nup53 RRM domain and the F172E/W203E mutant, which rendered the domain monomeric. The green dots relate to the secondary axis and show the molecular weight of the eluting particles. (B) HeLa cells were co-transfected with myc- and HA-tagged Xenopus Nup53 wild-type protein and/or dimerization mutant as indicated (●). Proteins were immunoprecipitated from cell lysates with α-myc or α-HA antibodies, and analysed by western blotting. Ten per cent of the start materials are loaded as input. To exclude complex formation after cell lysis, extracts from single transfected myc–Nup53 and HA–Nup53 cell batches were mixed and processed for immunoprecipitation ( ). Under these conditions, no co-precipitation was observed. (C) Full-length Xenopus Nup53 and the respective F172E/W203E mutant (RRM mutant) were analysed for liposome binding as in Figure 1. The right panel shows the quantitation of liposome binding analysed by western blotting and normalized to the levels of the wild-type protein (one out of two independent experiments). (D) Western blot analysis of mock, Nup53-depleted (ΔNup53) and Nup53-depleted extracts supplemented with recombinant wild-type protein (Nup53) or the dimerization mutant (Nup53 RRM mutant), respectively. Recombinant proteins were added to approximate endogenous Nup53 levels (judged by the full-length protein, please note for both endogenous and recombinant Nup53 proteins C-terminal degradation products (*)). The recombinant Nup53 migrated slightly faster than the endogenous protein due to absence of eukaryotic post-translational modifications. The Nup93 antibody recognizes a slightly slower migrating cross-reactivity by western blotting (*). (E) Nuclei were assembled in mock, Nup53-depleted extracts or Nup53-depleted extracts supplemented with wild-type protein (Nup53) or the dimerization mutant for 120 min, fixed with 4% PFA and analysed with Nup53 antibodies (red) and mAb414 (green). Chromatin was stained with DAPI. Bar: 10 μm. (F) Nuclei were assembled as in (E), fixed with 2% PFA and 0.5% glutaraldehyde and analysed for chromatin (blue: DAPI) and membrane staining (red: DiIC18, bar: 10 μm). Right panel shows the quantitation of chromatin substrates with a closed nuclear envelope (averages of three independent experiments with >300 randomly chosen chromatin substrates per sample, error bars represent the range).
). Under these conditions, no co-precipitation was observed. (C) Full-length Xenopus Nup53 and the respective F172E/W203E mutant (RRM mutant) were analysed for liposome binding as in Figure 1. The right panel shows the quantitation of liposome binding analysed by western blotting and normalized to the levels of the wild-type protein (one out of two independent experiments). (D) Western blot analysis of mock, Nup53-depleted (ΔNup53) and Nup53-depleted extracts supplemented with recombinant wild-type protein (Nup53) or the dimerization mutant (Nup53 RRM mutant), respectively. Recombinant proteins were added to approximate endogenous Nup53 levels (judged by the full-length protein, please note for both endogenous and recombinant Nup53 proteins C-terminal degradation products (*)). The recombinant Nup53 migrated slightly faster than the endogenous protein due to absence of eukaryotic post-translational modifications. The Nup93 antibody recognizes a slightly slower migrating cross-reactivity by western blotting (*). (E) Nuclei were assembled in mock, Nup53-depleted extracts or Nup53-depleted extracts supplemented with wild-type protein (Nup53) or the dimerization mutant for 120 min, fixed with 4% PFA and analysed with Nup53 antibodies (red) and mAb414 (green). Chromatin was stained with DAPI. Bar: 10 μm. (F) Nuclei were assembled as in (E), fixed with 2% PFA and 0.5% glutaraldehyde and analysed for chromatin (blue: DAPI) and membrane staining (red: DiIC18, bar: 10 μm). Right panel shows the quantitation of chromatin substrates with a closed nuclear envelope (averages of three independent experiments with >300 randomly chosen chromatin substrates per sample, error bars represent the range).
To confirm that the dimerization occurs also in vivo, we performed co-transfection experiments in HeLa cells using HA- and myc-tagged Xenopus Nup53. Either α-HA or α-myc antibodies immunoprecipitated both HA- and myc-tagged wild-type Nup53 indicating that the proteins formed a complex (Figure 2B, lanes 5 and 10). If cells were transfected with either construct separately before they were mixed for protein extraction, then no co-immunoprecipitation was observed (Figure 2B, lanes 9 and 14) demonstrating that complex formation cannot occur under the conditions of the immunoprecipitation. Co-transfections of RRM mutants as well as RRM mutants and wild-type protein did not result in complex formation (Figure 2B, lanes 6–8 and 11–13) indicating that the F172E/W203E exchange inhibited dimerization/oligomerization.
Next, we tested the effect of these mutations on membrane binding. In the context of the full-length protein, these mutations decreased liposome binding by 70% (Figure 2C) suggesting that the dimerization of Nup53 is important for its membrane interaction.
As Nup53 is essential for postmitotic NPC formation (Hawryluk-Gara et al, 2008) we examined the relevance of Nup53 membrane binding for this. We employed Xenopus laevis egg extracts to study nuclear reformation in vitro (Lohka, 1998). With antibodies against Nup53 we depleted the endogenous protein without co-depletion of other nucleoporins including the other members of the Nup93 complex: Nup93, Nup155, Nup205 and Nup188 (Figure 2D). These depleted extracts were incubated with sperm heads serving as chromatin template. In the absence of Nup53, NPC formation was blocked (Figure 2E) as reported (Hawryluk-Gara et al, 2008). This was indicated by the absence of immunofluorescent signal on the chromatin surface for mab414, an antibody recognizing several nucleoporins that represent a major subfraction of the NPC. Addition of recombinantly expressed wild-type Nup53 to the depleted extracts at levels similar to the endogenous protein (Figure 2D) restored NPC formation. The recombinant protein was integrated into NPCs as indicated by immunostaining with a Nup53 antibody. In contrast, the dimerization and membrane binding defective Nup53 mutant (1–320 F172E/W203E) was unable to substitute for the endogenous protein in NPC formation (Figure 2E).
Individual depletion of several nucleoporins essential for NPC assembly from Xenopus egg extracts also blocks formation of a closed NE. These nucleoporins include POM121, NDC1, Nup155, Nup93 (Antonin et al, 2005; Franz et al, 2005; Mansfeld et al, 2006; Sachdev et al, 2012) and Nup53 (Hawryluk-Gara et al, 2008). Upon Nup53 depletion, membrane vesicles bound to the chromatin surface but did not fuse to form a closed NE (Figure 2F; Hawryluk-Gara et al, 2008). This phenotype was rescued by the wild-type Nup53 protein, but not by the dimerization defective mutant. Together with the liposome-binding assay, this suggests that Nup53 membrane binding could be important for NPC assembly and formation of a closed NE. However, we cannot exclude that the RRM mutant also affects interaction of Nup53 with other nucleoporins. Indeed, in GST pull-down assays we observed a slight reduction of Nup93, Nup205 and Nup155 binding to Nup53 in the context of the RRM mutant as compared to the wild-type protein (Supplementary Figure S1A). In contrast, NDC1 binding to Nup53 was unaffected by the dimerization mutant (Supplementary Figure S1B).
Nup53 possesses two membrane binding regions
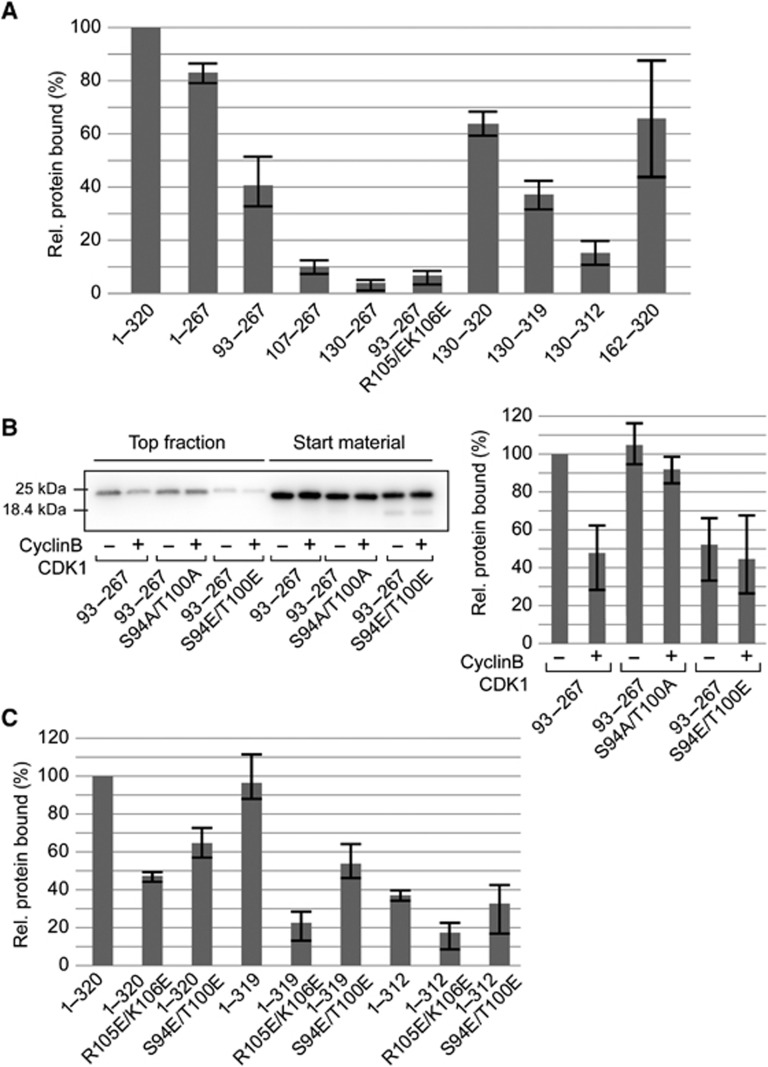
These results reveal a crucial role for Nup53 dimerization via its RRM domain in NE reformation. However, the RRM domain alone did not bind to membranes. We tested different Nup53 truncations for liposome binding to map the membrane interaction sites (Figure 3A; Supplementary Figure S2 shows the purity of all recombinant proteins used in the different liposome experiments). An N-terminal fragment of Nup53 including the RRM domain (aa 1–267) bound to liposomes with only a slightly decreased binding efficiency, 83% of the levels of the full-length protein. Further truncations from the N-terminus revealed a minimal membrane binding region between aa 93 and 107 as indicated by a four-fold decrease in liposome binding upon removal of these 15 amino acids. This region comprises a patch of basic residues, which as in other membrane binding proteins might be important for the interaction with negatively charged lipids. Indeed, changing two residues to negatively charged residues (R105E/K106E) abolished membrane binding (Figure 3A, 83% reduction as compared to the 93–267 fragment).
Figure 3.
Nup53 possesses two independent membrane binding regions. (A) Full-length protein (1–320) and different fragments of Xenopus Nup53 were quantitatively analysed for liposome binding as in Figure 2B (normalized to the full-length protein, three independent experiments). (B) A fragment comprising the first membrane binding region and the RRM domain (93–267) as well as a phosphomimetic (93–267 S94E/T100E) and a non-phosphorylatable mutant (93–267 S94A/T100A) was treated with CyclinB/CDK1. Samples were tested for liposome binding as in Figure 1A and analysed by western blotting (left panel) and quantified (right panel: two independent experiments, normalized to liposome binding of wild-type fragment without CDK1 pretreatment). (C) Mutants/truncations affecting the N- (1–320 R105E/K106E, 1–320 S94E/T100E), the C-terminal (1–319, 1–312) as well as both (1–319 R105E/K106E, 1–319 S94E/T100E, 1–312 R105E/K106E, 1–312 S94E/T100E) membrane binding sites of Nup53 were quantitatively assayed for liposome binding in the context of full-length protein (normalized to wild-type protein (1–320), average of three independent experiments, error bars represent the range).
Interestingly, the N-terminal membrane binding region of Nup53 contains one of two regions that were differentially phosphorylated depending on the cell-cycle stage (Supplementary Figure S3; Supplementary Table S1). Two amino acids identified, Serine 94 and Threonine 100, are phosphorylated during mitosis, most likely by cdk 1 as they possess a consensus site for this kinase (Blethrow et al, 2008). To investigate the impact of this modification on membrane binding, we generated mutants mimicking the phosphorylated (S94E/T100E) and the unphosphorylated (S94A/T100A) state of the protein. The phosphomimetic mutant was impaired in liposome binding (Figure 3B, reduced by 50% compared to the 93–267 fragment) while the S94A/T100A control bound to liposomes with efficiency comparable to the wild type. Furthermore, in vitro phosphorylation by CyclinB/CDK1 reduced liposome binding of the wild-type protein by 50%, levels similar to the phosphomimetic mutant (S94E/T100E), but did not affect the S94A/T100A mutant, suggesting that mitotic phosphorylation regulates the membrane binding of Nup53.
The C-terminal part of Nup53 also interacts with membranes, an activity that requires the presence of the RRM domain (Figure 1B). Fragments comprising both regions showed efficient binding to liposomes (aa 130–320 and 162–320) (Figure 3A). The second membrane binding region was mapped to the absolute C-terminus of Nup53 as deletion of the last amino acid reduced liposome binding by 42% (aa 130–319) and removal of the last eight amino acids abolished liposome binding (aa 130–312).
These data suggest that Nup53 possesses two independent membrane binding sites. Consistently, in the context of the full-length protein mutations in the N-terminal site (R105E/K106E as well as S94E/T100E) reduced liposome binding by 50 and 40% (Figure 3C). Deletion of the C-terminal amino acid had a less prominent effect, but removal of the last eight amino acids reduced liposome binding by 60%. The combination of mutations and truncations affecting both binding sites showed an additive effect supporting the view that each site is individually capable of membrane binding. Our data also suggest that the N-terminal membrane interaction is mediated via a pair of basic amino acids. The N-terminal binding site is additionally dependent on membrane curvature, the fragment binding less efficiently to more highly curved membranes (Supplementary Figure S4). Conversely, the C-terminal membrane binding site is less sensitive to membrane curvature and seems to largely depend on the last amino acid, a hydrophobic tryptophan. This could indicate that the two membrane interaction sites operate via different binding mechanisms. In both cases, the dimerization of Nup53 via the RRM domain is important: mutations in the individual membrane binding fragments (93–267 F172E/W203E and 130–320 F172E/W203E) rendering the RRM domain monomeric reduced their membrane interaction (Supplementary Figure S5).
Nup53 membrane binding is necessary for NPC formation
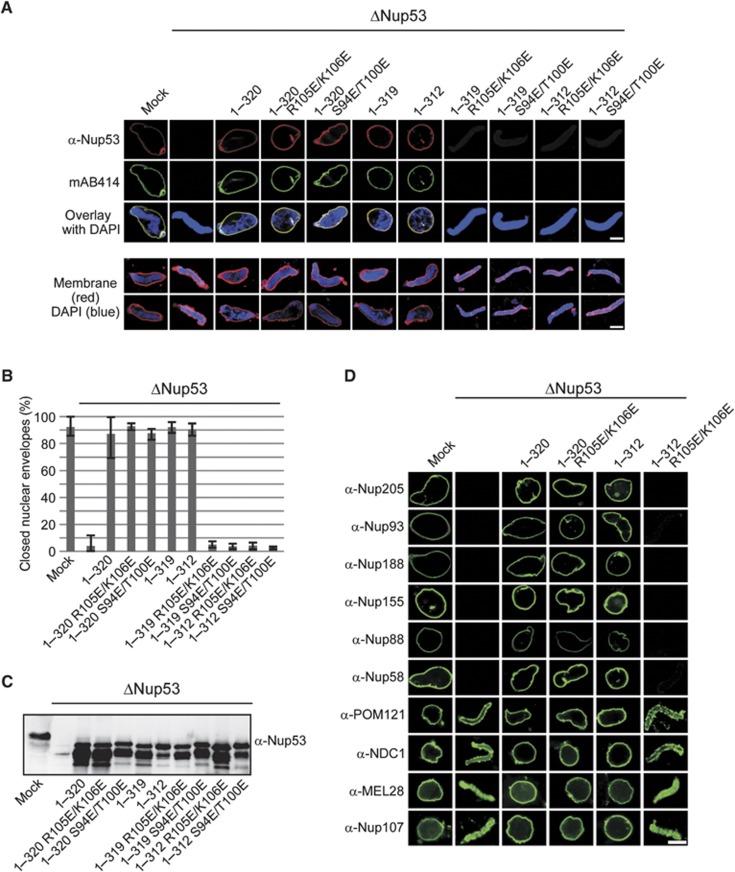
The Nup53 mutant defective in RRM dimerization, which showed reduced membrane binding, was unable to substitute for the endogenous protein in nuclear assembly (Figure 2). We therefore analysed the contribution of each of the membrane interaction sites to NPC assembly by substituting endogenous Nup53 with constructs defective in the N-terminal membrane interaction site, lacking the C-terminal membrane interaction site, or comprising a combination of both deficient sites (Figure 4A). Surprisingly, mutants of the N-terminal membrane binding site (1–320 R105E/K106E and 1–320 S94E/T100E) supported NPC formation as indicated by mAB414 staining (Figure 4A). They also supported formation of a closed NE (Figure 4A and B). Correspondingly, mutations in the N-terminal binding region did not alter the NE localization of any other nucleoporins (Figure 4D). These nucleoporins include members of the Nup93 complex (Nup93, Nup188, Nup205 and Nup155) as well as the transmembrane nucleoporins NDC1 and POM121. Accordingly, interactions of these mutants with Nup93 and Nup205, which bind the N-terminal part of Nup53 (Fahrenkrog et al, 2000; Hawryluk-Gara et al, 2008), were unaffected as shown by GST pull downs (Supplementary Figure S1A).
Figure 4.
Either of the two membrane binding regions of Nup53 is sufficient for postmitotic NPC assembly. (A) Nuclei were assembled in mock, Nup53-depleted extracts or Nup53-depleted extracts supplemented with wild-type Nup53 (1–320), constructs featuring mutations in the N-terminal membrane binding site (1–320 R105E/K106E, 1–320 S94E/T100E), constructs lacking the C-terminal membrane binding site (1–319, 1–312) or both (1–319 R105E/K106E, 1–319 S94E/T100E, 1–312 R105E/K106E, 1–312 S94E/T100E), respectively. Samples were fixed after 120 min with 4% PFA and analysed with Nup53 antibodies (red) and mAb414 (green, upper panel) or with 2% PFA and 0.5% glutaraldehyde and analysed for chromatin (blue: DAPI) and membrane staining (red: DiIC18, lower panel). Bars: 10 μm. (B) Quantitation of chromatin substrates with a closed NE was done as in Figure 2F. (C) Western blot analysis of extracts used in (A) showing the re-addition of the recombinant proteins to approximately endogenous Nup53 levels. (D) Nuclei were assembled as in (A), fixed with 4% PFA and analysed with respective antibodies. Bar: 10 μm.
The fragment lacking the C-terminal tryptophan (1–319) also supported NPC assembly and formation of a closed NE. Deletion of this tryptophan did not interfere with Nup53 binding to NDC1 or Nup155 (Supplementary Figure S1B), two binding partners interacting with the C-terminal region. The truncation lacking the last eight C-terminal amino acids (1–312) also allowed for NPC assembly and formation of a closed NE. All tested nucleoporins were located at the NE in these nuclei (Figure 4D). Notably, this truncation did not bind to NDC1 (Supplementary Figure S1B), supporting the view that the Nup53–NDC1 interaction is not required for postmitotic NPC formation (Hawryluk-Gara et al, 2008). These observations suggest that a single Nup53 membrane binding region is sufficient for NPC assembly at the end of mitosis.
In contrast to the Nup53 mutants and truncations that abrogate one membrane binding region, mutants affecting both membrane binding sites (1–319 R105E/K106E, 1–319 S94E/T100E, 1–312 R105E/K106E and 1–312 S94E/T100E) did not support NPC assembly and NE reformation (Figure 4A). Under these conditions, MEL28 (also referred to as ELYS) as well as Nup107, which both bind early to chromatin during the NPC assembly process (Galy et al, 2006; Rasala et al, 2006; Franz et al, 2007), as well as the transmembrane nucleoporins, NDC1 and POM121, were detected on the chromatin (Figure 4D). In contrast, Nup205, Nup188, Nup155 and Nup93 were not recruited. However, this lack of recruitment as well as the block in NPC assembly is unlikely to be caused by disrupting Nup53 binding to these interaction partners as the mutations introduced did not affect binding to Nup93, Nup205 and Nup155 in GST pull downs (Supplementary Figure S1A). It is also unlikely that the loss of the Nup53–NDC1 interaction caused this phenotype as the Nup53 fragment 1–312 still allowed for postmitotic NPC assembly but was not able to bind NDC1 (Supplementary Figure S1B). Nup58 representing an FG-containing nucleoporin of the central channel as well as the peripheral nucleoporin Nup88 was also absent on chromatin in the presence of a Nup53 construct that lacked both membrane binding regions (1–312 R105E/K106E) (Figure 4D). These data support the conclusion that the direct Nup53 membrane interaction is important for postmitotic NPC formation.
Interphasic NPC assembly requires the C-terminal membrane binding site of Nup53
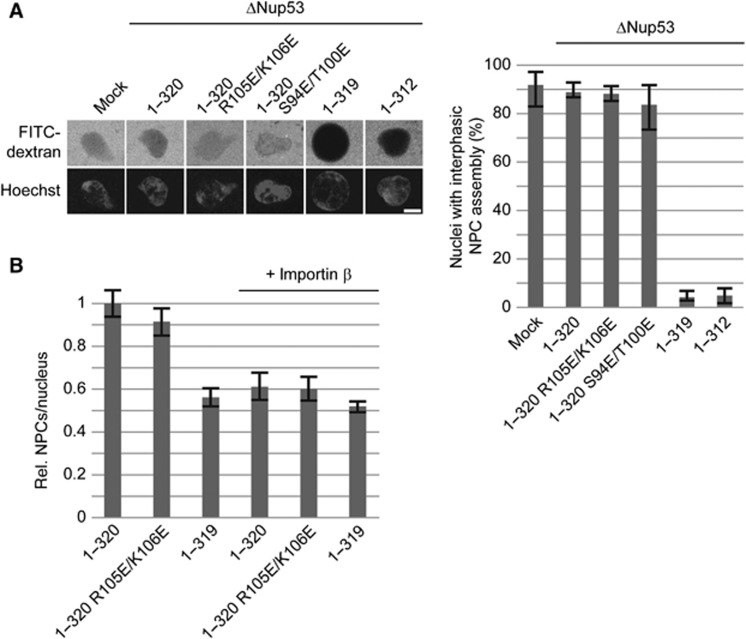
Metazoan NPC assembly occurs in two different stages of the cell cycle: at the end of mitosis when NPCs assemble concomitantly with the reforming NE and during interphase when new NPCs are assembled and integrated into the intact NE (Antonin et al, 2008; Doucet and Hetzer, 2010). Recent data suggest that there are different requirements for these two possible modes of NPC assembly (Doucet et al, 2010). The in vitro nuclear assemblies described up to now reflect the situation at the end of mitosis. We therefore assayed whether the mutants that support postmitotic NPC assembly also support NPC formation during interphase. In an experimental set-up developed by the Hetzer laboratory (Dawson et al, 2009), nuclei with newly integrated NPCs can be visualized by an influx of dextrans when nucleoporins forming the permeability barrier of newly formed NPCs are depleted. Under these conditions, mutants defective in the N-terminal membrane binding site of Nup53 (1–320 R105E/K106E and 1–320 S94E/T100E) supported interphasic NPC formation (Figure 5A). Conversely, Nup53 truncations affecting the C-terminal membrane binding site did not substitute for the wild-type protein in this mode of NPC assembly. This indicates that, in contrast to postmitotic NPC assembly where both membrane binding regions individually support NPC formation, only the C-terminal membrane binding site of Nup53 is required for interphasic NPC formation.
Figure 5.
The C-terminal Nup53 membrane binding site is essential for interphasic nuclear pore complex (NPC) formation. (A) Nuclei were preassembled in mock or Nup53-depleted extracts supplemented with wild-type full-length protein (1–320), constructs with a mutated N-terminal membrane binding site (1–320 R105E/K106E, 1–320 S94E/T100E), or constructs lacking the C-terminal membrane interaction site (1–319, 1–312), respectively. After 90 min, the samples were supplemented with cytosol depleted of Nup53 and FG-containing nucleoporins. After 60 min, FITC-labelled 70-kDa dextran and Hoechst was added. Bar: 10 μm. The right panel shows the quantitation of three independent experiments with >300 randomly chosen chromatin substrates per sample. Error bars represent the range. (B) Nuclei were assembled in Nup53-depleted extracts supplemented with wild-type full-length protein (1–320), a construct with a mutated N-terminal membrane binding site (1–320 R105E/K106E), or a construct lacking the last C-terminal amino acid (1–319), respectively, for 120 min. Where indicated, de novo NPC assembly was blocked by the addition of 2 μM importin β after 50 min and nuclei were further incubated for 70 min. For each construct, total NPC numbers per nucleus identified by mAB414 immunofluorescence were quantified from 20 nuclei in 2 independent experiments and normalized to the wild-type full-length protein. Error bars represent the s.e. of the mean.
Interestingly, the Nup53 truncation lacking the C-terminal tryptophan (1–319) did not support interphasic NPC assembly in the dextran influx assay despite the fact that membrane interaction is only slightly reduced (Figure 3C). We confirmed these findings in an independent assay directly counting NPCs identified by mAB414 immunostaining (D'Angelo et al, 2006; Theerthagiri et al, 2010). NPC numbers were determined on nuclei where NPCs assembled in the postmitotic and interphasic mode and nuclei where interphasic NPC formation was specifically blocked by addition of 2 μM importin β (D'Angelo et al, 2006; Theerthagiri et al, 2010). The NPC numbers of nuclei assembled for 120 min in the presence of recombinant wild-type Nup53 as well as the Nup53 mutant defective in the N-terminal membrane binding site (1–320 R105E/K106E) were reduced by importin β addition after 50 min, i.e., when a closed NE with intact NPC was formed (Figure 5B). In contrast, the Nup53 truncation lacking the C-terminal tryptophan showed after 120 min a lower number of NPCs, which was not sensitive to importin β addition, indicating that in this condition interphasic NPC assembly did not occur.
Nup53 can deform membranes
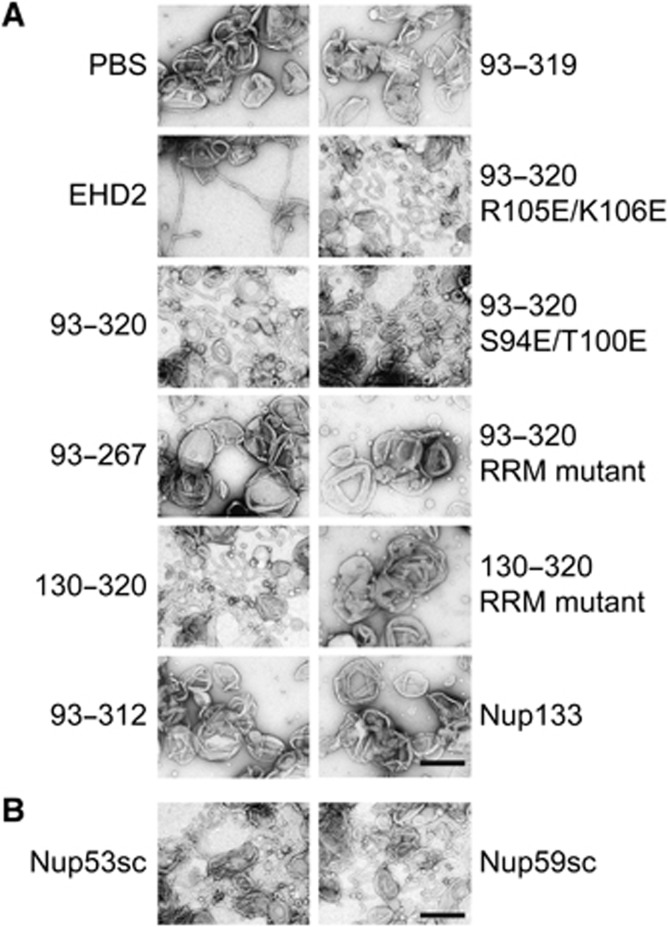
Many membrane binding proteins induce membrane deformation. Such a function is of special interest in the context of NPC formation as NPCs are integrated in the NE at sites where both nuclear membranes are deformed and fused to a pore. We therefore analysed the morphology of Nup53-bound liposomes using electron microcopy. As reported (Daumke et al, 2007), tubulation of liposomes is induced by the EH domain-containing protein EHD2 (Figure 6A). Interestingly, a Nup53 fragment containing both membrane binding sites (93–320) strongly induced liposome tubulation indicating a membrane deformation capability. A shorter fragment containing only the N-terminal membrane binding site and including the RRM domain (93–267) did not induce membrane tubulation, despite the fact that it was able to bind liposomes (Figure 3A). In contrast, a fragment comprising the RRM domain and the full C-terminus of Nup53 (130–320) strongly induced membrane tubulation indicating that the second, C-terminal membrane binding domain deforms membranes. Accordingly, fragments lacking the last eight residues (130–312), and therefore the second membrane binding site or only the last C-terminal tryptophan (130–319) did not cause membrane tubulation. Consistently, fragments mutated in the first, or N-terminal, membrane binding region of Nup53 (93–320 R105E/K106E and 93–320 S94E/T100E) induced membrane deformation. Efficient membrane binding of Nup53 depends not only on the individual membrane binding domains but also on dimerization (Figure 2C; Supplementary Figure S5). To determine the importance of the dimerization of Nup53 for membrane tubulation the RRM mutants of fragments that contained either both (93–320 RRM mutant) or only the C-terminal binding site (130–320 RRM mutant) were tested. Neither of the two fragments induced membrane tubulation emphasizing that membrane interaction is indeed required for this phenotype. The membrane binding fragment of Nup133 did not induce detectable tubulation, emphasizing that membrane binding alone does not account for membrane deformation.
Figure 6.
The C-terminus of Nup53 binds and deforms membranes. (A) Folch fraction I liposomes were incubated where indicated with 3 μM recombinant Nup53 fragments containing both (93–320), the N-terminal (93–267) or C-terminal (130–320) membrane binding sites including the RRM domain as well as fragments and mutants where the C-terminal (93–312, 93–319), the N-terminal (93–320 R105E/K106E, 93–320 S94E/T100E) membrane interaction site or the dimerization (93–320 RRM mutant and 130–320 RRM mutant) is compromised. The liposome deforming protein EHD2 (aa 1–543) and a fragment of Nup133 were used as positive and negative control, respectively. (B) 3 μM recombinant yeast Nup53 and Nup59 protein was incubated with liposomes and analysed. Bars: 400 nm.
Similar to Xenopus Nup53 both yeast isoforms—Nup53p and Nup59p—induced membrane tubulation (Figure 6B). This indicates that, in addition to membrane binding (Figure 1A), the membrane bending activity of Nup53 is conserved during evolution which suggests an important role for Nup53-induced membrane deformation in NPC formation and/or function.
Discussion
Here, we show that Nup53 binds membranes directly and independently of other proteins. We demonstrate that dimerization of the protein via its RRM domain is necessary for membrane interaction and identify two separate membrane binding regions within the protein. Binding of Nup53 to membranes is important for NPC assembly. Although either of the two membrane interaction regions is sufficient for postmitotic NPC formation, NPC assembly in interphase specifically requires the C-terminal membrane binding site, probably because of its capacity to induce membrane deformation.
Our results support the view that Nup53 is crucial for postmitotic NPC assembly in Xenopus egg extracts (Hawryluk-Gara et al, 2008). Depletion of Nup53 blocks NPC assembly and the formation of a closed NE. This phenotype is rescued by the addition of recombinant Nup53, confirming the specificity of the depletion (Hawryluk-Gara et al, 2008; Figures 2 and 4). In agreement with the cell-free data, RNAi-mediated depletion of Nup53 in HeLa cells results in severe nuclear morphology defects and reduced levels of Nup93, Nup205 and Nup155 at the nuclear rim, suggestive of defects in NPC assembly (Hawryluk-Gara et al, 2005). In C. elegans, RNAi knockdown of Nup53 as well as a deletion within the protein blocks postmitotic nuclear reformation and results in embryonic lethality (Galy et al, 2003; Rodenas et al, 2009), suggesting that Nup53 function is conserved in metazoa. Notably, Nup53 is found in all eukaryotic supergroups indicating that it is part of the NPC in the last common ancestor of eukaryotes (Neumann et al, 2010). However, its absence in some eukaryotic organisms shows that its loss can be compensated (DeGrasse et al, 2009; Neumann et al, 2010). Double deletion of both S. cerevisiae orthologues, Nup53p and Nup59p, is viable (Marelli et al, 1998; Onischenko et al, 2009). However, these strains exhibit growth defects and Nup53 becomes essential when interacting nucleoporins, including integral membrane proteins, are deleted (Marelli et al, 1998; Miao et al, 2006; Onischenko et al, 2009).
NPC assembly is a highly ordered process. In metazoa the NE and NPCs break down and reform during each round of mitosis. Postmitotic reassembly occurs on the decondensing chromatin. The earliest step involves the recruitment of the Nup107–160 subcomplex to the chromatin surface by MEL28/ELYS (Galy et al, 2006; Rasala et al, 2006; Franz et al, 2007), a DNA-binding protein that acts as a seeding point for NPC assembly. Membranes are subsequently recruited to chromatin causing an enrichment of NE/NPC-specific membrane proteins, including the transmembrane nucleoporins POM121 and NDC1 (Antonin et al, 2005; Mansfeld et al, 2006; Anderson et al, 2009). The order of these initial steps has been defined using both in vitro experiments and live-cell imaging (Dultz et al, 2008); however, less is known about the order of nucleoporin assembly following these events. MEL28 and Nup107 as well as POM121 and NDC1 containing membranes are detectable on the chromatin in Nup53-depleted nuclear assemblies (Figure 4D). The same pattern was seen in Nup93-depleted extracts (Sachdev et al, 2012). Our results suggest that Nup53, which is part of the Nup93 complex, is a key determinant for the recruitment of the other members of this complex. In the absence of Nup53, the chromatin recruitment of Nup155, Nup205, Nup188 and Nup93 was impaired (Figure 4D). Similarly, C. elegans Nup53 is necessary for the efficient accumulation of Nup155 and Nup58 but not Nup107 at the NE (Rodenas et al, 2009). This is also supported by live-cell imaging experiments in HeLa cells, which capture the recruitment of Nup58 slightly after Nup93 (Dultz et al, 2008). Accordingly, we have found that upon depletion of the two Nup93 containing subcomplexes, Nup93–Nup188 and Nup93–205, the two other members of the complex, Nup155 and Nup53, are still detectable, albeit at reduced levels on the assembling NPCs (Sachdev et al, 2012). Recruitment of the Nup62 complex to the chromatin template is prevented in the absence of both Nup53 (see lack of a Nup58 immunostaining, which is a constituent of the Nup62 complex, in Figure 4D) and Nup93 (Sachdev et al, 2012) consistent with the notion that Nup93 is a key determinant in recruiting the Nup62 complex during vertebrate NPC assembly at the end of mitosis (Sachdev et al, 2012). Taken together, these data suggest that after the binding of the Nup107–160 complex and nuclear membranes to the chromatin surface, Nup53 recruitment is the next decisive step in NPC assembly. Nup93 (Nup93–Nup188/Nup93–Nup205) binding and the subsequent recruitment of the Nup62 complex follow.
Nup53 interacts with a number of other nucleoporins, including NDC1, Nup155 and Nup93 (Lusk et al, 2002; Hawryluk-Gara et al, 2005, 2008; Mansfeld et al, 2006; Sachdev et al, 2012) a feature that is conserved in yeast (Fahrenkrog et al, 2000; Onischenko et al, 2009; Amlacher et al, 2011). As previously reported (Hawryluk-Gara et al, 2008), the interaction of Nup53 with NDC1 is not necessary for postmitotic NPC assembly in Xenopus egg extracts (Figure 4A). A possible explanation might be that Nup53 can interact directly with membranes and that one of its two membrane binding regions is sufficient for postmitotic NPC formation. In addition, Nup53 interaction with other nucleoporins such as Nup155, which in turn binds POM121 (Mitchell et al, 2010; Yavuz et al, 2010) could be a possible mechanism linking Nup53 to the pore membrane which might compensate for the loss of the direct Nup53–NDC1 interaction.
The interaction of Nup53 with Nup155 is thought to be important for NPC assembly. A previous study found that after depleting Nup53 from Xenopus egg extracts, only fragments capable of binding to Nup155 allow for NPC formation (Hawryluk-Gara et al, 2008). However, in this case all fragments that rescued the NE/NPC assembly defect included the RRM domain and all fragments defective in NPC assembly and the Nup155 interaction lacked the intact RRM domain. Similarly, a deletion in C. elegans Nup53 blocked NE assembly also impaired the RRM domain (Rodenas et al, 2009). We demonstrate here that the RRM domain is important for Nup53 dimerization and in turn for membrane binding. Therefore, we currently cannot rule out that the primary cause for the previously described defects was a loss of the Nup53 membrane interaction.
Nup93 binds directly Nup53 (Hawryluk-Gara et al, 2005; Sachdev et al, 2012) and the interaction domain resides in the N-terminal half of Nup53 (Hawryluk-Gara et al, 2008). This interaction was previously considered to be dispensable for NPC assembly as a fragment lacking the N-terminal region as well as the C-terminal 26 amino acids replaced endogenous Nup53 in nuclear assemblies in Xenopus egg extracts (Hawryluk-Gara et al, 2008). This is quite surprising in the light of the results presented here, as this fragment also lacked both membrane binding regions identified in this study and does not allow for Nup93 recruitment which is an essential factor for postmitotic NPC assembly (Sachdev et al, 2012). Using a number of different Nup53 fragments that lacked the Nup93 binding region we were not able to replace endogenous Nup53 in NPC assembly (Supplementary Figure S6). Currently, we cannot rule out that this discrepancy is due to different Nup53 depletion efficiencies. In fact, we found that a small percentage of floated membrane preparations used in the nuclear assembly reactions contained minor amounts of Nup53, and we were careful to exclude these from our experiments.
Nup53 is also known as mitotic phosphoprotein of 44 kDa (Stukenberg et al, 1997). Indeed, Nup53 from mitotic extracts migrates significantly slower in SDS–PAGE compared to Nup53 from interphasic extracts (Supplementary Figure S3A). We identified a number of mitosis-specific phosphorylation sites on Nup53, of which a subset were consensus sites for CDK1. These findings are consistent with the fact that Nup53 has been identified as a CDK1 target in both yeast and humans (Lusk et al, 2007; Blethrow et al, 2008). The N-terminal membrane binding region of Nup53 is phosphorylated during mitosis. Phosphomimetic mutations and in vitro phosphorylation experiments suggest that CDK1-mediated phosphorylation renders this region incompetent for membrane interaction (Figure 3B). It is therefore possible that this mitotic phosphorylation weakens the interaction of Nup53 with the pore membrane to facilitate NPC disassembly during prophase.
Proteins without integral membrane regions can associate with cellular membranes by a variety of mechanisms (Cho and Stahelin, 2005). First, they can be covalently attached to a lipid moiety. However, we have no indication that Nup53 is modified in such a way. Second, peripheral membrane proteins are recruited to the lipid bilayer by specific protein–lipid interactions that involve binding to particular lipid head groups. In this regard, lipid arrays performed to date have not demonstrated an affinity of Nup53 for any specific lipid (unpublished observation). Furthermore, recombinant Nup53 binds to liposomes prepared from different lipid sources like pure DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), a lipid mixture mimicking the ER/NE lipid composition (Franke et al, 1970; Supplementary Figure S7) or folch fraction I (unpublished observation). Third, some proteins are recruited to membranes via electrostatic interactions. As a fraction of the lipid head groups are negatively charged this involves positively charged clusters on the protein surface. Indeed, we found that the first membrane binding region of Nup53 contains such a cluster. Replacing these positive with negative residues as well as introducing negative charge by phosphorylation rendered this site incapable of membrane interaction (Figure 3A and B). Finally, peripheral membrane proteins can associate with lipid bilayers via hydrophobic regions. It has been suggested that the C-terminal region of Nup53 contains an amphipathic helix, which could serve as a hydrophobic module (Marelli et al, 2001; Patel and Rexach, 2008). In fact, the C-terminus of Nup53 contains a membrane binding region and deleting the last eight amino acids abolished its membrane interaction. Deletion of the C-terminal tryptophan only slightly reduced membrane binding of Nup53 in the full-length protein (Figure 3C), but inhibited interphasic NPC assembly (Figure 5) suggesting that this residue fulfills an important additional function. Indeed, the insertion of hydrophobic parts of a protein into a membrane is one of several mechanisms by which proteins can deform membranes (McMahon and Gallop, 2005). Our data suggest that the C-terminal membrane binding region and especially the last tryptophan of Nup53 fulfills this task (Figure 6A). Interestingly, in vivo intranuclear tubular membranes are induced upon overexpression of yeast Nup53p which is dependent on its C-terminus (Marelli et al, 2001). Thus, Nup53 might not only have an important function in binding to the pore membrane in turn promoting the recruitment of other nucleoporins, especially members of the Nup93 complex, but may additionally function to deform the NE membrane into a pore. In the latter instance, the insertion of the Nup53 C-terminus into the hydrophobic phase of the membrane would result in displacement of lipid head groups and reorientation of the hydrophobic lipid side chains, bending the lipid bilayer into a convex shape and inducing membrane curvature necessary to form and stabilize the pore (Antonin et al, 2008). The doughnut-like shape of the pore requires likewise stabilization of a concave curvature in the plane of the pore in addition to the convex curved membrane connection between outer and inner nuclear membrane (see Figure 7). This curvature might be induced and stabilized by a number of at least partially redundant mechanisms, such as formation of a coat-like structure by the Nup107–160 complex (Devos et al, 2004; Mans et al, 2004; Brohawn et al, 2008) or oligomerization of pore membrane proteins, although it is not clear how the different proteins contribute to the different modes of membrane bending. Similarly, in the context of the protein interaction network of an assembled NPC the N-terminal membrane binding region of Nup53, in addition to membrane interaction, might induce and/or stabilize curved membranes. Indeed, Nup53 is only functional when it dimerizes probably because this increases the avidity of the Nup53 membrane interaction. In addition, all the factors might also impose geometrical constrains to the membranes supporting a pore structure.
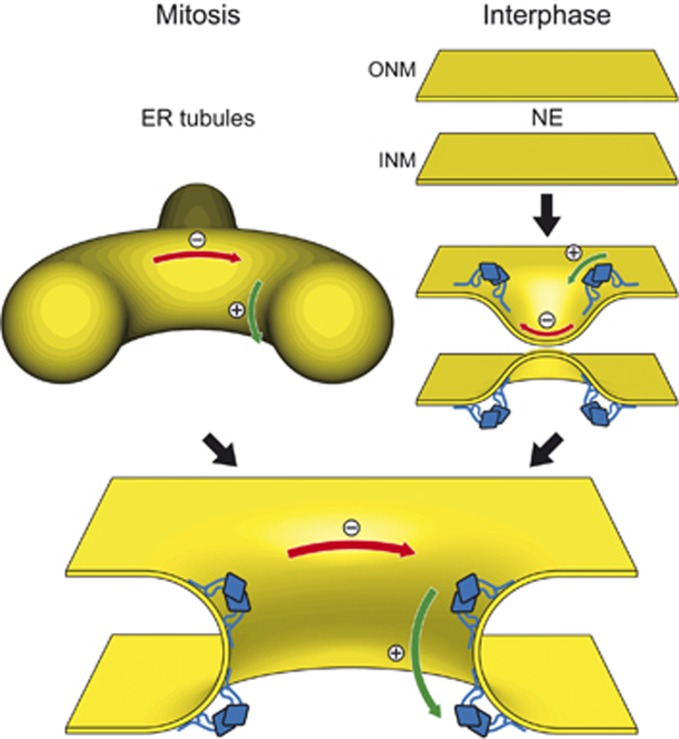
Figure 7.
The role of Nup53 in NPC assembly. Schematic drawing of the postmitotic and interphasic modes of nuclear pore assembly focused on the membrane interacting function of Nup53. For the sake of clarity other membrane associated and integral proteins, including nucleoporins, participating in this process are omitted. Left pathway: after mitosis, outgrowing ER tubules (yellow) surround assembling NPCs providing negative/concave (red) and positive/convex (green) curvature which is stabilized by membrane binding Nup53 dimers (blue) for pore formation. Right pathway: in interphase, the intact nuclear envelope membranes (yellow) are deformed by the C-terminal membrane binding site of Nup53 introducing a convex membrane curvature for the approximation and following fusion of the two membranes leading to pore formation.
Although either one of the two membrane binding regions of Nup53 is sufficient for postmitotic NPC assembly and stability, the C-terminal site is specifically required for NPC assembly during interphase. It is a matter of debate whether NPC assembly during these different cell-cycle phases occurs by distinct mechanisms (Doucet et al, 2010; Dultz and Ellenberg, 2010; Lu et al, 2011). At the end of mitosis, NPC assembly occurs concomitantly with formation of a closed NE. It is possible that this mode of NPC formation does not require fusion between the outer and inner nuclear membrane to form a nuclear pore, in contrast to interphasic NPC assembly. Postmitotic pore assembly could rather arise by the enclosement of the assembling NPCs on the chromatin surface by an outgrowing ER network. In this scenario, Nup53 would stabilize the membrane curvature provided by the ER tubules rather than induce membrane deformation (see Figure 7, left pathway). The different requirements for Nup53 membrane binding regions in postmitotic and interphasic NPC assembly support the view of two different mechanistic pathways. A loss of the membrane deforming capability of Nup53 in postmitotic NPC assembly and NPC stability might be compensated by other factors such as the Nup107–160 complex or integral membrane proteins. However, during metazoan interphasic NPC assembly Nup53-mediated membrane deformation might be crucial for the initial approximation and/or fusion of both membrane layers (see Figure 7, right pathway). Interestingly, ER bending proteins of the reticulon family that induce convex membrane curvature (Hu et al, 2008) were shown to be important for NPC assembly into the intact NE both in yeast and in vertebrates (Dawson et al, 2009). Currently, it is unknown whether these proteins do also contribute to postmitotic NPC assembly. As their effect on ER membrane reorganization at the end of mitosis is a prerequisite for NE reformation (Anderson and Hetzer, 2008) it is difficult to separate these two functions. Finally, how the fusion of outer and inner nuclear membranes is achieved is largely unclear but our results suggest that Nup53, importantly its C-terminal membrane binding region, is critical for this process.
Materials and methods
Antibodies against POM121 and GP210 (Antonin et al, 2005), NDC1 (Mansfeld et al, 2006), Nup155 (Franz et al, 2005), MEL28/ELYS (Franz et al, 2007), Nup107 (Walther et al, 2003), Nup53 and Nup58 (Sachdev et al, 2012) as well as Nup188, Nup205, Nup98 and Nup53 (Theerthagiri et al, 2010) have been described. mAB414 and Nup88 antibodies were from Babco or BD Bioscience, respectively. For quantitation of Nup53 liposomes binding the antibody was affinity purified with a fragment comprising the RRM domain as this domain is included in all tested fragments (please see Supplementary Table S2 for list of all DNA constructs used in this study).
Nuclear assemblies
Nuclear assemblies and immunofluorescence (Theerthagiri et al, 2010), generation of affinity resins, sperm heads and floated membranes (Franz et al, 2005) as well as prelabelled membranes (Antonin et al, 2005) were done as described. Interphasic NPC assembly using dextran influx was monitored as described (Dawson et al, 2009) except that mock or Nup53-depleted extracts were incubated with 1.5 vol WGA-Agarose (Sigma) for 40 min. Counting of NPCs was performed on mAB414-labelled nuclei as described (Theerthagiri et al, 2010).
Protein expression and purification
Constructs for full-length Xenopus Nup53 and fragments, S. cerevisiae Nup53 and Nup59 were generated from a synthetic DNA optimized for codon usage in E. coli (Geneart, see Supplementary data) and cloned into a modified pET28a vector with a yeast SUMO solubility tag followed by a TEV site upstream of the Nup53 fragments. Proteins were expressed in E. coli, purified using Ni-agarose, His6 and SUMO tags were cleaved by TEV protease, concentrated using VIVASPIN columns (Sartorius) and purified by gel filtration (Superdex200 10/300 GL or Superdex200 PC 3.2/30, GE Healthcare) either in PBS for liposome binding experiments or in sucrose buffer (Theerthagiri et al, 2010) for nuclear assemblies, respectively. Nup53 fragments aa 162–320 and aa 254–320 were purified by size exclusion chromatography without removal of the tags to retain stability. Fragments of Xenopus Nup98 (aa 676-863) as well as human Nup133 (aa 67–514) (Berke et al, 2004) were expressed from modified pET28a vectors with a His6-NusA or His6 tag, which was cleaved of by thrombin or precision protease, respectively.
Liposome generation and flotation
E. coli polar lipid extract with 0.2 mol% 18:1–12:0 NBD-PE (1-oleoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphoethanolamine) (Avanti polar lipids) were dissolved in ethanol at 45°C. To form liposomes, the mixture was diluted 10-fold into PBS resulting in a final lipid concentration of 6.7 mg/ml while gently agitating. Liposomes were passed 21 × through Nuclepore Track-Etched Membranes (Whatman) with defined pore sizes (50, 100, 200, 400, 800 nm) at 45°C using the Avanti Mini-Extruder. To remove ethanol, liposomes were dialysed against PBS using Spectra/Por 2 dialysis tubing (MWCO 12-14 kDa). Liposome sizes were determined by light scattering using the AvidNano W130i. For quantitation of liposome binding, fluorescence intensity of the protein/liposome mixture and the top fraction was determined using a Molecular Imager VersaDoc MP 4000 Imaging System and ImageJ.
Folch fraction I lipids (Sigma) dissolved in chloroform were dried on a rotary evaporator and overnight under vacuum. PBS buffer was gently added to result in a final lipid concentration of 10 mg/ml. After 2 h of incubation at 37°C to allow spontaneous liposome formation the flask was agitated to dissolve residual lipids. After 10 cycles of freeze/thawing, liposomes were diluted 10-fold in PBS and extruded as described before.
Immunoprecipitation
Xenopus Nup53 as well as the RRM dimerization mutant (F172E/W203E) was cloned with N-terminal myc or HA tag, respectively, into a pSI vector (Promega). HeLa cells were transfected using Fugene 6 (Roche) following manufacturer’s instructions, harvested 24 h post transfection and solubilized in 1% Triton X-100 in PBS supplemented with protease inhibitors (2 μg/ml leupeptin, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 0.1 mg/ml AEBSF final concentration) for 10 min at 4°C. After centrifugation for 10 min at 15 000 g samples were diluted five-fold in PBS and employed for immunoprecipitation using α-myc or α-HA antibodies (Roche).
Miscellaneous
For in vitro phosphorylation, 3 μM proteins were incubated with 0.33 U/μl CDK1-CyclinB (NEB), 1 mM ATP, 10 mM MgCl2 and 1 mM EDTA in PBS for 1 h at 30°C.
For liposome tubulation copper grids filmed with pioloform and carbon-coated were glow discharged before usage. Proteins were incubated with 1 mg/ml folch fraction I liposomes for 7 min on grids, washed with buffer (10 mM Hepes, 150 mM NaCl, 4.5 mM KCl) and stained with 2% UAc for 2 min and examined on a FEI Technai spirit 120 kV microscope.
Supplementary Material
Acknowledgments
We thank M Flötenmeyer and S Würtenberger for help with the tubulation assay, B Uluvar for support in protein purification and M Lorenz for critical reading of the manuscript.
Author contributions: BV and WA designed and performed experiments and wrote the manuscript. AS quantified interphasic NPC assembly, RS and NE performed pull-down experiments, AMS performed the size determinations of the RRM domains. CS cloned constructs and prepared Xenopus laevis extracts. JM and BM performed mass spec analysis, UG performed measurements of different liposome sizes.
Footnotes
The authors declare that they have no conflict of interest.
References
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E (2011) Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146: 277–289 [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW (2008) Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 182: 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW (2009) Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol 186: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E (2008) Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett 582: 2004–2016 [DOI] [PubMed] [Google Scholar]
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW (2005) The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell 17: 83–92 [DOI] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU (2004) Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol 167: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU (2012) 3D ultrastructure of the nuclear pore complex. Curr Opin Cell Biol 24: 86–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM (2008) Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci USA 105: 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU (2008) Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Stahelin RV (2005) Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct 34: 119–151 [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Anderson DJ, Richard E, Hetzer MW (2006) Nuclear pores form de novo from both sides of the nuclear envelope. Science 312: 440–443 [DOI] [PubMed] [Google Scholar]
- Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT (2007) Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449: 923–927 [DOI] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR (2009) ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol 184: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT (2009) Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP (2004) Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Hetzer MW (2010) Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma 119: 469–477 [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW (2010) Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B (2007) A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol 14: 138–146 [DOI] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J (2010) Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol 191: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J (2008) Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol 180: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B, Hubner W, Mandinova A, Pante N, Keller W, Aebi U (2000) The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol Biol Cell 11: 3885–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Sali A, Rout MP (2011) Evolution: on a bender--BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol 193: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Deumling B, Baerbelermen, Jarasch ED, Kleinig H (1970) Nuclear membranes from mammalian liver. I. Isolation procedure and general characterization. J Cell Biol 46: 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW (2005) Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 24: 3519–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W (2007) MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW (2006) MEL-28, a novel nuclear envelope and kinetochore protein essential for zygotic nuclear envelope assembly in C. elegans. Curr Biol 16: 1748–1756 [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P (2003) Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell 14: 5104–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N, Kukimoto-Niino M, Akasaka R, Kishishita S, Murayama K, Terada T, Inoue M, Kigawa T, Kose S, Imamoto N, Tanaka A, Hayashizaki Y, Shirouzu M, Yokoyama S (2006) The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain. J Mol Biol 363: 114–124 [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW (2008) Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell 19: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW (2005) Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell 16: 2382–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G (2011) The structure of the nuclear pore complex. Annu Rev Biochem 80: 613–643 [DOI] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Voss C, Shemesh T, Li Z, Coughlin M, Kozlov MM, Rapoport TA, Prinz WA (2008) Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science 319: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Liu HL, De Souza CP, Osmani AH, Osmani SA (2009) The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell 20: 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka MJ (1998) Analysis of nuclear envelope assembly using extracts of Xenopus eggs. Methods Cell Biol 53: 367–395 [DOI] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T (2011) Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol 194: 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, Makhnevych T, Marelli M, Aitchison JD, Wozniak RW (2002) Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J Cell Biol 159: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CP, Waller DD, Makhnevych T, Dienemann A, Whiteway M, Thomas DY, Wozniak RW (2007) Nup53p is a target of two mitotic kinases, Cdk1p and Hrr25p. Traffic 8: 647–660 [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV (2004) Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3: 1612–1637 [DOI] [PubMed] [Google Scholar]
- Mansfeld J, Guttinger S, Hawryluk-Gara LA, Pante N, Mall M, Galy V, Haselmann U, Muhlhausser P, Wozniak RW, Mattaj IW, Kutay U, Antonin W (2006) The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol Cell 22: 93–103 [DOI] [PubMed] [Google Scholar]
- Marelli M, Aitchison JD, Wozniak RW (1998) Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J Cell Biol 143: 1813–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW (2001) A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J Cell Biol 153: 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438: 590–596 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Mills IG (2004) COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol 16: 379–391 [DOI] [PubMed] [Google Scholar]
- Miao M, Ryan KJ, Wente SR (2006) The integral membrane protein Pom34p functionally links nucleoporin subcomplexes. Genetics 172: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW (2010) Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol 191: 505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM (2010) Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 5: e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K (2009) Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol 185: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Weis K (2011) Nuclear pore complex-a coat specifically tailored for the nuclear envelope. Curr Opin Cell Biol 23: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Rexach MF (2008) Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol Cell Proteomics 7: 121–131 [DOI] [PubMed] [Google Scholar]
- Prufert K, Vogel A, Krohne G (2004) The lamin CxxM motif promotes nuclear membrane growth. J Cell Sci 117: 6105–6116 [DOI] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ (2006) ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA 103: 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas E, Klerkx EP, Ayuso C, Audhya A, Askjaer P (2009) Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev Biol 327: 399–409 [DOI] [PubMed] [Google Scholar]
- Sachdev R, Sieverding C, Flotenmeyer M, Antonin W (2012) The C-terminal domain of Nup93 is essential for assembly of the structural backbone of nuclear pore complexes. Mol Biol Cell 23: 740–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Hulsmann BB, Spang A, Hartmann E, Cordes VC, Gorlich D (2006) NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol 173: 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT, Lustig KD, McGarry TJ, King RW, Kuang J, Kirschner MW (1997) Systematic identification of mitotic phosphoproteins. Curr Biol 7: 338–348 [DOI] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W (2010) The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 189: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP (2010) The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR (1998) cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell 9: 2839–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B (1993) NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol 122: 743–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz S, Santarella-Mellwig R, Koch B, Jaedicke A, Mattaj IW, Antonin W (2010) NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett 584: 3292–3298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.