Abstract
Lyme disease Borrelia are invasive, nontoxigenic, persistent pathogens, and little is known about their mechanisms of pathogenesis. In our laboratory, a signature-tagged mutagenesis (STM) library of over 4,000 Borrelia burgdorferi transposon mutants has been constructed and is being screened for infectivity in mice. In this manner, a global view of the virulence determinants (factors required for full infectivity) is being developed. Additionally, the mechanisms of immune evasion involving the VMP-like system (vls) are under analysis, and cryo-electron microscopy is providing a detailed view of the three-dimensional structure of B. burgdorferi. These approaches will contribute to the improved understanding of how Lyme disease Borrelia cause disease.
Keywords: Lyme borreliosis, Borrelia burgdorferi, virulence determinants, transposon mutagenesis, antigenic variation.
Bacteria have been in existence for at least 3.5 billion years [1] and, until multicellular eukaryotes appeared ~1 billion years ago, were essentially free-living organisms. The advent of multicellar organisms provided a new environmental niche in which bacteria have continued to coevolve with their larger counterparts. Prokaryotes can be free-living, commensal (do no harm), symbiotic (do good), opportunistic pathogens (do harm, but only in compromised hosts), and frank pathogens (do harm in healthy as well as compromised hosts). In addition, pathogenic bacteria may be only briefly associated with the host, establish long-term infections, or require the host for survival (obligate pathogens). This relationship is continually evolving (toward or away from pathogenicity or host dependence), with the adaptation of both the bacterium and the host. This process is continuing today, as much as in any time during evolution.
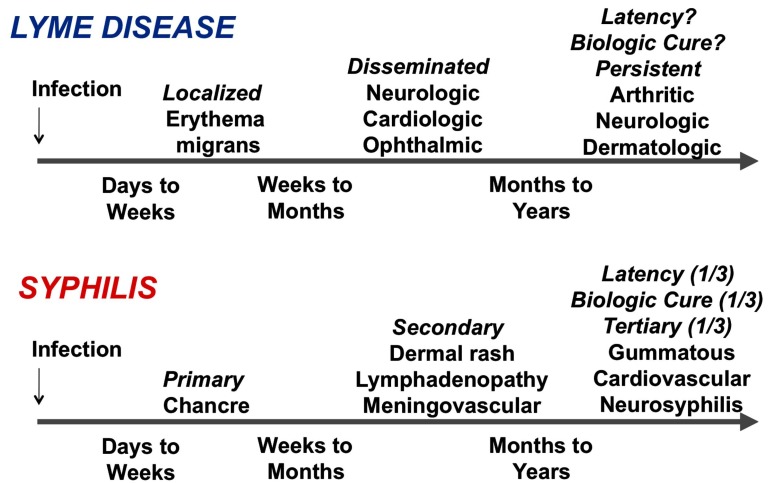
Bacteria of the genus Borrelia are host-dependent spirochetes that are reliant on continual transmission between vertebrate hosts (most commonly mammals and birds) and arthropod vectors (ticks and lice). The genus bifurcated into two forms that cause Lyme borreliosis (LB) and relapsing fever (RF) millions of years ago (based on genetic evidence), but these groups are still quite closely related. LB spirochetes are transmitted by slow-feeding, hard-bodied ticks of the genus Ixodes. The many species that cause endemic RF are predominantly carried by fast-feeding, soft-bodied Ornithodoros ticks, whereas B. recurrentis, the causative agent of epidemic RF, is passed between humans by the human body louse, Pediculus humanus subsp. humanus. Of the 10 species and 2 proposed species of LB spirochetes, B. burgdorferi, B. garinii, and B. afzelii commonly cause Lyme disease in Eurasia, but only B. burgdorferi is known to cause human disease in North America. Following transmission, LB infections typically have an indolent course with loosely defined localized, disseminated, and persistent stages; symptoms can persist over a period of months to years if untreated. This pattern thus has some similarity to infection with another invasive, persistent spirochetal pathogen, Treponema pallidum, which causes the sexually transmitted disease syphilis (Fig. 1). In contrast, the course of RF is typified by short, 3-day, recurrences of high level bacteremia (up to 108 bacteria/ml of blood) with high fever and other constitutional symptoms. No toxins have been identified for Borrelia species or T. pallidum, and it is thought that the manifestations of their corresponding diseases are due primarily to their ability to invade and propagate within the host and to stimulate of host inflammatory reactions.
Fig. (1).
Similarities of the time course of two invasive, persistent spirochetal infections, Lyme borreliosis and syphilis.
One of the main projects in our laboratory is the global identification and characterization of virulence determinants in B. burgdorferi. Virulence determinants represent a broader category than virulence factors. Virulence determinants include any gene product that enhances the ability of the organism to cause disease, e.g. metabolic pathways that are required for the survival and growth of the pathogen in the host. Virulence factors are more narrowly defined as bacterial products that are specifically involved in the pathogenic properties of the organism. Examples of virulence factors include adhesins, toxins, and degradative enzymes involved in tissue invasion, as well as protective activities against the immune system such as capsules, antigenic variation systems, and antibody or complement inactivation mechanisms. Because LB spirochetes produce no known toxins, it is not possible to simply purify a protein and demonstrate that it has a lytic or cytopathic activity. Instead, the screening for genes and gene products that play a role in infectivity and pathogenesis typically requires the whole organism; exceptions are adhesins, which can be identified ex vivo by their binding to host cells or extracellular matrix (ECM) components [2].
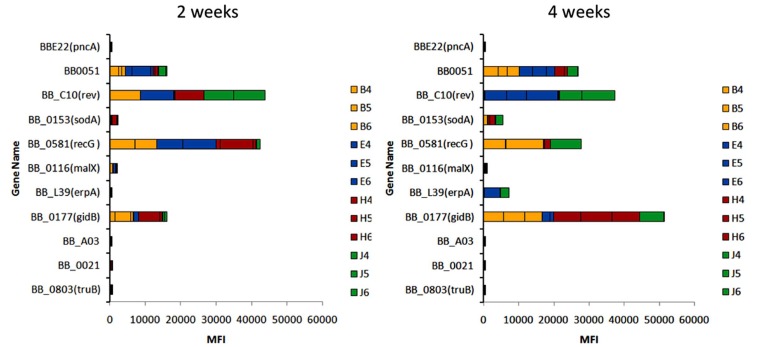
The system that we have chosen for virulence determinant analysis is transposon mutagenesis with the addition of DNA sequence ‘tags’, a technique called signature tag mutagenesis (STM) [3]. In our studies, we have utilized derivatives of the Mariner transposon vector called pGKT, which was developed by Dr. Philip Stewart specifically for use in LB organisms [4]. Mariner transposons insert in DNA at any occurrence of the dinucleotide TA, resulting in a library of transposon insertions that are randomly located throughout the genome. Dr. Tao Lin in our laboratory added 11 different 7-basepair ‘tags’ into pGKT, permitting the identification of individual clones in a mixture. The bacterial strain we utilized is B. burgdorferi B31 5A18NP1, which was derived by Dr. Hiroki Kawabata to provide an infectious yet moderately transformable LB strain [5]. By electroporating the pGK derivatives into 5A18NP1, we isolated 4,479 B. burgdorferi mutants in which the specific insertion sites and the plasmid content [6] were both determined. Analyzing the plasmid content was important, because this strain of B. burgdorferi has 19 plasmids. Most of these can be lost during in vitro culture, and at least three are required for full infectivity in mammals [7-10]. By identifying the insertion site of each transposon mutant, we were able to identify the genes that were inactivated by this insertion. This so-called ‘ordered’ library has two advantages. First, it permits the identification of nonessential genes, i.e. those that are not required for the growth of B. burgdorferi in the Barbour-Stoenner-Kelly medium that is standardly used for culturing Borrelia strains. Second, it allows the systematic examination of genes that may be involved in infectivity. To determine the infectivity of these mutants while minimizing the number of animals needed, we have inoculated mice with groups of 11 5A18NP1 transposon mutants, each marked with a different signature tag. After the mice have be infected for either 2 or 4 weeks, bladder, ear, heart, joint, and inoculation site tissue samples are obtained and subjected to a novel Luminex-based method that uses PCR of the signature tag region to specifically identify each of the 11 clones. Low median fluorescence intensity (MFI) values for a particular clone means that the gene disrupted by the transposon is required for infection or dissemination in the mouse model. An example of one of these experiments is shown in Fig. (2). For each time point, the MFI values obtained in 5 tissues in each of 3 mice are stacked to indicate the cumulative MFI value. The first two clones (with disruptions in the nicotinamidase gene pncA and the hypothetical protein gene BB0051, respectively) serve as negative and positive controls. Mutations in malX, bba06, bb0021, and truB yielded consistently low values, indicating loss of infectivity. sodA and erpA mutants yielded intermediate values, whereas rev, gidB, and recG mutants exhibited high levels on infection. The results from these studies thus serve as a screening method for determining which genes are most important in mammalian infection.
Fig. (2).
An example of signature tagged mutagenesis (STM) analysis of B. burgdorferi transposon mutant infectivity. In the experiment shown, C3H/HeN mice were inoculated with 11 different B. burgdorferi mutants, each with a different 7 bp ‘signature tag’. After 2 and 4 weeks, groups of three mice were euthanized, and five tissues were analyzed for the presence of each mutant using a PCR-based method with Luminex detection of the resulting fluorescent signal, called the Median Fluorescent Intensity (MFI). The genes mutated in each of the clones tested in this experiment is shown on the Y axis. The number at the end of each designation is the insertion ratio, i.e. the distance in bp of the insertion from the 5’ end of the gene divided by the full length of the gene. The bars represent the cumulative MFI values for each of the 5 tissues (Bladder, Ear, Heart, Joint, and Skin [inoculation site]) in the three mice (numbered 1-3). Short bars indicate little or no infectivity, whereas long bars correspond to full infectivity. The first mutant, in the pncA (nicotinamidase) gene, is noninfectious in mice and serves as a negative control. The second mutant, in the conserved hypothetical protein gene BB0051, has intermediate infectivity.
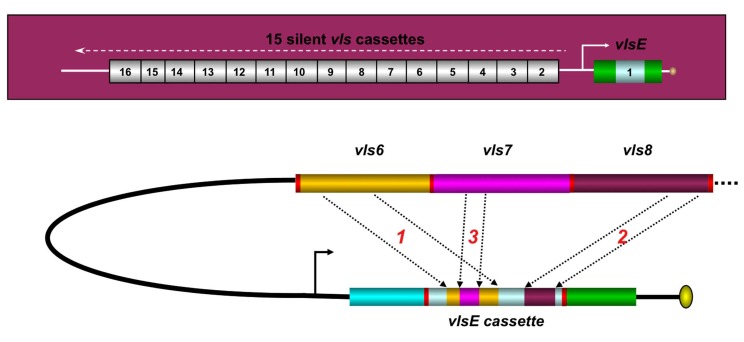
A second area of study in our laboratory is the vls antigenic variation systems of LB spirochetes. Humans and other mammals can be infected with LB organisms for a period of months to years. In experimentally infected C3H/HeN mice, B. burgdorferi can be routinely cultured from 2 mm skin biopsies throughout the lifetime of the animal (~2 years). It is thought that Lyme Borrelia utilize several mechanisms to evade the immune response and cause persistent infection; these include antigenic variation, complement cascade inactivation, and invasion of protective niches [11]. In B. burgdorferi B31, the VMP-like sequence (vls) antigenic variation system is harbored on a 28 kilobase (kb) linear plasmid called lp28-1 [12]. This elaborate system consists of a single expression site (vlsE) as well as a contiguous array of 15 silent cassettes (Fig. 3). These silent cassettes share >90% sequence identity with the central region of the vlsE gene, but have six variable regions that encode different amino acids. During mammalian infection, segments of the vls silent cassettes are transferred via gene conversion events into the vlsE central cassette region. This genetic mechanism results in changes in the amino acid sequence of the encoded surface lipoprotein VlsE, which in turn alters the epitopes present. Thus antibodies directed against previous versions of VlsE will not be effective in eliminating organisms expressing the new variants of VlsE. This process keeps the spirochete one step ahead of the immune response and hence contributes to immune evasion.
Until recently, vlsE recombination had only been observed during infection of mammals; it had not been detected in ticks or during in vitro culture. This result indicated that some condition present in mammalian tissue (a ‘signal’) activated the gene conversion process. Diane Edmondson et al. (manuscript in preparation) have now shown that B. burgdorferi invades and proliferates to high levels in mouse tissue explants, and that vlsE recombination events can be detected in the spirochetes present in these explants. The recombination events appear to occur at low frequency, but their detection indicates that at least some of the environmental signals required for the activation of this process are present in this ex vivo system. The availability of this model system may permit detailed examination of the mechanisms of vlsE recombination and the regulation of this activity.
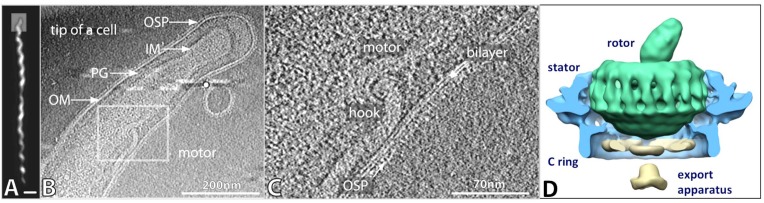
Structural analysis of Lyme disease Borrelia provides another potential avenue for exploring how these spirochetes interact with vertebrate and arthropod hosts. Dr. Jun Liu in our department has begun a series of studies of B. burgdorferi and other spirochetes using a technique called cryoelectron tomography (CryoET). CryoET is a form of electron microscopy (EM) in which viable organisms are flash frozen without fixation on EM grids and then imaged at different angles relative to the electron beam. Using Fourier transformation, these images are then averaged to create a three dimensional model of the organisms. These 3D models resemble microscopic CT scans and contain a remarkable amount of structural information. Thus far, CryoET has been utilized to study the inner and outer membranes, peptidoglycan layer, flagellar motor and filament, and chemotaxis receptor structures of B. burgdorferi and other organisms (Fig. 4). Discrete structures such as the flagellar motor and filament and the chemotaxis receptor can be extracted and averaged to form more detailed molecular models of their architecture. Comparison of mutant bacteria lacking one or more proteins with their ‘wild type’ parent strains provides valuable information about the structural location and hence the functional roles of the proteins under study. This approach has begun to unravel the mechanisms of motility and chemotaxis, both of which are important in the pathogenesis of Lyme borreliosis and many other infectious diseases.
Fig. (4).
Structure of B. burgdorferi B31, as revealed by cryoelectron tomography (CryoET). A) Full length view of B. burgdorferi B31, as seen by darkfield microscopy. Bar = 1 µm. B) One CryoET ‘slice’ through the three-dimensional model of a B. burgdorferi cell, depicting the outer membrane (OM) with outer surface proteins (OSP), peptidoglycan layer (PG), inner membrane (IM), and a flagellar motor and filament. C) Closer view of the inset shown in panel B, showing a flagellar motor, the hook structure connecting the motor to the flagellar filament, the lipid bilayer of the OM, and the OSP layer. D) a 3D cutaway model of the flagellar motor, produced through the compilation of data from 1280 individual motors. The predicted locations of the rotor, stator, C-ring, and export apparatus are indicated. Adapted from Ref. [14].
Many mysteries remain with regard to the ability of Lyme Borrelia to colonize, proliferate, disseminate, persist, and cause disease in humans and other vertebrate hosts. However, the powerful combination of genetic, molecular, structural, and pathobiologic approaches has begun to ‘chip away’ at the underlying processes in the infection cycle, and promises to lead to further improvements in Lyme borreliosis diagnosis, treatment, and prevention in the coming years.
Fig. (3).
The vlsE antigenic variation system of Lyme disease Borrelia. The vls locus of B. burgdorferi B31 (top of figure) consists of the vlsE gene (that encodes the surface lipoprotein VlsE) and a contiguous series of 15 vls silent cassettes (that are not expressed). Each of the silent cassettes are highly homologous to the central cassette region of vlsE, but encode different amino acids in each of 6 variable regions. During mammalian infection, segments of the silent cassettes replace the corresponding region in the vlsE expression site, as shown in the hypothetical example in the lower part of the figure. This process results in vlsE sequence variants that have different amino acid sequences and hence different epitopes. vlsE recombination occurs continuously during mammalian infection, and contributes to immune evasion and long-term survival of Lyme disease Borrelia in humans and other vertebrate hosts. Adapted from Ref. [13].
ACKNOWLEDGEMENTS
I gratefully acknowledge the involvement of the following individuals in the research described: STM analysis -Tao Lin, Lihui Gao, Chuhua Zhang, and Evelyn Odeh; VlsE studies - Diane G. Edmondson and Sabitha Prabhakaran; CryoET - Jun Liu and his group. Work described was supported by NIH Award Numbers R01AI037277, R01AI55449, and R01AI087946 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Allergy And Infectious Diseases or the National Institutes of Health.
Footnotes
Based on a presentation with the same title at the Annual Meeting of the Deutsche Borreliose-Gesellschaft on April 8-10, 2011 in Wuppertal, Germany.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1. Schopf JW. Solution to Darwin's dilemma: Discovery of the missing Precambrian record of life. Proc Natl Acad Sci USA. 2000;97(13 ):6947–53. doi: 10.1073/pnas.97.13.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coburn J, Chege W, Magoun L, Bodary SC, Leong JM. Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library. Mol Microbiol. 1999;34(5 ):926–40. doi: 10.1046/j.1365-2958.1999.01654.x. [DOI] [PubMed] [Google Scholar]
- 3. Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269(5222 ):400–3. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 4. Stewart PE, Rosa PA. Transposon mutagenesis of the Lyme disease agent Borrelia burgdorferi. Methods Mol Biol. 2008;431:85–95. doi: 10.1007/978-1-60327-032-8_7. [DOI] [PubMed] [Google Scholar]
- 5. Kawabata H, Norris SJ, Watanabe H. BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect Immun. 2004;72(12 ):7147–54. doi: 10.1128/IAI.72.12.7147-7154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Norris SJ, Howell JK, Odeh EA, Lin T, Gao L, Edmondson DG. High-throughput plasmid content analysis of Borrelia burgdorferi B31 by using Luminex multiplex technology. Appl Environ Microbiol. 2011;77(4 ):1483–92. doi: 10.1128/AEM.01877-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2000;97(25 ):13865–70. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Labandeira-Rey M, Seshu J, Skare JT. The absence of linear plasmid 25 or 28-1 of Borrelia burgdorferi dramatically alters the kinetics of experimental infection via distinct mechanisms. Infect Immun. 2003;71(8 ):4608–13. doi: 10.1128/IAI.71.8.4608-4613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grimm D, Eggers CH, Caimano MJ, et al. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect Immun. 2004;72(10 ):5938–46. doi: 10.1128/IAI.72.10.5938-5946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jewett MW, Lawrence K, Bestor AC, et al. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol. 2007;64(5 ):1358–74. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Norris SJ, Coburn J, Leong JM, Hu LT, Höök M. Pathobiology of Lyme disease Borrelia. In: Samuels DS, Radolf JD, editors. Borrelia: Molecular and cellular biology. Hethersett, Norwich, UK: Caister Academic Press; 2010. pp. 299–331. [Google Scholar]
- 12. Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89(2 ):275–85. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 13. Norris SJ. Antigenic variation with a twist - the Borrelia story. Mol Microbiol. 2006;60(6 ):1319–22. doi: 10.1111/j.1365-2958.2006.05204.x. [DOI] [PubMed] [Google Scholar]
- 14. Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191(16 ):5026–36. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]