Abstract
The degree to which genes and environment determine variations in brain structure and function is fundamentally important to understanding normal and disease-related patterns of neural organization and activity. We studied genetic contributions to the midsagittal area of the corpus callosum (CC) in pedigreed baboons (68 males/112 females) to replicate findings of high genetic contribution to area of the CC, reported in humans, and to determine if the heritability of the CC midsaggital area in adults was modulated by fetal development rate. Measurements of callosal area were obtained from high-resolution MRI scans. Heritability was estimated from pedigree based maximum likelihood estimation of genetic and non-genetic variance components as implemented in SOLAR. Our analyses revealed significant heritability for the total area of the CC and all of its subdivisions, with h2 = 0.46 for the total CC and h2 = .54, .37, .62, .56, and 0.29 for genu, anterior midbody, medial midbody, posterior midbody and splenium, respectively. Genetic correlation analysis demonstrated that the individual subdivisions shared between 41% and 98% of genetic variability. Combined with previous research reporting high heritability of other brain structures in baboons, these results reveal a consistent pattern of high heritability for brain morphometric measures in baboons.
Keywords: corpus callosum, heritability, baboons, genetics, imaging
Introduction
Genetic differences account for a significant proportion of neuroanatomic variability in humans (Hulshoff Pol, et al., 2006; Pennington, et al., 2000; Pfefferbaum, Sullivan, Swan, & Carmella, 2000). While several studies have considered heritable influences on total brain volume (Cheverud, et al., 1990; Posthuma, et al., 2002; Rogers, et al., 2007; Rogers, et al., 2010; Thompson, et al., 2001; Toga & Thompson, 2005), little is known about genetic influences on regional structures such as the corpus callosum [CC]. The CC is the largest commissural white matter tract in the brain, and is essential for interhemispheric integration of sensory, motor and higher-order cognitive information. Numerous genetic disorders affect the morphology of CC, producing specific regional abnormalities (Di Rocco, Biancheri, Rossi, Filocamo, & Torotori-Donati, 2004; Kochunov, et al., 2005). Disruptions in the structural integrity of the CC during aging or as a result of specific disorders are associated with impairments in problem solving and working memory (Zahr, Rohlfing, Pfefferbaum, & Sullivan, 2009), bimanual movement, or interhemispheric transfer (Bonzano, et al., 2008). Neuropsychiatric conditions including schizophrenia (Wang, et al., 2010) and major depression (Korgaonkar, et al., 2010) are associated with changes in CC. Given the importance of the CC to various cognitive functions, understanding the genetic mechanisms that influence variation in the size and shape of this structure will likely have important clinical implications.
Using MRI, human twin studies have suggested a high heritability of the midsaggital CC area (Scamvougeras, Kigar, Jones, Weinberger, & Witelson, 2003). Analyses in a small number of mono- (N=10) and dizygotic (N=7) twin pairs estimated heritability to be at 94.4% for the size of the CC. Pfefferbaum et al. (2000) reported similarly high heritability (85%) for the CC size in another small (N=85) twin sample. More recent investigations into the regional heritability of the CC partitions using DTI (diffusion tensor imaging) have reported that the degree of contributions by genetic factors was variable among midsaggital CC sections and the sources of this variability remained unknown (Brouwer, et al., 2010; Chiang, et al., 2011; Kochunov, Glahn, Lancaster, et al., 2010). Some developmental biologists have suggested that the rate of development may modulate the degree of genetic contribution and earlier developing structures will be more tightly controlled by genetic factors during development, thus leading to higher heritability. However this assertion is not consistent with results that demonstrate that the earliest developing structures are clearly evolvable through the course of the evolutionary history and may be susceptible to environmental perturbations (Raff, 1996). This was further demonstrated by recent studies that showed that the heritability of white matter increases with age (Peper, Brouwer, Boomsma, Kahn, & Pol, 2007) and the brain regions associated with more complex reasoning become increasingly more heritable with development (Lenroot & Giedd, 2008).
Comparative studies of animal models can provide unique insights into the biological processes that underlie human neurobiology and neurodevelopment. Previous studies of baboons have documented significant genetic effects on brain structure (Rogers et al. 2007; Kochunov et al 2010) and shown that there are unanticipated parallels in the architecture of genetic effects on cortical folding and brain volume in humans and baboons (Rogers et al 2010). In this latter paper, we showed that an inverse relationship between genetic effects on brain size and cortical folding is conserved in humans and baboons. Thus, the baboon results provide both a confirmation of an unexpected finding in human neurogenetics and demonstrate that there are long-term evolutionary genetic relationships that are shared across primate clades. Significant results concerning brain structure and the genetics of brain evolution have also come from studies of macaques, vervet monkeys and chimpanzees (e.g., (Fears, et al., 2009; Lyn, et al., 2011; Semendeferi, Lu, Schenker, & Damasio, 2002; Sherwood, et al., 2010). In addition to comparative analyses of nonhuman primate and human brain structure, researchers have also successfully used nonhuman primates to study genetic influences on brain function and metabolism (Oler, et al., 2010). Between-species differences in gene expression within the brain have also informed our understanding of human brain function (Konopka, et al., 2009).
We aimed to evaluate genetic influences on inter-subject variability in the midsagittal CC size and the degree to which the genetic heritability of regional CC variability was modulated by rate of development during fetal and early postnatal growth. The evaluation was performed in a non-human primate: baboons, Papio hamadryas. Papio baboons were chosen because they share several neurological characteristics with humans, including high heritability of brain volume, cortical surface area and cortical gyrification (Kochunov, Glahn, Fox, et al., 2010). Additionally, age-related changes in the development of the CC are consistent with the developmental course observed in humans (Phillips & Kochunov, in press). Therefore, the baboon holds great potential as a model for human brain development.
Methods
Subjects
One hundred-eighty adult baboons (Papio hamadryas) (68 males and 112 females) were selected from the large multi-generation pedigreed colony of more than 2000 baboons maintained by the Southwest National Primate Research Center (SNPRC) at the Texas Biomedical Research Institute in San Antonio, Texas. The average age of the study animals was 16.0 ± 4.2 years [range: 7–28 years]. This age range was chosen to minimize the effects of development or senescence based on studies of cerebral ontogeny (Leigh, 2004; Leigh, Shah, & Buchanan, 2003). The genealogical relationships among study animals included 414 parent-offspring pairs, 51 full sib pairs, 645 half-sib pairs, and a large number of more distant kinship relationships. Captive male baboons are sexually mature at 5 and fully adult at 6. Female baboons s start to cycle at between 3–4 years and are fully grown around 5 years.
We measured the CC in utero and during the early postnatal period to estimate developmental rate. In utero imaging of 13 normally developing fetuses was performed covering the period of gestational week 17 through birth (gestational week 28); postnatal imaging was performed on 16 baboons between postnatal weeks 1 and 32. The details for the in-utero and early postnatal imaging and animal handling protocols are described elsewhere (Kochunov, Castro, et al., 2010; Kochunov & Duff Davis, 2009b) and (K. Phillips & Kochunov, 2011).
Animal handling and MR imaging
Animals were transported from the SNPRC to the Research Imaging Institute, University of Texas Health Sciences Center at San Antonio for imaging. Handling and anesthesia procedures followed previously described procedures (Kochunov and Duff Davis, 2009; Rogers et al., 2007) and are briefly summarized here. Fifteen minutes prior to scanning, animals were immobilized with ketamine (10 mg/kg) and intubated with a MR-compatible endotracheal tube. Anesthesia was maintained with 5% isoflurane with a MR-compatible gas anesthesia machine. Animals remained anesthetized throughout the imaging procedure and respiration rate, heart rate, and oxygen consumption were continually monitored. This protocol and all animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the Texas Biomedical Research Center.
The imaging protocols used to acquire images from all subjects are detailed elsewhere (Kochunov & Duff Davis, 2009a; Rogers, et al., 2007). In short, high-resolution (isotropic 500μm), T1-weighted images were acquired using a 3D IR-TurboFlash sequence optimized for anatomical imaging of baboon brain. An adiabatic inversion recovery (IR) contrast pulse with linear phase encoding schema was employed primarily because it led to a uniform tissue contrast across the imaging volume (being less affected by B1-inhomogeneity/RF penetration artifacts). The sequence control parameters for the adult and postnatal subjects (FOV=128mm, TI=795ms, TE=3.04, TR1=5 ms, TR2=2000ms and flip angle=10 degrees) were modeled to produce GM/WM contrast of 25% based on the analytical solutions to Bloch equations (Deichmann, Good, Josephs, Ashburner, & Turner, 2000) and average measured values of T1, T2 and PD. The model-determined imaging sequence parameters were verified in a group of 5 animals, where group-average WM-GM contrast was calculated to be 25.2 ± 2% (range 22–26%). Image acquisition was performed using a retrospective motion-corrected protocol (Kochunov, et al., 2006). Under this protocol, six full resolution segments, 9 min long each, were acquired for a total sequence running time of ~54min. The sequence control parameters for the in utero subjects (FOV=180mm, T1=, TE=2.5ms, TR1=, TR2=, and flip angle=75 degrees) allow for rapid collection of 3D data phase partition of 360 lines within a single respiration cycle as detailed in (Kochunov & Duff Davis, 2009b). This protocol allowed for a high-SNR and 3D and isotropic coverage of the fetal brain with good, regional gray matter –white matter tissue contrast.
Image processing and measurement of CC
The image processing pipeline consisted of the following steps: removal of non-brain tissue, correction for spatial variations in intensity due to scanner radio-frequency inhomogeneity and global spatial normalization to a population-based template to reduce global variability in brain size and orientation (Figure 1). The details of this processing are described elsewhere (Rogers, et al., 2007). In short, the removal of nonbrain tissue used both automatic (Smith SM, 2002) and manual detailing methods. The correction for RF-inhomogeneity was performed using the FMRIB automated segmentation tool (Smith, et al., 2004). A nine-parameter global spatial normalization procedure was used to reduce intersubject variability in global brain size, shape and orientation, and was performed using the FMRIB linear image registration tool (Smith et al., 2004). A population-based, pseudo-Talairach, median-geometry atlas served as the target brain for global spatial normalization. This atlas was created using methods previously described for humans (Kochunov, et al., 2002) and primates (Kochunov & Duff Davis, 2009b)
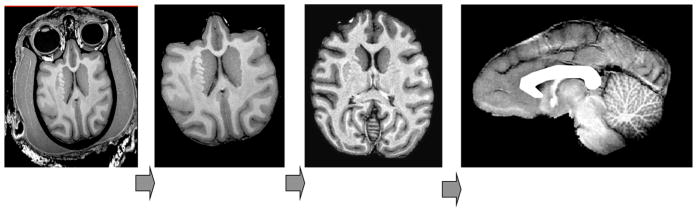
Figure 1.
Structural image processing pipeline, which allows for a simple automation of sequential processing steps. Our pipeline consists of the following steps: removal of non-brain tissue, correction for RF-inhomogeneity artifacts, global spatial normalization (A), hemispheric segmentation (B), tissue classification (C), extraction of the inner/outer cortical surfaces (D,E), extraction of cortical sulci (F), automated labeling of cortical sulci (H) and gyral segmentation (I).
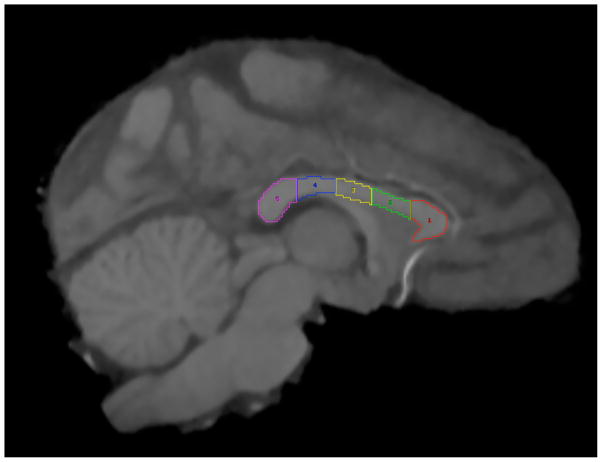
Measurements of CC area were then performed from the midsagittal section, where the CC can be readily identified, using methodology originally described by Biegon and colleagues (Biegon, et al., 1994) and later adapted to nonhuman primates (Sanchez, Hearn, Do, Rilling, & Herndon, 1998). In the original procedure, the anterior 20% of the CC was defined as the genu, the posterior 20% defined as the splenium, and the middle 60% defined as the body. In adapting this to nonhuman primates, Sanchez et al. (1998), Phillips et al. (2007), and Pierre et al. (2008) further delineated the body into three equal regions: anterior midbody, medial midbody, and caudal midbody. These subdivisions of the CC are believed to correspond to functional connectivity with cortical areas (Aboitiz, Scheibel, Fisher, & Zaidel, 1992; Alexander, et al., 2007; Hofer & Frahm, 2006). The anterior region of the genu connects higher-association areas of the frontal lobe; the anterior, medial, and caudal midbody connect primarily sensorimotor regions; the posterior region of the splenium integrates visuospatial regions of the cortex. Analyze 10.0 (Mayo Foundation for Medical Education and Research) was used to divide and measure the midsagittal area of the CC in mm2. To subdivide the CC, the entire length of the CC was first manually traced, then divided into five equally spaced sections (see Figure 2). Two individuals (KAP and EAB) performed measurements of the CC; there was a high degree of concordance in measures, r = .88. Details on the measurements of total CC area and CC subdivision area were provided in (K. Phillips & Kochunov, 2011). Regional development rates were estimated by fitting a linear regression to the dataset consisting of both in-utero and post-natal datapoints. In this way we estimated mm2/week of development in callosal subdivisions.
Figure 2.
Anatomical subdivision of the baboon corpus callosum from MRI sagittal view. The total midsagittal area was divided into five equally spaced subdivisions. 1 = genu; 2 = anterior midbody; 3 = medial midbody; 4 = caudal midbody; 5 = splenium.
Quantitative genetic analysis
Variance components methods, as implemented in the Sequential Oligogenic Linkage Analysis Routines (SOLAR) software package (http://solar.sfbrgenetics.org) (Almasy & Blangero, 1998), were used to estimate the heritability of measured traits. The algorithms in SOLAR employ maximum likelihood variance decomposition methods and are an extension of the strategy developed by (Amos, 1994). The covariance matrix Ω for a pedigree of individuals is given by equation 1:
| (1) |
where sg2 is the genetic variance due to the additive genetic factors, F is the kinship matrix representing the pair-wise kinship coefficients among all animals, se2 is the variance due to individual-specific environmental effects, and I is an identity matrix. The kinship matrix F was calculated based on the known breeding records and was verified by genetic, microsatellite marker based testing that confirmed parent-offspring relationships among baboons.
This produces a multigenerational pedigree that summarizes the genetic relationships among all individuals. For additional explanation of the variance components approach in this context, see (Almasy & Blangero, 1998) and (Blangero, Williams, & Almasy, 2001).
Heritability (h2), the portion of phenotypic variance (sp2) that is accounted for by additive genetic variance (Eq 1), is assessed by contrasting the observed phenotypic covariance matrix with the covariance matrix predicted by kinship. Significance of heritability is tested by comparing the likelihood of the model in which sg2 is constrained to zero with that of a model in which sg2 is estimated. Twice the difference between the two log likelihoods of these models yields a test statistic, which is asymptotically distributed as a 1/2:1/2 mixture of a χ12 variable and a point mass at zero. During testing for the significance of heritability, the phenotype values for each individual are adjusted for a series of covariates. In our analysis we used a polygenic model that estimated the influence of specific variables (additive genetic variation, and covariates including sex, age, age2, age x sex interaction, age2 x sex interaction and random unidentified environmental effects) calculating heritability and its significance (p-value) for each trait’s variance within this population. The level of significance for the heritability analysis for callosal subdivisions was set at p≤ .01 (Bonferroni correction) to reduce the probability of Type 1 errors associated with multiple (N=5) measurements.
Genetic correlation analyses
Bivariate genetic correlation analyses were performed to study the proportion of shared genetic variance between the subdivisions of the CC using methods implemented in the SOLAR software package. Bivarate genetic analysis calculates the magnitude and significance of genetic correlation coefficient (ρG), which is the proportion of variability due to shared genetic effects. The overall phenotypic correlation (ρP) between two traits A and B (equation 2) can be expressed using the correlation due to shared additive genetic effects (ρG) and the residual correlation (ρE) due to shared environmental effects.
| (2) |
Where, h2A and h2B denote the additive genetic heritabilities for each of the traits i.e. the proportion of the total phenotypic variance that is explained by additive genetic factors. If the genetic correlation coefficient (ρG) is significantly different from zero, then the traits are considered to be partially influenced by shared genetic factors (Almasy, Dyer, & Blangero, 1997).
Results
The mean area for callosal subdivisions in adult baboons is shown in Table 1. Quantitative genetic analyses revealed that the total area of the CC and all subdivisions is heritable, with h2 at 0.46±0.16 for the total CC, and ranging from 0.29±0.14 for the splenium to 0.62±0.17 for the medial midbody (see Table 2). Bivarate genetic correlation analysis demonstrated that the individual subdivisions shared between 41% and 98% of genetic variability (Table 2). There were no significant covariates for any of the phenotypes, which was likely a result of using global spatial normalization to correct for differences in head size.
Table 1.
Mean (SD) area, the rates of development (mm2/week), heritability (SD), the proportion of the total variance explained by covariates, and p-values shown for the corpus callosum and regional subdivisions in adult baboons. *Data taken from Phillips and Kochunov, in press
| CC Region | Area (mm2) | SD | Rates of Development* | h2 | SD | p | Significant covariates | % variance explained by covariance |
|---|---|---|---|---|---|---|---|---|
| Total CC | 143.07 | 22.11 | 0.46 | 0.16 | 0.00005 | None | 0.5 | |
| Genu | 34.43 | 5.57 | 0.368 | 0.54 | 0.18 | 8.6·10−6 | None | 0.2 |
| Anterior Midbody | 22.74 | 4.38 | 0.33 | 0.37 | 0.17 | 0.0006 | None | 0.3 |
| Medial Midbody | 24.31 | 4.71 | 0.31 | 0.62 | 0.17 | 2.1·10−6 | None | 0.2 |
| Caudal Midbody | 26.68 | 5.84 | 0.34 | 0.56 | 0.19 | 0.0001 | None | 1.3 |
| Splenium | 34.94 | 5.98 | 0.51 | 0.29 | 0.14 | 0.0007 | None | 0.1 |
Table 2.
Genetic correlations between the subdivisions of the corpus callosum.
| ρP ;ρG;ρE (p) | Genu | Anterior Midbody | Medial Midbody | Caudal Midbody | Splenium |
|---|---|---|---|---|---|
| Genu | 1 | .75; .97; .62 (p=10−34; 10−3; 0.01) | .57; .57; .59 (p=10−13; 0.01; .04) | .59; .57; .59 (p=10−14; 10−3; .15) | .55; .41; .72 (p=10−12; .10; 10−3) |
| Anterior Midbody | 1 | .65; .63; .72 (p=10−18; .02; 0.01) | .75; .97; .62 (p=10−34; 10−3; 0.01) | .58; .81; .52 (p=10−15; .09; 0.01) | |
| Medial Midbody | 1 | .78; .98; .48 (p=10−38; 10−6; 0.14) | .54; .96; .46 (p=10−15; .01; .03) | ||
| Caudal Midbody | 1 | .60; .98; .50 (p=10−19; .01; 0.02) | |||
| Splenium | 1 |
Note: The overall phenotypic correlation (ρP) between two traits is expressed using the correlation due to shared additive genetic effects (ρG) and the residual correlation (ρE) due to shared environmental effects.
To test if the rate of cerebral development was predictive of the level of heritability in adulthood, we plotted the degree of genetic contribution to regional variability (i.e. heritability in size) in the CC subdivisions versus the regional development rates during the fetal and early postnatal period. These rates were determined from brain images of 29 normally developing fetuses, covering the period of gestational week 17 through birth (gestational week 28). Imaging was also performed on 16 baboons up to postnatal week 32. The regional estimates of heritability were negatively correlated with the rates of change in midsagittal CC area during this developmental period (r=0.74, p=0.08) (Table 1).
Discussion
Our study demonstrated significant heritability for the size of the CC and its subdivisions in adult baboons. To our knowledge, this is the first investigation of heritability of the CC subdivisions performed in a pedigree of non-human primates. Heritability of individual callosal subdivisions varied from 0.29 (± 0.14) for the splenium to 0.62 (± 0.17) for the medial midbody, suggesting callosal subdivisions may differ in the degree of genetic contribution to the individual variation but the large standard errors we obtained suggest that in this dataset the heritabilities are not statistically different. Overall, the estimates of heritability in baboons were about half of these reported in humans (Pfefferbaum et al., 2001; Scamvougeras et al., 2003). This discrepancy can potentially be explained by the methodological differences since the human studies did not correct for the intersubject differences in brain volume. The total brain volume is highly heritable in both humans and baboons (Rogers, et al., 2007; Rogers, et al., 2010) and this study aimed to measure genetic contribution of the morphology of CC independent of brain size. The global intersubject differences in brain size and shape can be corrected by spatial normalization which transforms brains into a standard reference frame using a nine-parameter (three: translations, rotations and scaling) where they are adjusted to the same external dimensions (Rogers, et al., 2007). This normalization step was also shown to remove the effects of body weight and sex (Kochunov, et al., 2009; Rogers, et al., 2007). After spatial normalization, variability in brain structure chiefly reflects individual variability in structure’s shape such as the curvature, length and width. Additionally, the investigations of CC heritability in humans were performed in a small number of twin-pairs and used a simplified estimate of heritability that did not model the shared environmental effects which are known for overestimation of the heritability values (Keller, Medland, & Duncan, 2010).
As brain regions associated with more complex reasoning were reported to become increasingly heritable with maturation (Lenroot & Giedd, 2008), we expected higher heritability in subdivisions of the CC connecting higher association areas. The subdivisions of the CC are associated with functional connectivity to cortical regions (Alexander, et al., 2007; Hofer & Frahm, 2006). The anterior regions of the genu and anterior midbody connect primarily higher-order cognitive regions; the medial and caudal midbody connect primarily sensorimotor and motor regions; the posterior region of the splenium integrates visuospatial regions of the cortex. Thus, as the genu and splenium are involved in higher-association cognitive tasks, we expected these subdivisions to have the greatest heritability in adult baboons. When considering heritability of the CC in adults, the genu and splenium did not display the highest heritability rates. The value for the genu, which connects prefrontal regions, is higher than the average across regions, and higher than the value for the total CC, but the value of heritability for the splenium is lower than all other values. As the splenium is involved in visuospatial integration, perhaps experiences in coordination of visual and motor activities are important factors in influencing the variability in size of the splenium. When looking at heritability rates across fetal and early postnatal development, a similar pattern exits. The splenium showed the largest degree of variation due to environmental effects. The results of the genetic correlation analyses revealed that the degree of shared genetic contribution among CC subdivisions is complex and suggested that subdivisions may differ in the degree of genetic contribution. Heritability among subdivisions did not vary along the anterior-posterior direction but spatially adjacent subdivisions shared more genetic variability than more distal regions.
The second aim in this study was to investigate whether genetic contributions to inter-subject variability were modulated by the rate of development. Previously we calculated the regional rates of increase in midsagittal CC areas from fetal and early postnatal baboons (K. Phillips & Kochunov, 2011). Our findings of negative correlation between regional heritability values and the rate of development may suggest that the genetic contributions to regional CC size are negatively correlated with rate of development. Cheverud and colleagues (Cheverud, et al., 1990) drew a connection between developmental factors such as prenatal neurohormonal environment and the genetic versus environmental contributions to variability in the length of cortical sulci. They found lower heritability estimates for the length of the primary sulci that appear later in cerebral development (Cheverud, et al., 1990). This implied that lower heritability for later appearing sulci may be due to higher contributions of environmental factors to the overall phenotypic variance. They suggested that higher environmental contribution to sulcal morphology could be due to changes in prenatal hormone-mediated neurohumoral environment and tissue receptivity, which become progressively more variable during development (Cheverud, et al., 1990). A trend toward higher heritability values for primary cortical structures appearing earlier in development was also reported in humans (Brun, et al., 2008; Chiang, et al., 2008; Le Goualher, et al., 2000; Lohmann, von Cramon, & Colchester, 2007; Lohmann, von Cramon, & Steinmetz, 1999). Our result is supportive of this hypothesis; however, the negative relationship between heritability and the rate of development is primarily driven by the splenium, which had the highest rate of development and the lowest heritability estimate among the CC regions. The value of the correlation coefficient is greatly diminished if splenium is removed from the analysis (r=−0.11 vs. −0.74, respectively).
Understanding how genes and environmental variation determine brain structure and function is fundamentally important to understanding normal and disease-related patterns of neural structure and function. Significant genetic effects have been reported for other brain structures in baboons, including total brain volume and shape, and regions of motor cortex and the superior temporal gyrus (Rogers, et al., 2007). Thus, a consistent pattern of high heritability for brain morphometric measures is seen in baboons. The results of the present study further indicate that Papio baboons are a valuable model for translational neurologic genetic research.
Acknowledgments
This work was supported, in part, by the National Institute of Neurological Disorders and Stroke (R15 #NS070717) to KAP, and the National Institute of Biomedical Imaging and Bioengineering (K01 #EB006395) to PK.
References
- Aboitiz F, Scheibel AB, Fisher RS, Zaidel E. FIber composition of the human corpus callosum. 1992:143–153. doi: 10.1016/0006-8993(92)90178-c. [DOI] [PubMed] [Google Scholar]
- Alexander AL, Lee JE, Lazar M, Boudos R, Dubray MB, Oakes TR, et al. Diffusion tensor imaging of the corpus callosum in autism. NeuroImage. 2007;34:61–73. doi: 10.1016/j.neuroimage.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14(6):953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54(3):535–543. [PMC free article] [PubMed] [Google Scholar]
- Biegon A, Eberling JL, Richardson BC, Roos MS, Wong STS, Reed BR, et al. Human corpus callosum in aging and Alzheimer’s disease: A magnetic resonance imaging study. Neurobiology of Aging. 1994;15(4):393–397. doi: 10.1016/0197-4580(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Variance component methods for detecting complex trait loci. Adv Genet. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- Bonzano L, Tacchino A, Roccatagliata L, Abbruzzese G, Mancardi GL, Bove M. Callosal contributions to simultaneous bimanual finger movements. The Journal of Neuroscience. 2008;28:3227–3233. doi: 10.1523/JNEUROSCI.4076-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer RM, Mandl RCW, Peper JS, Baal GCMv, Kahn RS, Boomsma DI, et al. Heritability of DTI and MTR in nine-year-old children. NeuroImage. 2010;53:1085–1092. doi: 10.1016/j.neuroimage.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Brun C, Lepore N, Pennec X, Chou YY, Lee AD, Barysheva M, et al. A tensor-based morphometry study of genetic influences on brain structure using a new fluid registration method. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 2):914–921. doi: 10.1007/978-3-540-85990-1_110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Falk D, Vannier M, Konigsberg L, Helmkamp RC, Hildebolt C. Heritability of brain size and surface features in rhesus macaques (Macaca mulatta) J Hered. 1990;81(1):51–57. doi: 10.1093/oxfordjournals.jhered.a110924. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Lee AD, Madsen S, Klunder AD, Toga AW, et al. Brain fiber architecture, genetics, and intelligence: a high angular resolution diffusion imaging (HARDI) study. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2008;11(Pt 1):1060–1067. doi: 10.1007/978-3-540-85988-8_126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, McMahon KL, Zubicaray GId, Martin NG, Hickie I, Toga AW, et al. Genetics of white matter development: A DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage. 2011;54:2308–2317. doi: 10.1016/j.neuroimage.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deichmann R, Good C, Josephs O, Ashburner J, Turner R. Optimization of 3-D MP-RAGE sequences for structural brain imaging. Neuroimage. 2000;12(1):112–127. doi: 10.1006/nimg.2000.0601. [DOI] [PubMed] [Google Scholar]
- Di Rocco M, Biancheri R, Rossi A, Filocamo M, Torotori-Donati P. Genetic disorders affecting white matter in the pediatric age. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2004;129B:85–93. doi: 10.1002/ajmg.b.30029. [DOI] [PubMed] [Google Scholar]
- Fears S, Melega WP, Service SK, Lee C, Chen K, Tu Z, et al. Identifying heritable brain phenotypes in an extended pedigree of vervet monkeys. Journal of Neuroscience. 2009;29(9):2867–2875. doi: 10.1523/JNEUROSCI.5153-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer S, Frahm J. Topography of the human corpus callosum revisited - Comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. NeuroImage. 2006;32:989–994. doi: 10.1016/j.neuroimage.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Hulshoff Pol HE, Schnack HG, Posthuma D, Mandl RC, Baare WF, van Oel C, et al. Genetic contributions to human brain morphology and intelligence. Journal of Neuroscience. 2006;26:10235–10242. doi: 10.1523/JNEUROSCI.1312-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC, Medland SE, Duncan LE. Are extended twin family designs worth the trouble? A comparison of the bias, precision, and accuracy of parameters estimated in four twin family models. Behavior Genetics. 2010;40(3):377–393. doi: 10.1007/s10519-009-9320-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Castro C, Davis D, Dudley D, Brewer J, Zhang Y, et al. Mapping primary gyrogenesis during fetal development in primate brains: high-resolution in utero structural MRI of fetal brain development in pregnant baboons. Front Neurosci. 2010;4:20. doi: 10.3389/fnins.2010.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Duff Davis M. Development of structural MR brain imaging protocols to study genetics and maturation. Methods. 2009a doi: 10.1016/j.ymeth.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Duff Davis M. Development of structural MR brain imaging protocols to study genetics and maturation. Methods. 2009b;50(3):136–146. doi: 10.1016/j.ymeth.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Fox PT, JLL, Saleem K, Shelledy W, et al. Genetics of primary cerebral gyrification: Heritability of length, depth and area of primary sulci in an extended pedigree of Papio baboons. Neuroimage. 2010;53:1126–1134. doi: 10.1016/j.neuroimage.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Fox PT, Lancaster J, Saleem K, Shelledy W, et al. Genetics of primary cerebral gyrification: Heritability of length, depth and area of primary sulci in an extended pedigree of Papio baboons. Neuroimage. 2009;15(53):1126–1132. doi: 10.1016/j.neuroimage.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Lancaster J, Wincker P, Smith S, Thompson P, et al. Genetics of microstructure of cerebral white matter using diffusion tensor imaging. Neuroimage. 2010;15(53):1109–1116. doi: 10.1016/j.neuroimage.2010.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Lancaster J, Hardies J, Thompson PM, Woods RP, Cody JD, et al. Mapping structural differences of the corpus callosum in individuals with 18q deletions using targetless regional spatial normalization. Hum Brain Mapp. 2005;24(4):325–331. doi: 10.1002/hbm.20090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Lancaster J, Thompson P, Toga AW, Brewer P, Hardies J, et al. An optimized individual target brain in the Talairach coordinate system. Neuroimage. 2002;17(2):922–927. [PubMed] [Google Scholar]
- Kochunov P, Lancaster J, Glahn DC, Purdy D, Laird AR, Gao F, et al. A Retrospective Motion Correction Protocol for High-Resolution Anatomical MRI. Human Brain Mapping. 2006 doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar AK, Grieve SM, Koslow SH, Gabrieli JD, Gordon E, Williams LW. Loss of white matter integrity in major depressive disorder: Evidence using tract-based spatial statistical analysis of diffusion tensor imaging. Human Brain Mapping. 2010 doi: 10.1002/hbm.21178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goualher G, Argenti AM, Duyme M, Baare WF, Hulshoff Pol HE, Boomsma DI, et al. Statistical sulcal shape comparisons: application to the detection of genetic encoding of the central sulcus shape. Neuroimage. 2000;11(5 Pt 1):564–574. doi: 10.1006/nimg.2000.0559. [DOI] [PubMed] [Google Scholar]
- Leigh SR. Brain growth, life history, and cognition in primate and human evolution. Am J Primatol. 2004;62(3):139–164. doi: 10.1002/ajp.20012. [DOI] [PubMed] [Google Scholar]
- Leigh SR, Shah NF, Buchanan LS. Ontogeny and phylogeny in papionin primates. J Hum Evol. 2003;45(4):285–316. doi: 10.1016/j.jhevol.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Lenroot RK, Giedd JN. The changing impact of genes and environment on brain development during childhood and adolescence: Initial findings from a neuroimaging study of pediatric twins. Dev Psychopathol. 2008;20(4):1161–1175. doi: 10.1017/S0954579408000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann G, von Cramon DY, Colchester AC. Deep Sulcal Landmarks Provide an Organizing Framework for Human Cortical Folding. Cereb Cortex. 2007 doi: 10.1093/cercor/bhm174. [DOI] [PubMed] [Google Scholar]
- Lohmann G, von Cramon DY, Steinmetz H. Sulcal variability of twins. Cereb Cortex. 1999;9(7):754–763. doi: 10.1093/cercor/9.7.754. [DOI] [PubMed] [Google Scholar]
- Lyn H, Pierre P, Bennett AJ, Fears S, Woods R, Hopkins WD. Planum temporale grey matter asymmetries in chimpanzees (Pan troglodytes), vervet (Chlorocebus aethiops sabaeus), rhesus (Macaca mulatta) and bonnet (Macaca radiata) monkeys. Neuropsychologia. 2011 doi: 10.1016/j.neuropsychologia.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler JA, Fox AS, Shelton SE, Rogers J, Dyer TD, Davidson RJ, et al. Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature. 2010;466(7308):864–868. doi: 10.1038/nature09282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington BF, Filipek PA, Lefly D, Chhabildas N, Kennedy DN, Simon JH, et al. A twin MRI study of size variations in the human brain. Journal of Cognitive Neuroscience. 2000;12:223–232. doi: 10.1162/089892900561850. [DOI] [PubMed] [Google Scholar]
- Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Pol HEH. Genetic influences on human brain structure: A review of brain imaging studies in twins. Human Brain Mapping. 2007;28:464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferbaum A, Sullivan EV, Swan GE, Carmella D. Brain structure in men remains highly heritable in the seventh and eighth decades of life. Neurobiology of Aging. 2000;21:63–74. doi: 10.1016/s0197-4580(00)00086-5. [DOI] [PubMed] [Google Scholar]
- Phillips K, Kochunov P. Tracking Development of the Corpus Callosum in Fetal and Early Postnatal Baboons Using Magnetic Resonance Imaging. The Open Neuroimaging Journal. 2011;5 doi: 10.2174/1874440001105010179. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Sherwood CC, Lilak AL. Corpus callosum morphology in capuchin monkeys is influenced by sex and handendess. PLosONE. 2007;2(8):1–7. doi: 10.1371/journal.pone.0000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre PJ, Hopkins WD, Taglialatela JP, Lees CJ, Bennett AJ. Age-related neuroanatomical differences from the juvenile period to adulthood in mother-reared macaques (Macaca radiata) Brain Research. 2008;126:56–60. doi: 10.1016/j.brainres.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJ, Baare WF, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nature Neuroscience. 2002;5:83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- Raff M. Neural development: mysterious no more? Science. 1996;274(5290):1063. doi: 10.1126/science.274.5290.1063. [DOI] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Lancaster J, Shelledy W, Glahn D, Blangero J, et al. Heritability of brain volume, surface area and shape: An MRI study in an extended pedigree of baboons. Hum Brain Mapp. 2007;28(6):576–583. doi: 10.1002/hbm.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Zilles K, Shelledy W, Lancaster J, Thompson P, et al. On the genetic architecture of cortical folding and brain volume in primates. Neuroimage. 2010;53(3):1103–1108. doi: 10.1016/j.neuroimage.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MM, Hearn EF, Do D, Rilling JK, Herndon JG. Differential rearing affects corpus callosum size and cognitive function of rhesus monkeys. Brain Research. 1998;812:38–49. doi: 10.1016/s0006-8993(98)00857-9. [DOI] [PubMed] [Google Scholar]
- Scamvougeras A, Kigar DL, Jones D, Weinberger DR, Witelson SF. Size of the human corpus callosum is genetically determined: an MRI study in mono and dizygotic twins. Neurosci Lett. 2003;338(2):91–94. doi: 10.1016/s0304-3940(02)01333-2. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damasio H. Humans and great apes share a large frontal cortex. Nature Neuroscience. 2002;5:272–276. doi: 10.1038/nn814. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Raghanti MA, Stimpson CD, Spocter MA, Uddin M, Boddy AM, et al. Inhibitory interneurons of the human prefrontal cortex display conserved evolution of the phenotype and related genes. Proceedings of the Royal Society B. 2010;277:1011–1020. doi: 10.1098/rspb.2009.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Jenkinson M, Woolrich M, Beckmann C, Behrens T, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, et al. Genetic influences on brain structure. Nature Neuroscience. 2001;4(12):1–6. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson P. Genetics of brain structure and intelligence. Annual Review of Neuroscience. 2005;28:1–23. doi: 10.1146/annurev.neuro.28.061604.135655. [DOI] [PubMed] [Google Scholar]
- Wang Q, Deng W, Huang C, Li M, Ma X, Wang Y, et al. Abnormalities in connectivity of white-matter tracts in patients with familiar and non-familial schizophrenia. Psychol Med. 2010 doi: 10.1017/S0033291710002412. [DOI] [PubMed] [Google Scholar]
- Zahr NM, Rohlfing T, Pfefferbaum A, Sullivan EV. Problem solving, working memory, and motor correlates of association and commissural fiber bundles in normal aging: a quantitative fiber tracking study. Neuroimage. 2009;44:1050–1062. doi: 10.1016/j.neuroimage.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]