Abstract
The reduction of 12-oxophytodienoic acid (OPDA) to 3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid is catalyzed by 12-oxophytodienoate-10,11-reductase (OPR). Analysis of the isomer preference of OPR has indicated that the activity is composed of two isoenzymes exhibiting different stereoselectivities. The two isoforms of OPR have been separated, using protein extracts of Rock Harlequin (Corydalis sempervirens) as the starting material. OPRI, the enzyme reported earlier from the same species and corresponding to the cloned OPR from Arabidopsis, utilized 9R,13R-OPDA >> 9S,13R-OPDA but not the 13S-configured isomers, whereas the new activity, OPRII, effectively reduced all four OPDA isomers, including the natural 9S,13S-OPDA (cis-[+]-OPDA). OPRII activity is characterized in detail. The enzyme's enzymatic, biochemical, and immunological properties prove that it is a close relative of OPRI. The roles of OPRI and OPRII in octadecanoid biology are discussed.
The biosynthesis of octadecanoids, cyclic metabolites derived from the C18 fatty acid α-linolenic acid (Vick and Zimmerman, 1984), proceeds in three phases that occur in separate cellular compartments. In phase I, α-linolenic acid is converted to its 13(S)-hydroperoxide, the substrate for the AOS/AOC reaction yielding OPDA. In green tissues these reactions occur in the chloroplast (Bell et al., 1995; Blée and Joyard, 1996; Laudert et al., 1996; for review, see Weiler, 1997). In phase II, OPDA is reduced to OPC-8:0 by a flavoprotein reductase, OPR (EC 1.3.1.42), a soluble, cytosolic protein that was characterized for the first time from corn (Vick and Zimmerman, 1986), was later purified to homogeneity from Rock Harlequin (Corydalis sempervirens; Schaller and Weiler, 1997a), and was finally cloned from Arabidopsis (Schaller and Weiler, 1997b). In phase III, OPC-8:0 is subjected to β-oxidation to yield JA (Vick and Zimmerman, 1984). Since β-oxidation in plants occurs only in peroxisomes and glyoxysomes, it is believed that phase III of the octadecanoid biosynthetic pathway is associated with these organelles, although this has not yet been proven.
OPR plays an important role in the biosynthesis of octadecanoids in that the enzyme is one of the factors that determines the pool size of OPDA and provides the substrate OPC-8:0 for the synthesis of JA. Meanwhile, it has been shown several times that plants are able to regulate the levels of OPDA and JA independently of each other (Parchmann et al., 1997; Stelmach et al., 1998). Furthermore, evidence is accumulating that OPDA is a signaling compound in its own right, such as in White Bryony (Bryonia dioica) mechanotransduction (Weiler et al., 1994; Stelmach et al., 1998), and that structural requirements for signaling through a JA pathway are quite different from those required for signaling through an OPDA pathway (S. Blechert and E.W. Weiler, unpublished data). Thus, OPR could be a decisive factor determining the relative abundances of OPDA versus JA in octadecanoid signaling.
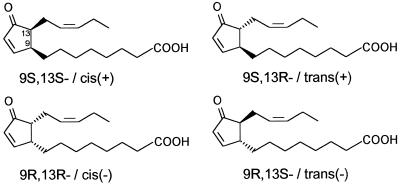
OPDA extracted from tissues is the cis-(+) isomer (Laudert et al., 1997), for which the 9S,13S configuration has been determined at the two stereocenters (Crombie and Mistry, 1988; Grieco and Abood, 1989; Laudert et al., 1997; Fig. 1). Some enzyme preparations, such as flax, may yield a substantial amount of the cis-(−) isomer (9R,13R-OPDA) in addition to cis-(+)-OPDA, and recombinant AOS, in the absence of AOC, gives rise to racemic-cis-OPDA (Laudert et al., 1997). Enolization leads to a slow conversion of cis-OPDA isomers to the trans-isomers, a process also reported for JA (Mueller and Brodschelm, 1994). It is very difficult to analyze the in vivo situation, because thermal isomerization occurs during GC, and a workup of the samples under acidic conditions may lead to enolization as well. Nevertheless, OPDA extracted from tissue is predominantly the cis isomer. This is in contrast to the situation for JA.
Figure 1.
Structures of the four stereoisomers of OPDA.
From control tissue predominantly the trans isomer (−)-JA is extracted (which is at or near thermodynamic equilibrium with the cis isomer [+]-7-iso-JA; Sembdner and Klose, 1985; Mueller and Brodschelm, 1994). When tissue is induced to produce additional JA, such as after wounding or elicitation, the percentage of (+)-7-iso-JA relative to (−)-JA increases (Mueller and Brodschelm, 1994), suggesting that biosynthesis initially yields (+)-7-iso-JA, which then enolizes to some extent in situ and/or during workup and analysis to (−)-JA. However, it cannot be excluded at present that, whereas (+)-7-iso-JA is being synthesized from cis-(+)-OPDA, (−)-JA, at least in control tissue, may originate directly from 9S,13R-OPDA. It is obvious that this process depends on the isomer preference of OPR. The availability of a chiral GC-MS technique for the analysis of enantiomeric composition of OPDA (Laudert et al., 1997) has now allowed the determination of the stereoselectivity of OPR. It quickly became clear that the OPR activity in a plant extract is a composite of two activities representing enzymes of different isomer preference. These isoenzymes were separated and their characteristics determined.
MATERIALS AND METHODS
Materials
Soybean (Glycine max L.) lipoxygenase type I-S, α-linolenic acid, and NADPH were from Sigma, and DEAE-cellulose (DE-52) was obtained from Whatman. Immobiline DryStrip gels, chromatofocusing gel PBE-94, and Polybuffer-74 were from Pharmacia. Nitrocellulose was obtained from Schleicher & Schuell.
Plant Material
A cell-suspension culture of Corydalis sempervirens was grown in Linsmaier and Skoog medium (Linsmaier and Skoog, 1965) at 25°C and 3.8 μmol photons m−2 s−1 illumination on a rotary incubator (100 rpm) for 6 to 7 d. Arabidopsis (race Columbia) plants were grown in a greenhouse in standard soil under short days (8-h photoperiods).
Assay of OPR Activity
Racemic cis-OPDA was obtained from 13(S)-hydroperoxylinolenic acid, as described by Laudert et al. (1997), and purified according to the method of Graff et al. (1990). The (+) enantiomer of cis-OPDA, 9S,13S-OPDA, was prepared using a coupled assay of recombinant AOS and partially purified potato tuber AOC (Laudert et al., 1997). Enantiomeric excess of 9S,13S-OPDA over 9R,13R-OPDA in the final products was 99%. The trans isomers were obtained from the corresponding cis forms through alkaline enolization and HPLC purification (Hamberg and Hughes, 1988). The chemical purity, determined by GC-MS, exceeded 98% for all octadecanoids used as the substrates or standards in this work.
For the determination of OPR enzyme activity, an appropriate amount of enzyme preparation (300 μg of protein from crude plant extracts, 5 μg of protein containing enriched OPRI, or 13 μg of protein containing enriched OPRII, see Results) was incubated in 50 mm potassium phosphate buffer containing 0.1 mm OPDA substrate and 1.0 mm NADPH, pH 7.5, in a total volume of 0.5 mL. The reaction proceeded at a constant rate for at least 60 min at 25°C. Substrate consumption and product formation were determined by capillary GC-MS, as described by Schaller and Weiler (1997a, 1997b). To achieve this the reaction mixtures at the end of the incubation period were acidified (pH 3.0), extracted with peroxide-free diethyl ether, and, after removal of the ether, treated with base (1 n NaOH) for 1 h to convert the cis isomers into their trans forms (this step was omitted when trans-OPDA was used as the substrate). After reextraction with ether, the octadecanoids were converted to the methyl esters by treatment with ethereal diazomethane. The methylated fractions were finally redissolved in CHCl3, and aliquots were subjected to chiral capillary GC-MS (Laudert et al., 1997).
Chiral Capillary GC-MS
Separations were made on a β-Dex120 column (30 m × 0.25 mm × 0.15 μm stationary-phase thickness) coated with 20% permethyl-β-cyclodextrin in SPB-35 (Supelco, Deisenhofen, Germany) using a gas chromatograph (model HP 5890, Hewlett-Packard). Mass spectra were recorded on a TSQ-7000 quadrupole mass spectrometer (Finnigan MAT, Bremen, Germany) operated in electron-impact ionization mode (70 eV). The carrier gas used was helium, and the column was operated isothermically at 190°C. Details of the technique have been reported (Laudert et al., 1997). Quantitation of substrate conversions were based on substrate-to-product ratios derived from integrations of reconstructed total ion current traces, as detailed by Schaller and Weiler (1997a). These ratios are independent of absolute recoveries. The overall absolute recovery of the procedure (from the extraction of OPDA or OPC-8:0 from enzymatic reactions until quantitation by GC-MS) was estimated to be 85% ± 5%.
Separation of OPDA-Reductase Isoforms
Workups of tissue, ammonium sulfate fractionation, and anion-exchange chromatography on Whatman DE-52 were performed as described previously (Schaller and Weiler, 1997a). The protein fractions with OPR activity obtained after anion-exchange chromatography were pooled and the buffer exchanged to 25 mm His, pH 5.5. This material (approximately 35 mg of protein) was loaded onto an HR 10/10 column (Pharmacia) filled with the chromatofocusing resin PBE-94. The column was then rinsed for 10 min with the His buffer, and proteins were eluted with 8-fold diluted Polybuffer-74 (pH 5.5) at a flow rate of 1 mL min−1. Fractions of 2 mL (or in some cases 4 mL) were collected for biochemical and enzymatic analyses.
Biochemical Methods
Blue native-PAGE was performed on 4% to 15% gradient gels (Schägger et al., 1994; Oecking et al., 1997) at 4°C to 7°C at 100 V until the sample had reached the separating gel. Separations were then carried out at 400 V. For the second dimension, the blue native-gel strips were soaked in SDS sample buffer (>1 h) and the denatured proteins in the gel strips were then subjected to SDS-PAGE, according to the method of Laemmli (1970), or they were soaked in 1% SDS and 1% β-mercaptoethanol for 1 h followed by electrophoresis, according to the method of Schägger and von Jagow (1987). For the determination of enzyme activity, proteins were eluted from the blue native gels by incubation two times for 2 h in 50 mm potassium phosphate buffer, pH 7.5.
The transfer of proteins from SDS-polyacrylamide gels to nitrocellulose was achieved electrophoretically either at 4°C and 64 mA for 16 h or at 200 mA for 2 h (Towbin et al., 1979). Immunolabeling of the OPDA reductases was performed as described by Schaller and Weiler (1997b).
IEF of proteins was performed on precast gels (Immobiline DryStrips) in a horizontal flat-bed chamber (Pharmacia) using the protocol of Görg et al. (1995). For second-dimension separations, the gel strips were attached to the sampling gel and a 12.5% separating polyacrylamide gel (Laemmli, 1970).
Protein was determined according to the method of Bradford (1976) using BSA as a standard.
RESULTS
Chiral GC-MS of OPDA and OPC-8:0 Isomers
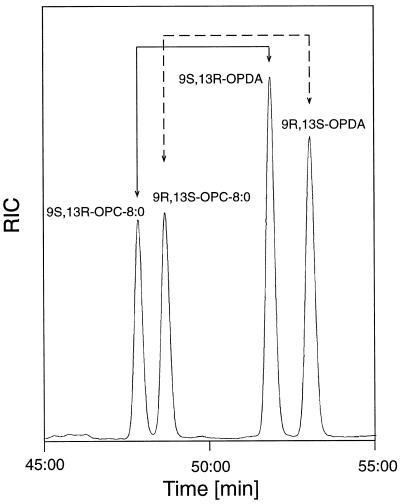
Both the cis and the trans enantiomers of OPDA can be separated on a permethyl-γ-cyclodextrin chiral stationary phase (Laudert et al., 1997); however, retention times for the cis enantiomers are exceedingly long. Under the same conditions, the cis enantiomers of OPC-8:0 were incompletely separated (data not shown). On the other hand, the trans enantiomers of OPDA and of OPC-8:0 could be separated on a single column with acceptable retention times when using permethyl-β-cyclodextrin as the stationary phase (Fig. 2). The identity of all compounds was verified by full-scan electron impact mass-spectral analysis (data not shown), and the assignments of the peaks to the enantiomeric forms were made using either racemic or optically pure trans-(+)-OPDA (9S,13R-OPDA) prepared from racemic cis-OPDA or cis-(+)-OPDA (9S,13S-OPDA) by base treatment. This treatment did not lead to any detectable racemization at C9 (data not shown). The assignment of the OPC-8:0 enantiomers was made using standards produced enzymatically from either racemic cis-OPDA or 9S,13S-OPDA. Again, reduction of OPDA using OPR did not lead to racemization at C9. Thus, if cis enantiomers of OPDA were used as the OPR substrates, conversion of the cis to the trans isomers was carried out on ether-extracted reaction mixtures prior to methylation and GC-MS analysis. The rates of substrate conversion were calculated from reconstructed ion current traces of full-scan electron impact mass spectra using the peak areas of individual, corresponding OPDA-OPC-8:0 pairs (total area of both peaks: 100%). Reaction rates were calculated using several discrete data points during the linear phase of the reaction collected during a reaction period of 1 h.
Figure 2.
Separation and analysis of the enantiomers of trans-OPDA (9R,13S-OPDA and 9S,13R-OPDA) and of the enantiomers of trans-OPC-8:0 (9R,13S-OPC-8:0 and 9S,13R-OPC-8:0) on a permethyl-β-cyclodextrin stationary phase by capillary GC-MS. RIC, Reconstructed total ion current. Retention times were as follows (minutes:seconds): 9S,13R-OPC-8:0, 42:42; 9R,13S-OPC-8:0, 43:20; 9S,13R-OPDA, 46:42; and 9R,13S-OPDA, 47:54.
Evidence for the Occurrence of Isoforms of OPR Exhibiting Different Isomer Preferences
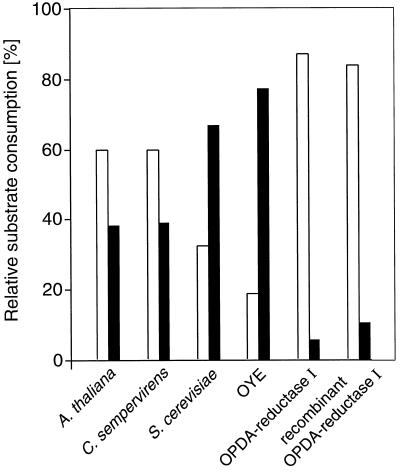
Crude extracts from Arabidopsis leaf tissue or C. sempervirens cell cultures, when given racemic-cis-OPDA, reduced both enantiomeric cis forms to the corresponding OPC-8:0 enantiomers with, in each case, a slight substrate preference for 9R,13R-OPDA (Fig. 3). OPR is a member of the OYE family of flavoprotein reductases (Schaller and Weiler, 1997b), and these authors showed that Saccharomyces cerevisiae OYE possessed OPR activity. It is interesting that both S. cerevisiae crude enzyme extracts and affinity-purified OYE prefer the 9S,13S enantiomer (i.e. the one naturally occurring in plants) as a substrate. On the other hand, OPR purified from the C. sempervirens cell culture, according to the method of Schaller and Weiler (1997a), shows a clear preference for 9R,13R-OPDA over 9S,13S-OPDA as a substrate, as does OPR cloned from Arabidopsis (Schaller and Weiler, 1997b; cf. Fig. 3). Taken together, the data in Figure 3 suggest that in both plant species, a second OPR isoenzyme must exist in crude extracts, which, in contrast to the purified and cloned enzymes, reduces 9S,13S-OPDA, the naturally occurring isomer.
Figure 3.
Conversion of racemic-cis-OPDA by plant and yeast enzyme preparations. Shown is the substrate consumption of 9S,13S-OPDA (black bars) relative to 9R,13R-OPDA (white bars) after a total reaction time of 1 h at 25°C (0.1 mm racemic-cis-OPDA, 1 mm NADPH) during which the reactions proceeded at a constant rate. Arabidopsis, C. sempervirens, and S. cerevisiae: 300 μg of total soluble protein; OYE, 7.5 μg of S. cerevisiae OYE purified by N-(4-hydroxybenzoyl)aminohexyl-Sepharose affinity chromatography; OPDA-reductase I, 4.9 μg of purified C. sempervirens OPR (Schaller and Weiler, 1997a); recombinant OPDA-reductase I, 20 μg of recombinant Arabidopsis OPR (Schaller and Weiler, 1997b) expressed in insect cells (Sf9; F. Schaller, unpublished data) and purified by N-(4-hydroxybenzoyl)aminohexyl-Sepharose affinity chromatography (Abramovitz and Massey, 1976). The amounts of enzyme used for each sample were adjusted to result in comparable absolute rates of enzymatic substrate conversion (100% = 2.8 ± 0.68 pkat).
Separation and Characterization of OPR Isoenzymes
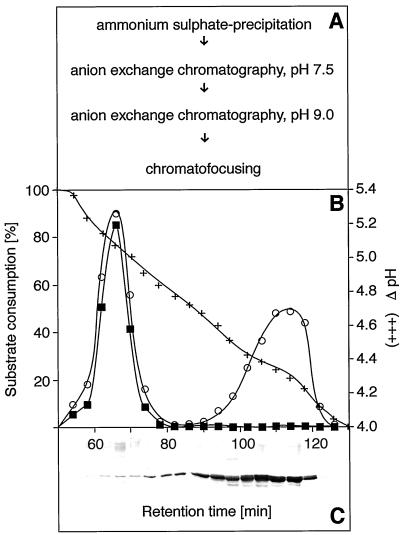
The results obtained for the enantiomer preference of OYE (compare Fig. 3) suggested that closely related OPR isoforms may exhibit different substrate specificities with respect to OPDA isomers. It was found that 9R,13R- and 9S,13S-converting activities initially copurified when the protocol of Schaller and Weiler was used (1997a). A modified purification scheme was therefore devised (Fig. 4A) during which separation of two OPR activities was achieved by chromatofocusing (Fig. 4B). One activity, eluting between 100 and 120 min, converted almost exclusively 9R,13R-OPDA and was associated with the occurrence of a strongly immunoreactive 42-kD band using an antiserum against C. sempervirens OPR (Schaller and Weiler, 1997a; Fig. 4C). This activity thus corresponded to the enzyme purified earlier (Schaller and Weiler, 1997a). A second peak of OPR activity eluted between 60 and 75 min from the chromatofocusing gel. The C. sempervirens OPR antiserum detected a minor immunoreactive band at approximately 42 kD in these fractions (compare Fig. 4C). This enzyme effectively converted both 9S,13S-OPDA and 9R,13R-OPDA to the corresponding OPC-8:0 enantiomers. The new activity is termed OPRII activity, whereas the enzyme described earlier is now designated OPRI.
Figure 4.
Purification protocol (A), separation by chromatofocusing (B), and immunoblot analysis (C) of C. sempervirens OPR isoforms. To determine OPR activity, 0.2-mL aliquots of the individual 4-mL fractions were reacted under standard conditions with either racemic-cis-OPDA (○) or optically active cis-(+)-OPDA (9S,13S-OPDA) (▪). Immunoblot analysis followed the procedure given by Schaller and Weiler (1997b). Substrate consumption: 100% = 100 μm substrate consumed in a total assay volume of 0.5 mL, equaling an absolute amount of 50 nmol of substrate converted. Note that reaction rates are only linear until about 50% of the substrate has been consumed (compare Fig. 7 for time courses).
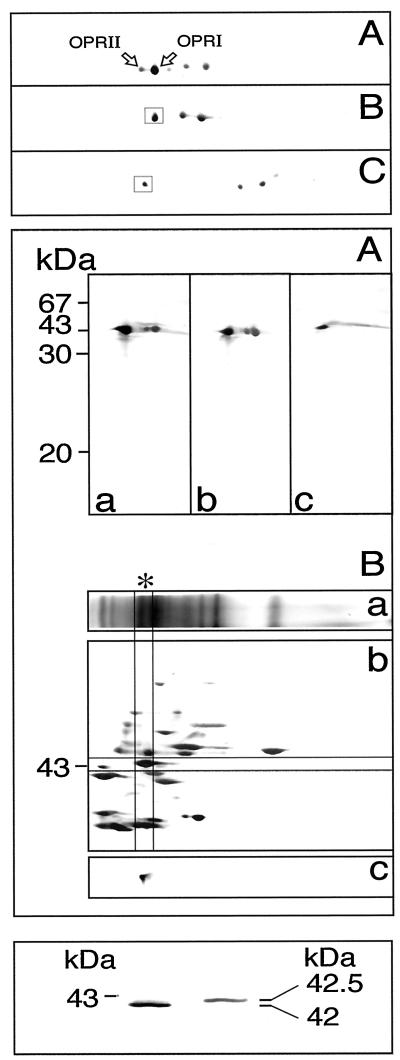
Protein from the OPRI and OPRII activity peaks was analyzed further by two-dimensional gel electrophoresis using different gel systems (Fig. 5). Two-dimensional separations using IEF as the first and SDS-PAGE as the second dimension followed by immunoblotting (Fig. 5, top) allowed the identification of the OPRI and OPRII polypeptides (spots in box), which were also clearly separated when the protein was analyzed prior to chromatofocusing (Fig. 5, top, A). OPRII exhibited a slightly higher pI than did OPRI. The remaining spots on the immunoblots represent cross-reacting polypeptides and/or degradation products of OPR. This could be shown using blue native-gel electrophoresis instead of IEF as the first dimension (Fig. 5, middle). On blue native gels, OPRI and II were incompletely separated (Fig. 5A, middle, lane a versus b versus c). The technique, however, allowed enzymatic activity to be determined after the first-dimension electrophoresis.
Figure 5.
Gel-electrophoretic analysis of OPRI and OPRII isoforms separated by chromatofocusing as in Figure 4B. For the analysis, OPRI was prepared by pooling fractions corresponding to fraction nos. 110 to 120 and OPRII corresponding to fraction nos. 62 to 70 in Figure 4B. Top, Two-dimensional analysis using IEF as the first and SDS-PAGE as the second dimension. A, Immunoblot of the protein fraction prior to separation of OPRI and OPRII by chromatofocusing. B, Immunoblot analysis of the OPRI fraction. C, Immunoblot analysis of the OPRII fraction. Middle, Two-dimensional analysis using blue native-gel electrophoresis as the first and SDS-PAGE as the second dimension. A, Immunoblot analysis of two-dimensional gels with SDS-PAGE as the second dimension. Lane a, Protein fraction prior to separation of OPRI and OPRII by chromatofocusing; lane b, OPRI fraction; lane c, OPRII fraction. B, Blue native-PAGE analysis of the OPRII fraction. Lane a, One-dimensional blue native-PAGE of the OPRII fraction, Coomassie-stained gel. The asterisk marks the zone of OPR activity. Lane b, Blue native-PAGE followed by SDS-PAGE (Laemmli, 1970) silver-stained gel. Lane c, As in lane b, but immunoblot (only relevant sector shown). Bottom, One-dimensional SDS-PAGE (Laemmli, 1970) using a 12.5% separating gel of the OPRI and OPRII fraction, followed by immunoblotting. Left, Position of the nearest marker in kD. OPRI and OPRII exhibit slightly different apparent molecular masses of 42 and 42.5 kD, respectively.
A single zone of activity was detected (Fig. 5, middle, B, lane a, asterisk) that coincided with the major immunoreactive polypeptides in Figure 5, middle, A. Thus, the additional immunoreactive bands associated with the OPRI and OPRII fractions (Fig. 5, middle, A, lane a versus b versus c) were devoid of enzymatic activity. When OPRII was first separated from OPRI by chromatofocusing and then subjected to blue native-gel electrophoresis, the OPRII activity zone, after SDS-PAGE separation in the second dimension (Fig. 5, middle, B, lane b), gave a single immunoreactive spot when the C. sempervirens antiserum raised against OPR was used (Fig. 5, middle, B, lane c). One-dimensional SDS-PAGE of the two fractions separated by chromatofocusing followed by immunoblotting revealed OPRII to be slightly larger (42.5 kD) than OPRI (42 kD; Fig. 5, bottom). This difference in apparent molecular masses was also evident when the immunoblots (Fig. 5, middle, A, lane b versus c) were compared. Together, the data allowed us to assign OPRI and OPRII activity unequivocally to the immunoreactive polypeptides, as identified in Figure 5, top, A.
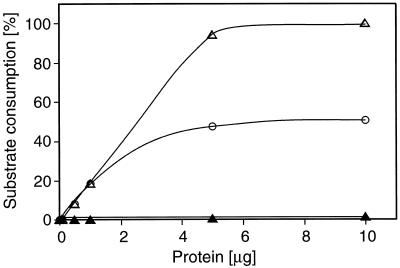
OPRII exhibits the same temperature optimum (35°C) but has a more acidic pH optimum (7.5–8.0) than OPRI (8.7; Schaller and Weiler, 1997a). Like OPRI, OPRII is active as the monomer and is retained on N-(4-hydroxybenzoyl)aminohexyl-Sepharose affinity columns (Schaller and Weiler, 1997b), a matrix originally devised to purify yeast OYE (Abramovitz and Massey, 1976). Since racemic-cis-OPDA had been used in earlier studies on OPRI, it was next analyzed to determine whether the lack of conversion of the natural 9S,13S-OPDA by OPRI was due to an inhibitory effect of 9R,13R-OPDA. For this, racemic-cis-OPDA was incubated in the presence of increasing amounts of OPRI. Figure 6 shows that even when 9R,13R-OPDA is completely consumed, 9S,13S-OPDA is not utilized by the enzyme. Thus, inhibitory effects of 9R,13R-OPDA on the conversion of 9S,13S-OPDA can be excluded. OPRI apparently does not utilize the natural 9S,13S-OPDA as a substrate. Likewise, no indications for one cis enantiomer affecting the rate of conversion of the other have been obtained for OPRII (data not shown). The substrate concentrations giving half-maximum rates of conversion by OPRII for the two enantiomers present in racemic-cis-OPDA were 21 μm (9R,13R-OPDA) and 15 μm (9S,13S-OPDA).
Figure 6.
Enantiomer consumption by OPRI using racemic-cis-OPDA as the substrate (0.1 mm) as a function of protein concentration. ○, Racemic-cis-OPDA; ▵, 9R,13R-OPDA; and ▴, 9S,13S-OPDA; 100%: 25 nmol OPDA, equivalent to quantitative conversion of cis-(−)-OPDA from the racemate.
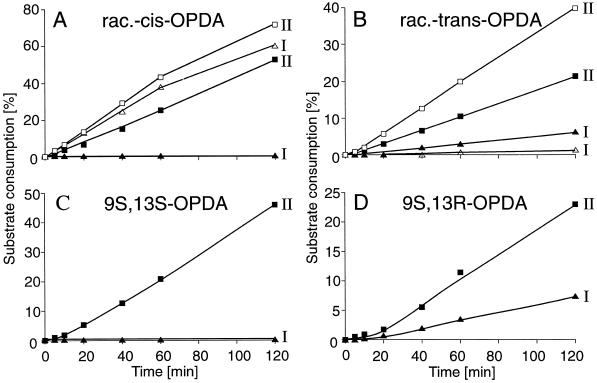
Finally, the conversions of the trans and cis enantiomers of OPDA by OPRI or OPRII were compared using either racemic-cis-OPDA versus 9S,13S-OPDA as the substrates (Fig. 7, A and C) or racemic-trans-OPDA versus 9S,13R-OPDA (the trans isomer produced from 9S,13S-OPDA by enolization and occurring in small amounts in plants; Fig. 7, B and D). Whereas OPRI does not convert 9S,13S-OPDA (Fig. 7C, triangles), the enzyme does convert the corresponding trans isomer, 9S,13R-OPDA (Fig. 7D, triangles), albeit at a lower rate compared with OPRII (Fig. 7D, squares). With respect to these enantiomers, conversion by both enzymes was comparable irrespective of whether optically pure substrate or the racemic substrates were used (compare closed symbols in Fig. 7, A versus C and B versus D). The conversion of the levorotatory cis enantiomer 9R,13R-OPDA by OPRI and OPRII was comparable (Fig. 7A, open symbols), whereas a sharp discrimination was again observed for the levorotatory trans isomer, 9R,13S-OPDA, which was not readily converted by OPRI but was effectively utilized by OPRII. Thus, OPRII is able to utilize all four isomers of OPDA effectively, whereas OPRI is more specific and utilizes 9R,13R-OPDA >> 9S,13R-OPDA, and the enzyme has a marginal to zero reactivity against the two 13S-configurated OPDA isoforms.
Figure 7.
Enantiomer selectivity of OPRI and OPRII from C. sempervirens. A, Substrate racemic-cis-OPDA: 9S,13S-/9R,13R-OPDA. B, Substrate racemic-trans-OPDA: 9S,13R-/9R,13S-OPDA. C, Substrate 9S, 13S-OPDA. D, Substrate 9S, 13R-OPDA. All substrates were tested under standard conditions at 0.1 mm. OPRI, Five micrograms of protein corresponding to pooled fractions 110 to 120 in Figure 4B; OPR II, 13 μg of protein corresponding to pooled fractions 62 to 70 in Figure 4B. ▵, Conversion of 9R,13R-OPDA or 9R,13S-OPDA by OPRI; □, conversion of 9R,13R-OPDA or 9R,13S-OPDA by OPRII; ▴, conversion of 9S,13S-OPDA and 9S,13R-OPDA by OPRI; ▪, conversion of 9S,13S-OPDA and 9S,13R-OPDA by OPRII. A linear rate of substrate consumption of 10% per hour corresponds to an absolute OPR activity of 1.4 pkat. I, OPRI; II, OPRII; rac., racemic.
DISCUSSION
Our data show that two isoenzymes of OPR occur in C. sempervirens (and, by inference from the data in Fig. 3, also in Arabidopsis). They are similar in their molecular masses, pI values, immunological reactivities, and overall biochemical and enzymatic properties, but they differ in their preference for OPDA isomers. The new activity described here (OPRII) is the only one that is able to convert the natural 9S,13S-OPDA to the corresponding 9S,13S-OPC-8:0, the precursor of (+)-7-epi-JA. The physiological role of OPRI remains to be shown. This enzyme does not utilize 9S,13S-OPDA as a substrate and has a moderate activity against 9S,13R-OPDA, the trans isomer originating from enolization of 9S,13S-OPDA. OPDA extracted from control plant tissues is predominantly the cis-(+) enantiomer, 9S,13S-OPDA, with a few of the corresponding trans forms (which might, however, arise from work-up and GC analysis; Laudert et al., 1997). On the other hand, JA isolated from control tissue is largely the trans form, (−)-JA. It seems possible that OPRI functions in removing 9S,13R-OPDA, which might otherwise accumulate from enolization of 9S,13S-OPDA. From this reaction product, 9S,13R-OPC-8:0, (−)-JA would be produced through β-oxidation.
Another function of OPRI might be the removal of 9R,13R-OPDA. It has been shown (Baertschi et al., 1988; Laudert et al., 1996) that AOS in vitro yields racemic-cis-OPDA. In the presence of AOC, 9S,13S-OPDA is being formed in vitro, depending on the activity of AOC. In some enzyme preparations, e.g. from flax, a significant by-product of 9R,13R-OPDA is always observed (Baertschi et al., 1988; Laudert et al., 1996). However, cis-OPDA extracted from plant tissues is exclusively the 9S,13S enantiomer. This could mean that (a) in vivo coupling of AOS and AOC is so effective as to avoid the formation of 9R,13R-OPDA or (b) this enantiomer is being removed by some other process. OPRI could be involved here.
It remains a possibility that OPRI is not an enzyme of octadecanoid biosynthesis but in vivo has a different, yet-unidentified substrate. This requires further work. On the other hand, the occurrence of two OPR isoforms, only one of which would convert the endogenously accumulating 9S,13S isomer (whereas the second isoenzyme would remove all other isomers except this one), bears some logic in that it would allow the cell to maintain a pool of pure 9S,13S-OPDA as a precursor for (+)-7-iso-JA and as a signal in its own right (Weiler et al., 1994; Parchmann et al., 1997; Stelmach et al., 1998) by removing unwanted isomers originating from side reactions. Control over OPRII activity would furthermore allow the cell to generate transients of 9S,13S-OPDA accumulation without concomitant transients in JA accumulation. Such a process has been observed repeatedly (Parchmann et al., 1997; Stelmach et al., 1998). Full appreciation of the roles of OPRI and OPRII will await cloning of OPRII and the analysis of both isoenzymes in transgenic plants. This work is in progress.
ACKNOWLEDGMENT
The authors wish to thank Dr. Claudia Oecking, Bochum, for introducing them to the technique of blue native-gel electrophoresis.
Abbreviations:
- AOC
allene oxide cyclase
- AOS
allene oxide synthase
- JA
jasmonic acid
- OPC-8:0
3-oxo-2(2′[Z]-pentenyl)-cyclopentane-1-octanoic acid
- OPDA
12-oxophytodienoic acid
- OPR
12-oxophytodienoate-10,11-reductase
- OYE
old yellow enzyme
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft, Bonn, and Fonds der Chemischen Industrie, Frankfurt (literature provision).
LITERATURE CITED
- Abramovitz AS, Massey V. Purification of intact old yellow enzyme using an affinity matrix for the sole chromatographic step. J Biol Chem. 1976;251:5321–5326. [PubMed] [Google Scholar]
- Baertschi SW, Ingram CD, Harris TM, Brash AR. Absolute configuration of cis-12-oxophytodienoic acid of flaxseed: implications for the mechanism of biosynthesis from the 13(S)-hydroperoxide of linolenic acid. Biochemistry. 1988;27:18–24. doi: 10.1021/bi00401a004. [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blée E, Joyard J. Envelope membranes from spinach chloroplasts are a site of metabolism of fatty acid hydroperoxides. Plant Physiol. 1996;110:445–454. doi: 10.1104/pp.110.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Crombie L, Mistry KM (1988) Synthesis of 12-oxophytodienoic acid. J Chem Soc Chem Commun 537–539
- Görg A, Boguth G, Obermaier C, Posch A, Weiss W. Two-dimensional polyacrylamide gel electrophoresis with immobilized pH gradients in the first dimension (IPG-Dalt): the state of the art and the controversy of vertical versus horizontal systems. Electrophoresis. 1995;16:1079–1086. doi: 10.1002/elps.11501601183. [DOI] [PubMed] [Google Scholar]
- Graff G, Anderson LA, Jaques LW. Preparation and purification of soybean lipoxygenase derived unsaturated hydroperoxy and hydroxy fatty acids and determination of molar absorptivities of hydroxy fatty acids. Anal Biochem. 1990;188:38–47. doi: 10.1016/0003-2697(90)90525-e. [DOI] [PubMed] [Google Scholar]
- Grieco PA, Abood N. Cycloalkenone synthesis via Lewis acid catalyzed retro Diels-Alder reactions of norbonene derivatives: synthesis of 12-oxophytodienoic acid. J Org Chem. 1989;54:6008–6010. [Google Scholar]
- Hamberg M, Hughes MA. Fatty acid allene oxides. III. Albumin-induced cyclization of 12,13(S)-epoxy-9(Z),11-octadecadienoic acid. Lipids. 1988;23:469–475. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laudert D, Hennig P, Stelmach BA, Müller A, Andert L, Weiler EW. Analysis of 12-oxo-phytodienoic acid enantiomers in biological samples by capillary gas chromatography-mass spectrometry using cyclodextrin stationary phases. Anal Biochem. 1997;246:211–217. doi: 10.1006/abio.1997.2012. [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Holländer-Czytko H, Weiler EW. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Linsmaier EM, Skoog F. Organic growth factor requirements of tobacco tissue cultures. Physiol Plant. 1965;18:100–127. [Google Scholar]
- Mueller MJ, Brodschelm W. Quantification of jasmonic acid by capillary gas chromatography-negative chemical ionization-mass spectrometry. Anal Biochem. 1994;218:425–435. doi: 10.1006/abio.1994.1202. [DOI] [PubMed] [Google Scholar]
- Oecking C, Piotrowski M, Hagermeier J, Hagemann K. Topology and target interaction of the fusicoccin-binding 14–3-3 homologs of Commelina communis. Plant J. 1997;12:441–453. [Google Scholar]
- Parchmann S, Gundlach H, Mueller MJ. Induction of 12-oxo-phytodienoic acid in wounded plants and elicited plant cell cultures. Plant Physiol. 1997;115:1057–1064. doi: 10.1104/pp.115.3.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F, Weiler EW. Enzymes of octadecanoid biosynthesis in plants. 12-oxo-phytodienoate 10,11-reductase. Eur J Biochem. 1997a;245:294–299. doi: 10.1111/j.1432-1033.1997.t01-1-00294.x. [DOI] [PubMed] [Google Scholar]
- Schaller F, Weiler EW. Molecular cloning and characterization of 12-oxophytodienoate reductase, an enzyme of the octadecanoid signaling pathway from Arabidopsis thaliana. J Biol Chem. 1997b;272:28066–28072. doi: 10.1074/jbc.272.44.28066. [DOI] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1–100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sembdner G, Klose C. (−)-Jasmonsäure–ein neues Phytohormon? Biol Rundsch. 1985;23:29–40. [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW. Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in mechanotransduction. Phytochemistry. 1998;47:539–546. doi: 10.1016/s0031-9422(97)00547-5. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. Biosynthesis of jasmonic acid by several plant species. Plant Physiol. 1984;75:458–461. doi: 10.1104/pp.75.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA, Zimmerman DC. Characterization of 12-oxo-phytodienoic acid reductase in corn. Plant Physiol. 1986;80:202–205. doi: 10.1104/pp.80.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler EW. Octadecanoid-mediated signal transduction in higher plants. Naturwissenschaften. 1997;84:340–349. [Google Scholar]
- Weiler EW, Kutchan TM, Gorba T, Brodschelm W, Niesel U, Bublitz F. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 1994;345:9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]