Abstract
Background
The 2-oxoglutarate dependent superfamily is a diverse group of non-haem dioxygenases, and is present in prokaryotes, eukaryotes, and archaea. The enzymes differ in substrate preference and reaction chemistry, a factor that precludes their classification by homology studies and electronic annotation schemes alone. In this work, I propose and explore the rationale of using substrates to classify structurally similar alpha-ketoglutarate dependent enzymes.
Findings
Differential catalysis in phylogenetic clades of 2-OG dependent enzymes, is determined by the interactions of a subset of active-site amino acids. Identifying these with existing computational methods is challenging and not feasible for all proteins. A clustering protocol based on validated mechanisms of catalysis of known molecules, in tandem with group specific hidden markov model profiles is able to differentiate and sequester these enzymes. Access to this repository is by a web server that compares user defined unknown sequences to these pre-defined profiles and outputs a list of predicted catalytic domains. The server is free and is accessible at the following URL ( http://comp-biol.theacms.in/H2OGpred.html).
Conclusions
The proposed stratification is a novel attempt at classifying and predicting 2-oxoglutarate dependent function. In addition, the server will provide researchers with a tool to compare their data to a comprehensive list of HMM profiles of catalytic domains. This work, will aid efforts by investigators to screen and characterize putative 2-OG dependent sequences. The profile database will be updated at regular intervals.
Keywords: Hidden Markov Model, Facial triad, Ferryl, Dioxygenase
Background
Dioxygenases, which include pterin- and 2-OG-dependent, Rieske di-hydroxylases and extradiol dioxygenases, have a conserved triad (His-Asp/Glu-His) of amino acids that are responsible for product formation [1]. The 2-OG dependent subgroup comprises members that are non-haem in character, require iron (II), and 2-oxoglutarate as a co-substrate for catalysis. Members of this superfamily are ubiquitous in nature, possess a DSBH fold (Double Stranded Beta-Helical), and the major coordinating amino acids are (HX[DE]XnH). Iron interacts with the pair of histidine residues and aspartate/glutamate along one face of a distorted octahedral sphere, whilst, the other face is normally coordinated by three molecular waters. In the presence of 2-oxoglutarate (bi-dentate), the last dative covalent bond is with the substrate. The accepted general route to α-KG dependent catalytic conversion, requires, an increase in the oxidation state of iron (FeII → FeIII-superoxo → FeIV-peroxo) to a high-spin reactive ferryl intermediate (Fe1V = O) [2], proton abstraction, and substrate radical formation. The transformation itself could be an oxidative- introduction of a hydroxyl group, simultaneous removal of adjacent hydrogen atoms (desaturase activity), sulfate cleavage, and cyclopentane-, stereoisomer-, chlorinated adduct- formation [3-8]. These enzymes participate in hypoxic signaling, DNA repair, stress response mechanisms, lipid and growth factor metabolism, and biodegradation of herbicides [9-15].
Existent, publically accessible computational tools and databases use homology studies to cluster proteins with 2-OG dependent function. These, provide information on sequences with evidence of common ancestry (pairwise sequence identity > 30%). Hidden Markov Models, are theoretically sound formulations of stochastic processes, being used with increasing frequency in computational biology. The output of a HMM is, a markov chain of likely consecutive states, along with their associated transitional probabilities. This class of machine learning methods is well suited to comparing divergently evolved sequences (pairwise sequence identity ~ 10-25%). InterPro is a database of protein signatures that combines information from several sources, and is used as an automatic annotation tool for new sequences. There are a number of Hidden Markov Model based predictors of protein function and classification. Pfam is a repository of protein families formed by sequence and structural similarity, and organization of distant domain architectures; SMART searches protein sequences for pre-defined regulatory domain architectures using Pfam, signal peptides, trans-membrane helices, regions of low complexity, and internal repeats; SUPERFAMILY and Gene3D integrate fold and domain data with genomic and taxonomic information to provide a comprehensive resource for proteins of interest [16-20]. These algorithms, despite providing initial pointers to the reaction chemistry of novel 2-OG dependent sequences, are unable to segregate closely related proteins with reference to their substrate preferences. Other tools focus on factors that influence intra-cellular location, propensity for protein-protein interaction, organelle targeting, and sequence patterns, rather than active-site composition and catalysis (SMART, PROSITE) [18,21].
The utilities vide supra, are protein sequence/structure specific. In this work, I have used a reverse look-up strategy to infer function of related proteins from the nature and similarity of the substrates catalyzed. The prediction protocol uses, in-house coded PERL scripts in conjunction with existing protein analysis tools, to create a profile database. This, is then compared with user-defined sequences, and the presence/ absence of alpha-KG dependent function and a suitable catalytic profile are suggested.
Methods
Computational Tools used in this work
Structural data was downloaded from the RCSB PDB server (Research Consortium for Structural Bioinformatics Protein Data Bank) [22]. Pair wise analysis of structures was done using DaliLite [23]. Analysis of active site residues was done with the SPDBV (Swiss PDB viewer), alignments and cladograms were generated with the STRAP suite of programs (Structural Alignment of Proteins), and HMMER 3.0 was used for model building, analysis, database construction, and similarity studies with user defined input sequences [24-26]. All the above software was downloaded and installed locally. Sequence ids and information on predicted domains and were from UniProtKB in association with InterPro, Pfam, SMART, and PROSITE [16-18,21,27].
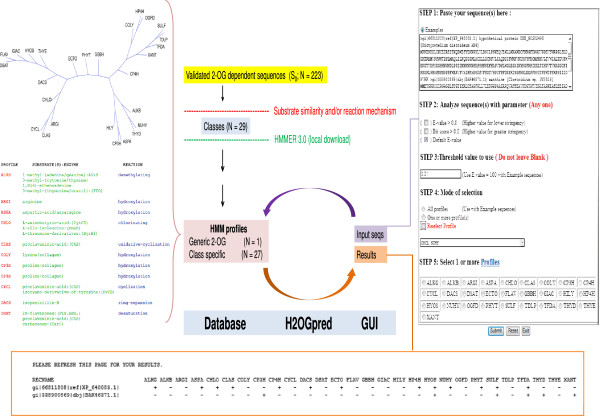
I coded the PERL scripts needed to interface the front- and back- ends of the server with HMMER-3.0 and perform other miscellaneous tasks. The GUI (Graphical User Interface) for input and the results page were coded and designed by me using HTML (Hyper Text Markup Language) and CSS (Cascading Style Sheets). A concise workflow, along with salient features of H2OGpred is presented (Figure 1).
Figure 1.
Outline of protocol used to predict catalytic domains in user-defined sequences. Salient features of H2OGpred: the HMM profiles used, formulation of the query required to search the database for suitable matches, and a summary of the profiles found in the sequence(s) of interest. Other details include (not shown): instructions for use, general scheme of the 2-OG dependent reaction, and detailed scores used by HMMER-3.0 for profile assignment.
Dataset creation and initial analysis
Several alpha-ketoglutarate enzymes have associated empirical data present in medical literature. This constitutes: demonstration of activity in vitro and in vivo (EC 1.14.11.x), transcript data with biochemical and/or physiological function, and the presence of a structure. These sequences (N = 223), were collated and comprised the template set (S0; Additional file 1: Table S1). This was divided into a training- (S1T, N = 81) and a validation- (S1V, N = 142) set of sequences. Early work to assess the catalytic profile of each member of S0, was done by searching for suitable domains in publically available databases (Additional file 2: Table S2). The feasibility of a substrate centric classification of αKG-dependent enzyme members was investigated subsequently. This was done by analyzing proteins with considerable structural similarity (Z score ≥ 20.0, Additional file 3: Table S3), and in complex with dissimilar preferred substrates and/or analogs. Differences in the amino acids that lined the substrate pocket were tabulated.
Construction of profile database and server
The 2-OG dependent enzymes are multi-functional catalysts. Clavaminate synthase (EC 1.14.11.21) transforms proclavaminate and/or analogs by introducing a hydroxyl group, double bond, and effecting a ring closure reaction [3]. The 2 S-flavanones, are similarly desaturated and hydroxylated by flavone-, flavonol-, and anthocyanidin- synthases (EC 1.14.11.x, x = 19, 22, 23) and flavanone 3-dioxygenase (EC 1.14.11. 9) [4]. Integrating prior information for each of the above enzymes (S0), such as reaction chemistry, participating macromolecules, simple organic compounds which include endogenous (amino acids, acyl-CoA molecules) and exogenous (herbicides, pesticides, detergents), and molecular and atomic level detail (transferred element or functional group), a secondary filter was set up. The resultant sub-clusters constituted overlapping members, were descriptively annotated, profiled as HMMs, and a sequence signature pattern composed of alignment specific identical amino acids, was assigned to each (Additional file 4: Table S4). In addition, class specific consensus sequences were generated and aligned. This data was used to create an unrooted cladogram (Figure 1).
The complete list of HMMs (N = 28), comprised, a superfamily (S1T) and group (S2; by analogy) specific models. The selection of sequences for the generic, αKG-profile (ALKG) was done to ensure adequate coverage and even sampling of S0. Classes with single enzyme members were excluded (ATSK; PTLH). The profile database created is available as (Additional file 5: Table S5; aKG-profile-database.hmm). Interface to this repository is through H2OGpred, a server that accepts user defined protein sequences, and predict domains specific to a particular substrate.
Findings
This study highlights and discusses the following characteristics of the 2-OG dependent superfamily. There are observable differences in the reaction mechanisms and/or substrates transformed in structurally related enzymes (Table 1, Figure 2). These variations are with reference to the amino acids that border the substrate binding pocket, interact with 2-OG, Fe(II), and participate in alpha-KG specific domain formation. A detailed analysis of predicted domains in previously collated sequences (S0, Additional file 2: Table S2), using publically accessible tools, revealed that, the TauD family (PF02668, sequences = 4205, non-redundant PDB ids = 8), consists of enzymes such as: taurine dioxygenase, alkylsulfataseK, asparagine oxygenase, carbapenem synthase C, L-arginine-beta-hydroxylase, and gamma-butyrobetaine hydroxylase among others. Similarly, the PhyH family (PF05721, sequences = 2319, non-redundant PDB ids = 3) encompasses activities of phytanoyl-CoA-dioxygenase, ectoine hydroxylase, and pentalenolactone synthase. Interestingly, all the above catalyze different substrates, clearly demonstrating the lack of discriminatory indices in current literature to delineate function in similar proteins.
Table 1.
Comparison between structurally similar 2-OG dependent proteins
| Taurine dioxygenase (TauD) | Alkylsulfatase (AtsK) | |
|---|---|---|
|
Organism |
Escherichia coli |
Pseudomonas putida |
|
alpha/beta content |
|
|
| DSBH (core) |
β (5–8,14-17) |
β (1–7;13–16) |
| Extended |
β (9–10), α (1–6) |
β (8–12), α (3–5) |
|
PDB id |
1GQW (Ref.
[229]) |
1OIK (Ref.
[227]) |
|
Sequence identity (%) |
41-42 |
|
|
Z score (rmsd) |
31.8-34.4 (1.2-1.3) |
|
|
Active site geometry (amino acid nos.) |
|
|
| Sphere of radius 5 A0 (Fe/2-OG/Substrate) |
18 |
17 |
| Identical residues |
13 |
|
|
Reaction catalyzed |
sulfate cleaving activity |
|
|
Substrate profile |
taurine (sulfonic acids) |
aliphatic sulfate esters |
| |
Phytanoyl -CoA hydroxylase(PAHX) |
Pentalenolactone hydrolase(PtlH) |
|
Organism |
Homo sapiens |
Streptomyces avermitilis |
|
alpha/beta content |
|
|
| DSBH (core) |
β (6,8-13) β (2-4, 7-10) |
|
| Extended |
β (1–2,5,15) β (1), a (1-6) |
|
|
PDB id |
2A1X (Ref.
[220]) |
2RDN (Ref.
[226]) |
|
Sequence identity (%) |
20 |
|
|
Z score (rmsd) |
17.1 (2.9) |
|
|
Amino acids within sphere of radius 5 A0(Fe/2-OG/Substrate) |
12 |
18 |
|
Reaction catalyzed |
hydroxylation |
ring closure |
| Substrate profile | phytanoyl CoA (medium chain) | 1-deoxypentalenic acid |
Figure 2.
Alignment and active site analysis of structurally similar pairs of proteins. Inter-molecular substrate modifying residues (1 or more atoms within 5 A0 of atom(s) of compound of interest) have been tabulated and compared. Color scheme for highlighting: Red – identity, Blue- protein specific, Black – amino acids with their side chains pointing away from the substrate, suggesting a structural role.
As an alternate approach to this problem, I, hypothesized that substrate interacting amino acids in the active site might be used to further classify structurally similar enzymes. To test this rationale, select pairs of the 2-OG dependent superfamily were analyzed and compared. The results indicate, that despite similarities in the composition of the active site, subtle differences exist in the nature of these additional substrate-modifying residues (Figure 2), which, in turn could correlate to differential catalytic behavior. The sub-classes formed by utilizing substrates as clustering parameters are evolutionarily diverse (Table 2, Figure 1). Nevertheless, the frequency of identical amino acids (iaa) in the signature patterns of the profiles (iaa = 0, N = 4; iaa = 1 – 2, N = 3; iaa > 2, N = 20), suggests, the existence of a conserved domain architecture for each group (Additional file 4: Table S4). These functional profiles compare favorably with classically annotated protein specific domains using as criteria: prediction of function, redundancy, and segregation of structurally related proteins (Table 3). A catalytic domain, in this work is defined, hereafter, as a HMM of a group of sequences with similar substrate preferences and/ or reaction chemistry.
Table 2.
Classification of 2-OG dependent dioxygenases
| Family (This work)* | |
|---|---|
|
REACTION |
|
| Demethylation |
ALKB, COLY |
| Chlorinating |
CHLO |
| Ring closure (cyclization) |
CYCL |
| Desaturation |
CLAS, CYCL, FLAV |
| Sulfate cleavage |
TDLP |
| Ring expansion |
DACS |
| Ether bond cleavage |
TFDA |
| Hydroxylation |
NUHY(THYD,THYE,XANT), CP3H,CP4H,HP4H,ARGI,ASPA,ECTO,PHYT,GBBH,HYOS, PTLH |
|
SUBSTRATE |
|
| Amino acid/protein/derivative |
|
| Arginine |
ARGI |
| Aspartyl; Asparagine |
ASPA |
| Lysine |
COLY, HILY |
| Proline |
CP3H,CP4H,HP4H |
| Sulfonic acids (taurine), isethionate, taurocholate |
SULF (TDLP) |
| gamma-butyrobetaine |
GBBH |
| Collagen |
CP3H,CP4H,COLY |
| Nucleotide/ nucleoside |
|
| Thymidine |
THYD |
| Thymine |
THYE |
| Xanthine |
XANT |
| Misc. organic |
|
| Pro-clavaminate |
CLAS |
| Deacetoxycephalosporin |
DACS |
| Ectoine |
ECTO |
| 2 S-flavanones |
FLAV |
| Gibberellins |
GIAC |
| Hyoscyamine |
HYOS |
| Phytanoyl-CoA |
PHYT |
| PAA based pesticides/ herbicides |
TFDA |
| Cyclopentane |
PTLH |
| Aliphatic sulfate esters |
ATSK |
| Translation in eukaryotes (eIF2α) | OGFD |
(*) Note: Details of protein sequences used in this analysis (uniprot & PDB ids, references) are included as Additional file1: Table S1.
Table 3.
Comparative analysis of catalytic domains of template sequences *
| InterPro | PFAM | SMART | PROSITE | |
|---|---|---|---|---|
|
Group 1 |
|
|
|
|
| FLAV |
Oxoglutarate/Fe-dep_oxygenase |
2OG-FeII_Oxy |
|
FE2OG_OXY |
| |
Isopenicillin-N_synthase |
|
|
|
| GIAC |
Oxoglutarate/Fe-dep_oxygenase |
2OG-FeII_Oxy |
|
FE2OG_OXY |
| |
Isopenicillin-N_synthase |
|
|
|
| HYOS |
Oxoglutarate/Fe-dep_oxygenase |
2OG-FeII_Oxy |
|
FE2OG_OXY |
| |
Isopenicillin-N_synthase |
|
|
|
| DACS |
Isopenicillin-N_synth_CS |
2OG-FeII_Oxy |
|
FE2OG_OXY |
| |
Oxoglutarate/Fe-dep_oxygenase |
|
|
IPNS_1 |
| |
|
|
|
IPNS_2 |
| THYE |
Oxoglutarate/Fe-dep_oxygenase |
2OG-FeII_Oxy |
|
FE2OG_OXY |
|
Group 2 |
|
|
|
|
| TDLP |
Taurine_dOase |
TauD |
|
|
| ATSK |
Taurine_dOase |
TauD |
|
|
| TFDA |
Taurine_dOase |
TauD |
|
|
| XANT |
Taurine_dOase |
TauD |
|
|
|
Group 3 |
|
|
|
|
| PHYT |
Phytyl_CoA_dOase |
PhyH |
|
|
| PTLH |
Phytyl_CoA_dOase |
PhyH |
|
|
| CP4H |
Oxoglutarate/Fe-dep_oxygenase |
2-OG-FeII_Oxy |
P4Hc |
FE2OG_OXY |
| |
Pro_4_hyd_alph |
P4Ha_N |
ShKT |
TPR |
| |
Pro_4_hyd_alph_N |
ShK |
|
TPR_REGION |
| |
ShK_toxin |
|
|
|
| |
TPR-contain |
|
|
|
| |
TPR-like_helical |
|
|
|
| |
TPR_repeat |
|
|
|
| HP4H |
Oxoglutarate/Fe-dep_oxygenase |
2OG-FeII_Oxy |
P4Hc |
FE2OG_OXY |
| |
Pro_4_hyd_alph |
Cupin_4 |
|
ZF_MYND_1 |
| |
Cupin_JmjC |
zf-MYND |
|
ZF_MYND_2 |
| |
Znf_MYND |
Ofd1_CTDD |
|
|
| Oxoglutarate/Fe-dep_Oase_C |
(*) Note: Complete domain analysis of template dataset is included as Additional file 2: Table S2.
Discussion
A fundamental detail of alpha-ketoglutarate dependent catalysis is the range of substrates transformed, and the distinct reaction mechanisms deployed. This remarkable feature is, despite the presence of several common structural features such as the presence of the jellyroll fold, active site composition, and the presence of the facial-triad of residues (Table 1). Several attempts to classify these enzymes have been made previously [28,29]. Both, sequence-based studies: location of the facial triad (central; C-terminal, flavanol synthase), number of amino acids between His-X-[Asp/Glu] and the terminal His (125 a. a, taurine dioxygenase, alkylsulfatase; 57 a. a, anthocyanidin synthase; 85 a. a, phytanoyl-CoA- hydroxylase), and reaction specific structural features such as the presence, location, composition, and conformational arrangements of conserved active site residues. These approaches, albeit informative are unable to account for the catalytic spectrum observed within sub-groups of the superfamily. This is attributed to subtle modifications in the distribution patterns of a few amino acids, and may, constitute an extended active site. Thus, the presence of small hydrophobic residues in alkylsulfatase K (V84, A104), as opposed to the corresponding charged residues in the related enzymes, i.e., taurine dioxygenase (Y73, D94, N95), ensure that sulfonic acids, modified amino and bile acids, are preferred over aliphatic sulfur esters (pair 1, Figure 2). Clavaminic acid synthase 1 and asparagine oxygenase share remarkable structural similarity. The presence of a glutamic acid residue (HEH, facial triad), and high Z score, notwithstanding, CAS1 is a tri-functional catalyst with a completely different set of preferred compounds (pair 2, Figure 2). Similarly, use of 2 S-flavanones (FLAV profile) in preference to isopenicillin N (deacetoxycephalosporin synthase, EC 1.14.11.26) is a function of a few specialized residues (pair 3, Figure 2). Characterizing these residues by homology alignments and subsequent mutagenesis experiments are currently the only known means to ascribe function.
2 – OG dependent enzymes catalyze the hydroxylation of their substrates. However, this step may also occur concomitantly with other reactions. In these cases, an intermediate substrate radical is the precursor for a subsequent catalytic event. Clearly, the notion of a substrate molecule as a passive transformant is passé, with increasing evidence of its role in modulating catalysis. Prediction, by existing tools, of product forming domains in an enzyme specific to a particular substrate is generic, with no information on substrate specificity for a number of families (Group 1, Table 3), whilst, the same catalytic domain for a reference sequence is assigned to a number of other enzymes (Groups −2 and 3, Table 3). However, by integrating the profiles it is possible to infer the function of an unknown protein. The HP4H (hypoxia inducible prolyl 4-hydroxylase, EC 1.14.11.29) domain in a protein, is an important indicator of a role in regulating downstream genes in response to hypoxic conditions, the same may be inferred from the ensemble of predicted domains (Pro_4_hyd_alph; Znf_MYND; Cupin_JmjC). Similarly, CP4H (collagen prolyl 4-hydroxylase, EC 1.14.11.2) function may be postulated by integrating its domain profile (Pro_4_hyd_alph; TPR_helical; TPR-contain).
These latter examples (Group 4, Table 3) suggest that absence of prior information will limit the utility of this substrate centric, profile assignment process. A sequence with no suitable profile matches might require a comparison by homology studies to existing/ computationally annotated protein sequences. Despite these constraints, this novel schema is able to categorize closely related protein sequences. As biochemical details of a greater number of enzymes emerge, it will be possible to develop improved docking algorithms and statistical models of the chemical signature of a substrate molecule. This could then predict active site conformers of a particular enzyme for an individual substrate.
Description of H2OGpred
The web server works by comparing sequences with each of these pre-defined HMM profiles (Figure 1). There is a brief introduction to the salient features of α-KG dependent enzymes, and general instructions of use. Users can paste their sequences of interest, select a threshold parameter and value and search the profile database. Output files comprise a tabular summary of suitably matched profiles, and detailed statistics with pair wise alignments. Details of the profiles are present as a hyperlink and combined with the result as a separate file. New users may utilize the examples option to analyze and view preliminary results. The server has been tested with approximately 250 sequences, pasted at once.
Validation of H2OGpred as a predictor 2-OG dependent catalysis
To verify functionality of the server, proteins that were not used to construct the generic profile (S1V), were analyzed further. The server was correctly able to predict the presence of a single 2-OG domain in all test sequences (N = 142). Further, two novel sequences, have been experimentally validated (unpublished data) with demonstration of catalytic activity towards their preferred substrates in concurrence with the top scoring profiles assigned by the server.
Conclusions
The 2-oxoglutarate dependent enzymes are amongst the largest group of non-haem dioxygenases, rivaling the more established mediators of xenobiotic metabolism, the cytochrome P450 family of haem monooxygenases. Current information on novel non-haem 2-OG dependent iron (II) enzymes is sparse, and relies on sequence/structure-based homology studies. In addition, complete biochemical characterization often necessitates prior knowledge of potential substrates. Here, I, have compiled a list of enzymes previously validated by several workers [30-236], and categorized them on the similarity of the reactions they catalyze, and/or, of the compounds they modify. The resulting HMMs are then used to construct a map of putative catalytic domains, thereby suggesting, a list of potential molecules that new, uncharacterized sequences might transform.
Enzyme members of the αKG-dependent superfamily are downstream mediators of a stimulus-induced-compensatory stress response in several organisms. This includes cycles of, cellular hypoxia and altered expression patterns of regulatory and effector genes, exposure to herbicides and arsenic with activation of catabolic pathways, and high salinity and thermal stress with overproduction of compatible solutes. Thus, an insight into the reaction chemistry of these proteins has the potential to aid development of newer classes of antimicrobials, bio-degradable compounds, and efficacious metabolic regulators.
Availability
Project URL: http://comp-biol.theacms.in/H2OGpred.html.
Usage: Free and no login required.
Abbreviations
2-OG, 2-oxoglutarate; HMM, Hidden Markov model; GUI, Graphical user interface.
Competing interests
The authors declare that they have no competing interests.
Author’s contribution
SK manually collated all the sequences and their references, carried out the computational analysis, constructed the server, designed the GUI, wrote all the code, and the manuscript.
Supplementary Material
Table S1. Uniprot and PDB ids of sequences used in this work.
Table S2. Comparative domain analysis of template sequences.
Table S3. Pair wise structural alignment of selected proteins.
Table S4. Analysis of HMM profiles with highlighted active site residues. (PDF 29 kb)
Table S5. Link to HMM profile database “profile-DB\aKG-profile-database.hmm”.
Acknowledgements
I wish to thank the editorial board and anonymous reviewers for suggestions on revising the manuscript.
I wish to thank Rajesh. S. Gokhale (Ph. D, Director – Institute of Genomics and Integrative Biology, New Delhi, INDIA), for introducing me to the scientific problem and providing several helpful suggestions during the course of this work.
References
- Koehntop KD, Emerson JP, Que L. The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes. Biol Inorg Chem. 2005;10(Suppl 2):87–93. doi: 10.1007/s00775-005-0624-x. [DOI] [PubMed] [Google Scholar]
- Price JC, Barr EW, Tirupati B, Bollinger JM, Krebs C. The first direct characterization of a high-valent iron intermediate in the reaction of an alpha-ketoglutarate-dependent dioxygenase: a high-spin FeIV complex in taurine/alpha-ketoglutarate dioxygenase (TauD) from Escherichia coli. Biochemistry. 2003;42(Suppl 24):7497–7508. doi: 10.1021/bi030011f. [DOI] [PubMed] [Google Scholar]
- Bredebach M, Matern U, Martens S. Three 2-oxoglutarate-dependent dioxygenases activities of Equisetum arvense L. forming flavone and flavonol from (2S)-naringenin. Phytochemistry. 2011;72(Suppl 7):557–563. doi: 10.1016/j.phytochem.2011.01.036. [DOI] [PubMed] [Google Scholar]
- Busby RW, Townsend CA. A single monomeric iron center in clavaminate synthase catalyzes three nonsuccessive oxidative transformations. Bioorg Med Chem. 1996;4(Suppl 7):1059–1064. doi: 10.1016/0968-0896(96)00088-0. [DOI] [PubMed] [Google Scholar]
- You Z, Omura S, Ikeda H, Cane DE. Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlH, a non-heme iron dioxygenase of Streptomyces avermitilis. J Am Chem Soc. 2006;128(Suppl 20):6566–6567. doi: 10.1021/ja061469i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M, Sandala GM, Smith DM, Schofield CJ, Easton CJ, Radom L. The unusual bifunctional catalysis of epimerization and desaturation by carbapenem synthase. J Am Chem Soc. 2004;126(Suppl 32):9932–9933. doi: 10.1021/ja047899v. [DOI] [PubMed] [Google Scholar]
- Vaillancourt FH, Vosburg DA, Walsh CT. Dichlorination and bromination of a threonyl-S-carrier protein by the non-heme Fe(II) halogenase SyrB2. Chembiochem. 2006;7(Suppl 5):748–752. doi: 10.1002/cbic.200500480. [DOI] [PubMed] [Google Scholar]
- Grzyska PK, Ryle MJ, Monterosso GR, Liu J, Ballou DP, Hausinger RP. Steady-state and transient kinetic analyses of taurine/alpha-ketoglutarate dioxygenase: effects of oxygen concentration, alternative sulfonates, and active-site variants on the FeIV-oxo intermediate. Biochemistry. 2005;44(Suppl 10):3845–3855. doi: 10.1021/bi048746n. [DOI] [PubMed] [Google Scholar]
- Chen YH, Comeaux LM, Eyles SJ, Knapp MJ. Auto-hydroxylation of FIH-1: an Fe(ii), alpha-ketoglutarate-dependent human hypoxia sensor. Chem Commun (Camb) 2008;39:4768–4770. doi: 10.1039/b809099h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419(Suppl 6903):178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- Bursy J, Pierik AJ, Pica N, Bremer E. Osmotically induced synthesis of the compatible solute hydroxyectoine is mediated by an evolutionarily conserved ectoine hydroxylase. J Biol Chem. 2007;282(Suppl 43):31147–31155. doi: 10.1074/jbc.M704023200. [DOI] [PubMed] [Google Scholar]
- Wehner KA, Schütz S, Sarnow P. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol Cell Biol. 2010;30(Suppl 8):2006–2016. doi: 10.1128/MCB.01350-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen GA, Mihalik SJ, Watkins PA, Moser HW, Jakobs C, Denis S, Wanders RJ. Phytanoyl-CoA hydroxylase is present in human liver, located in peroxisomes, and deficient in Zellweger syndrome: direct, unequivocal evidence for the new, revised pathway of phytanic acid alpha-oxidation in humans. Biochem Biophys Res Commun. 1996;229(Suppl 1):205–210. doi: 10.1006/bbrc.1996.1781. [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444(Suppl 1):11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- Saari RE, Hausinger RP. Ascorbic acid-dependent turnover and reactivation of 2,4-dichlorophenoxyacetic acid/alpha-ketoglutarate dioxygenase using thiophenoxyacetic acid. Biochemistry. 1998;37(Suppl 9):3035–3042. doi: 10.1021/bi972388p. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov EM. InterPro Consortium. InterPro--an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16(12):1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26(1):320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30(1):242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C. Assignment of Homology to Genome Sequences using a Library of Hidden Markov Models that Represent all Proteins of Known Structure. J Mol Biol. 2001;313(4):903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Lees J, Yeats C, Redfern O, Clegg A, Orengo C. Gene3D: merging structure and function for a Thousand genomes. NAR. 2009;38:D296–D300. doi: 10.1093/nar/gkp987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The RCSB PDB server. p. . http://www.rcsb.org/pdb/
- Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gille C, Frommel C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics. 2001;17(4):377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- The HMMER-3.0 site. p. . http://hmmer.janelia.org.
- The UniProtKB server. p. . http://www.uniprot.org/
- Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39(1):21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100(4):644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452(7190):961–965. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundheim O, Vågbø CB, Bjørås M, Sousa MM, Talstad V, Aas PA, Drabløs F, Krokan HE, Tainer JA, Slupphaug G. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25(14):3389–3397. doi: 10.1038/sj.emboj.7601219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q, Cheng W, Wang J, Feng Y, Chai J. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464(7292):1205–1209. doi: 10.1038/nature08921. [DOI] [PubMed] [Google Scholar]
- Westbye MP, Feyzi E, Aas PA, Vågbø CB, Talstad VA, Kavli B, Hagen L, Sundheim O, Akbari M, Liabakk NB, Slupphaug G, Otterlei M, Krokan H. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283(36):25046–25056. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PJ, Hollis T. Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching. PLoS One. 2010;5(1):e8680. doi: 10.1371/journal.pone.0008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E, Bekkelund A, Moen MN, Omelchenko MV, Klungland A, Falnes PØ. Bioinformatics and functional analysis define four distinct groups of AlkB DNA-dioxygenases in bacteria. Nucleic Acids Res. 2009;37(21):7124–7136. doi: 10.1093/nar/gkp774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ren J, Stammers DK, Baldwin JE, Harlos K, Schofield CJ. Structural origins of the selectivity of the trifunctional oxygenase clavaminic acid synthase. Nat Struct Biol. 2000;7(Suppl 2):127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- Helmetag V, Samel SA, Thomas MG, Marahiel MA, Essen LO. Structural basis for the erythro-stereospecificity of the L-arginine oxygenase VioC in viomycin biosynthesis. FEBS J. 2009;276(13):3669–3682. doi: 10.1111/j.1742-4658.2009.07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby RW, Chang MD, Busby RC, Wimp J, Townsend CA. Expression and purification of two isozymes of clavaminate synthase and initial characterization of the iron binding site. General error analysis in polymerase chain reaction amplification. J Biol Chem. 1995;270(9):4262–4269. doi: 10.1074/jbc.270.9.4262. [DOI] [PubMed] [Google Scholar]
- Janc JW, Egan LA, Townsend CA. Purification and characterization of clavaminate synthase from Streptomyces antibioticus. A multifunctional enzyme of clavam biosynthesis. J Biol Chem. 1995;270(10):5399–5404. doi: 10.1074/jbc.270.10.5399. [DOI] [PubMed] [Google Scholar]
- Hewitson KS, Holmes SL, Ehrismann D, Hardy AP, Chowdhury R, Schofield CJ, McDonough MA. Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH) J Biol Chem. 2008;283(Suppl 38):25971–25978. doi: 10.1074/jbc.M804999200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strieker M, Kopp F, Mahlert C, Essen L-O, Marahiel MA. Mechanistic and structural basis of stereospecific Cbeta-hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide. ACS Chem Biol. 2007;2:187–196. doi: 10.1021/cb700012y. [DOI] [PubMed] [Google Scholar]
- Krojer T, Kochan G, Pilka E, Hozjan V, Allerston CK, Bray J, Muniz JR, Chaikuad A, Gileadi O, Kavanagh K, von Delft F, Bountra C, Arrowsmith CH, Weigelt J, Edwards A, Oppermann U. Crystal structure of human aspartate beta-hydroxylase isoform a. p. . To be Published.
- Wang Q, Vandusen WJ, Petroski CJ, Garsky VM, Stern AM, Friedman PA. Bovine liver aspartyl beta-hydroxylase. Purification and characterization. J Biol Chem. 1991;266:14004–14010. [PubMed] [Google Scholar]
- Dinchuk JE, Henderson NL, Burn TC, Huber R, Ho SP, Link J, O'Neil KT, Focht RJ, Scully MS, Hollis JM, Hollis GF, Friedman PA. Aspartyl beta -hydroxylase (Asph) and an evolutionarily conserved isoform of Asph missing the catalytic domain share exons with junctin. J Biol Chem. 2000;275:39543–39554. doi: 10.1074/jbc.M006753200. [DOI] [PubMed] [Google Scholar]
- Silbermann E, Moskal P, Bowling N, Tong M, de la Monte SM. Role of aspartyl-(asparaginyl)-β-hydroxylase mediated notch signaling in cerebellar development and function. Behav Brain Funct. 2010;6:68. doi: 10.1186/1744-9081-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke RS, VanDusen WJ, Garsky VM, Jacobs JW, Sardana MK, Stern AM, Friedman PA. Aspartyl beta-hydroxylase: in vitro hydroxylation of a synthetic peptide based on the structure of the first growth factor-like domain of human factor IX. Proc Natl Acad Sci U S A. 1989;86(10):3609–3613. doi: 10.1073/pnas.86.10.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenflo J, Holme E, Lindstedt S, Chandramouli N, Huang LH, Tam JP, Merrifield RB. Hydroxylation of aspartic acid in domains homologous to the epidermal growth factor precursor is catalyzed by a 2-oxoglutarate-dependent dioxygenase. Proc Natl Acad Sci U S A. 1989;86(Suppl 2):444–447. doi: 10.1073/pnas.86.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neary JM, Powell A, Gordon L, Milne C, Flett F, Wilkinson B, Smith CP, Micklefield J. An asparagine oxygenase (AsnO) and a 3-ydroxyasparaginyl phosphotransferase (HasP) are involved in the biosynthesis of calcium-dependent lipopeptide antibiotics. Microbiology. 2007;153(Pt 3):768–776. doi: 10.1099/mic.0.2006/002725-0. [DOI] [PubMed] [Google Scholar]
- Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Crystal structure of the non- haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature. 2006;440(7082):368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- Wong C, Fujimori DG, Walsh CT, Drennan CL. Structural analysis of an open active site conformation of nonheme iron halogenase CytC3. J Am Chem Soc. 2009;131(13):4872–4879. doi: 10.1021/ja8097355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt FH, Yeh E, Vosburg DA, O'Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436(7054):1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- Vaillancourt FH, Yin J, Walsh CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII alpha-ketoglutarate- and O2-dependent halogenase. Proc Natl Acad Sci U S A. 2005;102(29):10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri M, Krojer T, Bagg EA, Webby CJ, Butler DS, Kochan G, Kavanagh KL, Oppermann U, McDonough MA, Schofield CJ. Crystal structure of the 2-oxoglutarate- and Fe(II)-dependent lysyl hydroxylase JMJD6. J Mol Biol. 2010;401:211–222. doi: 10.1016/j.jmb.2010.05.054. [DOI] [PubMed] [Google Scholar]
- Valtavaara M, Papponen H, Pirttila AM, Hiltunen K, Helander H, Myllylae R. Cloning and characterization of a novel human lysyl hydroxylase isoform highly expressed in pancreas and muscle. J Biol Chem. 1997;272:6831–6834. doi: 10.1074/jbc.272.11.6831. [DOI] [PubMed] [Google Scholar]
- Armstrong LC, Last JA. Rat lysyl hydroxylase: molecular cloning, mRNA distribution and expression in a baculovirus system. Biochim Biophys Acta. 1995;1264:93–102. doi: 10.1016/0167-4781(95)00130-9. [DOI] [PubMed] [Google Scholar]
- Mercer DK, Nicol PF, Wright MC. Robins SP Cloning and characterisation of lysyl hydroxylase isoforms 2 and 3 isolated from rat hepatic stellate cells. Biochem Biophys Res Commun. 2003;307:803–809. doi: 10.1016/S0006-291X(03)01262-2. [DOI] [PubMed] [Google Scholar]
- Ruotsalainen H, Sipila L, Kerkela E, Pospiech H, Myllylae R. Characterization of cDNAs for mouse lysyl hydroxylase 1, 2 and 3, their phylogenetic analysis and tissue-specific expression in the mouse. Matrix Biol. 1999;18:325–329. doi: 10.1016/S0945-053X(99)00016-5. [DOI] [PubMed] [Google Scholar]
- Norman KR, Moerman DG. The let-268 locus of Caenorhabditis elegans encodes a procollagen lysyl hydroxylase that is essential for type IV collagen secretion. Dev. 2000;227:690–705. doi: 10.1006/dbio.2000.9897. [DOI] [PubMed] [Google Scholar]
- Vaz FM, Ofman R, Westinga K, Back JW, Wanders RJA. Molecular and biochemical characterization of rat epsilon-N-trimethyllysine hydroxylase, the first enzyme of carnitine biosynthesis. J Biol Chem. 2001;276:33512–33517. doi: 10.1074/jbc.M105929200. [DOI] [PubMed] [Google Scholar]
- Passoja K, Rautavuoma K, Ala-Kokko L, Kosonen T, Kivirikko KI. Cloning and characterization of a third human lysyl hydroxylase isoform. Proc Natl Acad Sci USA. 1998;95:10482–10486. doi: 10.1073/pnas.95.18.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Granato M. Genomic structure and embryonic expression of zebrafish lysyl hydroxylase 1 and lysyl hydroxylase 2. Matrix Biol. 2007;26:12–19. doi: 10.1016/j.matbio.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Granato M. The myotomal diwanka (lh3) glycosyltransferase and type XVIII collagen are critical for motor growth cone migration. Neuron. 2006;50:683–695. doi: 10.1016/j.neuron.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Rautavuoma K, Takaluoma K, Passoja K, Pirskanen A, Kvist AP, Kivirikko KI, Myllyharju J. Characterization of three fragments that constitute the monomers of the human lysyl hydroxylase isoenzymes 1–3. The 30-kDa N-terminal fragment is not required for lysyl hydroxylase activity. J Biol Chem. 2002;277(25):23084–23091. doi: 10.1074/jbc.M112077200. [DOI] [PubMed] [Google Scholar]
- Clifton IJ, Hsueh LC, Baldwin JE, Harlos K, Schofield CJ. Structure of proline 3-hydroxylase. Evolution of the family of 2-oxoglutarate dependent oxygenases. Eur J Biochem. 2001;268:6625–6636. doi: 10.1046/j.0014-2956.2001.02617.x. [DOI] [PubMed] [Google Scholar]
- Fernandes RJ, Farnand AW, Traeger GR, Weis MA, Eyre DR. A role for prolyl 3-hydroxylase 2 in post-translational modification of fibril-forming collagens. J Biol Chem. 2011;286(35):30662–30669. doi: 10.1074/jbc.M111.267906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risteli J, Tryggvason K, Kivirikko KI. Prolyl 3-hydroxylase: partial characterization of the enzyme from rat kidney cortex. Eur J Biochem. 1977;73(2):485–492. doi: 10.1111/j.1432-1033.1977.tb11341.x. [DOI] [PubMed] [Google Scholar]
- Wassenhove-McCarthy DJ, McCarthy KJ. Molecular characterization of a novel basement membrane-associated proteoglycan, leprecan. J Biol Chem. 1999;274:25004–25017. doi: 10.1074/jbc.274.35.25004. [DOI] [PubMed] [Google Scholar]
- Jaernum S, Kjellman C, Darabi A, Nilsson I, Edvardsen K, Aaman P. LEPREL1, a novel ER and Golgi resident member of the Leprecan family. Biochem Biophys Res Commun. 2004;317:342–351. doi: 10.1016/j.bbrc.2004.03.060. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Sugihara T, Yoshida A, Nomura H, Wadhwa R. Gros1, a potential growth suppressor on chromosome 1: its identity to basement membrane-associated proteoglycan, leprecan. Oncogene. 2000;19:3576–3583. doi: 10.1038/sj.onc.1203696. [DOI] [PubMed] [Google Scholar]
- Vranka JA, Sakai LY, Bachinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004;279:23615–23621. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- Vranka J, Stadler HS, Bächinger HP. Expression of prolyl 3-hydroxylase genes in embryonic and adult mouse tissues. Cell Struct Funct. 2009;34(2):97–104. doi: 10.1247/csf.09002. [DOI] [PubMed] [Google Scholar]
- Tiainen P, Pasanen A, Sormunen R, Myllyharju J. Characterization of recombinant human prolyl 3-hydroxylase isoenzyme 2, an enzyme modifying the basement membrane collagen IV. J Biol Chem. 2008;283(28):19432–19439. doi: 10.1074/jbc.M802973200. [DOI] [PubMed] [Google Scholar]
- Koski MK, Hieta R, Hirsilä M, Rönkä A, Myllyharju J, Wierenga RK. The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J Biol Chem. 2009;284(37):25290–25301. doi: 10.1074/jbc.M109.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culpepper MA, Scott EE, Limburg J. Crystal structure of prolyl 4-hydroxylase from Bacillus anthracis. Biochemistry. 2010;49(1):124–133. doi: 10.1021/bi901771z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaakoski T, Veijola J, Vuori K, Rehn M, Chow LT, Taillon-Miller P, Kivirikko KI, Pihlajaniemi T. Structure and expression of the human gene for the alpha subunit of prolyl 4-hydroxylase. The two alternatively spliced types of mRNA correspond to two homologous exons the sequences of which are expressed in a variety of tissues. J Biol Chem. 1994;269:27847–27854. [PubMed] [Google Scholar]
- Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko KI. Cloning of the human prolyl 4-hydroxylase alpha subunit isoform alpha(II) and characterization of the type II enzyme tetramer. The alpha(I) and alpha(II) subunits do not form a mixed alpha(I)alpha(II)beta2 tetramer. J Biol Chem. 1997;272:17342–17348. doi: 10.1074/jbc.272.28.17342. [DOI] [PubMed] [Google Scholar]
- Nokelainen M, Nissi R, Kukkola L, Helaakoski T, Myllyharju J. Characterization of the human and mouse genes for the alpha subunit of type II prolyl 4-hydroxylase. Identification of a previously unknown alternatively spliced exon and its expression in various tissues. Eur J Biochem. 2001;268:5300–5309. doi: 10.1046/j.0014-2956.2001.02464.x. [DOI] [PubMed] [Google Scholar]
- Hieta R, Myllyharju J. Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J Biol Chem. 2002;277(26):23965–23971. doi: 10.1074/jbc.M201865200. [DOI] [PubMed] [Google Scholar]
- Tiainen P, Myllyharju J, Koivunen P. Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J Biol Chem. 2005;280(2):1142–1148. doi: 10.1074/jbc.M411109200. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Myllyharju J, Tu H, Hellman M, Kivirikko KI. Evidence for 4-hydroxyproline in viral proteins. Characterization of a viral prolyl 4-hydroxylase and its peptide substrates. J Biol Chem. 22131;274(32):22131–22134. doi: 10.1074/jbc.274.32.22131. [DOI] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K. Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 2005;41:81–94. doi: 10.1111/j.1365-313X.2004.02279.x. [DOI] [PubMed] [Google Scholar]
- Kukkola L, Koivunen P, Pakkanen O, Page AP, Myllyharju J. Collagen prolyl 4-hydroxylase tetramers and dimers show identical decreases in Km values for peptide substrates with increasing chain length: mutation of one of the two catalytic sites in the tetramer inactivates the enzyme by more than half. J Biol Chem. 2004;279(18):18656–18661. doi: 10.1074/jbc.M401514200. [DOI] [PubMed] [Google Scholar]
- Drake EJ, Gulick AM. Three-dimensional structures of Pseudomonas aeruginosa PvcA and PvcB, two proteins involved in the synthesis of 2-isocyano-6,7-dihydroxycoumarin. J Mol Biol. 2008;384(1):193–205. doi: 10.1016/j.jmb.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K, van Scheltinga AC, Lloyd MD, Hara T, Ramaswamy S, Perrakis A, Thompson A, Lee HJ, Baldwin JE, Schofield CJ, Hajdu J, Andersson I. Structure of a cephalosporin synthase. Nature. 1998;394(6695):805–809. doi: 10.1038/29575. [DOI] [PubMed] [Google Scholar]
- Coque JJR, Martin JF, Liras P. Characterization and expression in Streptomyces lividans of cefD and cefE genes from Nocardia lactamdurans: the organization of the cephamycin gene cluster differs from that in Streptomyces clavuligerus. Mol Gen Genet. 1993;236:453–458. doi: 10.1007/BF00277148. [DOI] [PubMed] [Google Scholar]
- Samson SM, Dotzlaf JE, Slisz ML, Becker GW, van Frank RM, Veal LE, Yeh WK, Miller JR, Queener SW, Ingolia TD. Cloning and expression of the fungal expandase/hydroxylase gene involved in cephalosporin biosynthesis. Biotechnology (N.Y.) 1987;5:1207–1214. doi: 10.1038/nbt1187-1207. [DOI] [Google Scholar]
- Baker BJ, Dotzlaf JE, Yeh WK. Deacetoxycephalosporin C hydroxylase of Streptomyces clavuligerus. Purification, characterization, bifunctionality, and evolutionary implication. J Biol Chem. 1991;266(8):5087–5093. [PubMed] [Google Scholar]
- Coque J, Enguita FJ, Cardoza RE, Martin JF, Liras P. Characterization of the cefF gene of Nocardia lactamdurans encoding a 3'-methylcephem hydroxylase different from the 7-cephem hydroxylase. Appl Microbiol Biotechnol. 1996;44:605–609. doi: 10.1007/BF00172492. [DOI] [PubMed] [Google Scholar]
- Lloyd MD, Lipscomb SJ, Hewitson KS, Hensgens CM, Baldwin JE, Schofield CJ. Controlling the substrate selectivity of deacetoxycephalosporin/deacetylcephalosporin C synthase. J Biol Chem. 2004;279(15):15420–15426. doi: 10.1074/jbc.M313928200. [DOI] [PubMed] [Google Scholar]
- Wilmouth RC, Turnbull JJ, Welford RW, Clifton IJ, Prescott AG, Schofield CJ. Structure and mechanism of anthocyanidin synthase from Arabidopsis thaliana. Structure. 2002;10(1):93–103. doi: 10.1016/S0969-2126(01)00695-5. [DOI] [PubMed] [Google Scholar]
- Clifton IJ, Doan LX, Sleeman MC, Topf M, Suzuki H, Wilmouth RC, Schofield CJ. Crystal structure of carbapenem synthase (CarC) J Biol Chem. 2003;278:20843–20850. doi: 10.1074/jbc.M213054200. [DOI] [PubMed] [Google Scholar]
- Martens S, Forkmann G, Matern U, Lukacin R. Cloning of parsley flavone synthase I. Phytochemistry. 2001;58:43–46. doi: 10.1016/S0031-9422(01)00191-1. [DOI] [PubMed] [Google Scholar]
- Martens S, Forkmann G, Britsch L, Wellmann F, Matern U, Lukacin R. Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenases in parsley. FEBS Lett. 2003;544:93–98. doi: 10.1016/S0014-5793(03)00479-4. [DOI] [PubMed] [Google Scholar]
- Wellmann F, Lukacin R, Moriguchi T, Britsch L, Schiltz E, Matern U. Functional expression and mutational analysis of flavonol synthase from Citrus unshiu. Eur J Biochem. 2002;269:4134–4142. doi: 10.1046/j.1432-1033.2002.03108.x. [DOI] [PubMed] [Google Scholar]
- Owens DK, Crosby KC, Runac J, Howard BA, Winkel BS. Biochemical and genetic characterization of Arabidopsis flavanone 3beta-hydroxylase. Plant Physiol Biochem. 2008;46(10):833–843. doi: 10.1016/j.plaphy.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Chua CS, Biermann D, Goo KS, Sim TS. Elucidation of active site residues of Arabidopsis thaliana flavonol synthase provides a molecular platform for engineering flavonols. Phytochemistry. 2008;69(1):66–75. doi: 10.1016/j.phytochem.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Britsch L, Dedio J, Saedler H, Forkmann G. Molecular characterization of flavanone 3 beta-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. Eur J Biochem. 1993;217:745–754. doi: 10.1111/j.1432-1033.1993.tb18301.x. [DOI] [PubMed] [Google Scholar]
- Holton TA, Brugliera F, Tanaka Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993;4:1003–1010. doi: 10.1046/j.1365-313X.1993.04061003.x. [DOI] [PubMed] [Google Scholar]
- Halbwirth H, Fischer TC, Schlangen K, Rademacher W, Schleifer K-J, Forkmann G, Stich K. Screening for inhibitors of 2-oxoglutarate-dependent dioxygenases: Flavanone 3 beta-hydroxylase and flavonol synthase. Plant Sci. 2006;171:194–205. doi: 10.1016/j.plantsci.2006.03.014. [DOI] [Google Scholar]
- Lin GZ, Lian YJ, Ryu JH, Sung MK, Park JS, Park HJ, Park BK, Shin JS, Lee MS, Cheon CI. Expression and purification of His-tagged flavonol synthase of Camellia sinensis from Escherichia coli. Protein Expr Purif. 2007;55(2):287–292. doi: 10.1016/j.pep.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Xu F, Li L, Zhang W, Cheng H, Sun N, Cheng S, Wang Y. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol Biol Rep. 2012;39(3):2285–2296. doi: 10.1007/s11033-011-0978-9. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Githiri SM, Hatayama K, Dubouzet EG, Shimada N, Aoki T, Ayabe S, Iwashina T, Toda K, Matsumura H. A single-base deletion in soybean flavonol synthase gene is associated with magenta flower color. Plant Mol Biol. 2007;63:125–135. doi: 10.1007/s11103-006-9077-z. [DOI] [PubMed] [Google Scholar]
- Fujita A, Goto-Yamamoto N, Aramaki I, Hashizume K. Organ-specific transcription of putative flavonol synthase genes of grapevine and effects of plant hormones and shading on flavonol biosynthesis in grape berry skins. Biosci Biotechnol Biochem. 2006;70:632–638. doi: 10.1271/bbb.70.632. [DOI] [PubMed] [Google Scholar]
- Xu F, Cheng H, Cai R, Li LL, Chang J, Zhu J, Zhang FX, Chen LJ, Wang Y, Cheng SH, Cheng SY. Molecular Cloning and Function Analysis of an Anthocyanidin Synthase Gene from Ginkgo biloba, and Its Expression in Abiotic Stress Responses. Mol Cells. 2008;26:536–547. [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RW, Yamazaki M, Saito K, Schofield CJ. Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta-hydroxylase. J Biol Chem. 2004;279(2):1206–1216. doi: 10.1074/jbc.M309228200. [DOI] [PubMed] [Google Scholar]
- Reuter K, Pittelkow M, Bursy J, Heine A, Craan T, Bremer E. Synthesis of 5-hydroxyectoine from ectoine: crystal structure of the non-heme iron(II) and 2-oxoglutarate-dependent dioxygenase EctD. PLoS One. 2010;5(5):e10647. doi: 10.1371/journal.pone.0010647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu J, Schauwecker F, Grammel N, Keller U, Bernhard M. Functional expression of the ectoine hydroxylase gene (thpD) from Streptomyces chrysomallus in Halomonas elongata. Appl Environ Microbiol. 2004;70:3130–3132. doi: 10.1128/AEM.70.5.3130-3132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl Environ Microbiol. 2008;74(23):7286–7296. doi: 10.1128/AEM.00768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Estepa R, Argandona M, Reina-Bueno M, Capote N, Iglesias-Guerra F, Nieto JJ, Vargas C. The ectD gene, which is involved in the synthesis of the compatible solute hydroxyectoine, is essential for thermoprotection of the halophilic bacterium Chromohalobacter salexigens. J Bacteriol. 2006;188:3774–3784. doi: 10.1128/JB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tars K, Rumnieks J, Zeltins A, Kazaks A, Kotelovica S, Leonciks A, Sharipo J, Viksna A, Kuka J, Liepinsh E, Dambrova M. Crystal structure of human gamma-butyrobetaine hydroxylase. Biochem Biophys Res Commun. 2010;398(4):634–639. doi: 10.1016/j.bbrc.2010.06.121. [DOI] [PubMed] [Google Scholar]
- Rueetschi U, Nordin I, Odelhoeg B, Joernvall H, Lindstedt S. Gamma-butyrobetaine hydroxylase. Structural characterization of the Pseudomonas enzyme. Eur J Biochem. 1993;213:1075–1080. doi: 10.1111/j.1432-1033.1993.tb17855.x. [DOI] [PubMed] [Google Scholar]
- Wen G, Kühne H, Rauer C, Ringseis R, Eder K. Mouse γ-butyrobetaine dioxygenase is regulated by peroxisome proliferator-activated receptor α through a PPRE located in the proximal promoter. Biochem Pharmacol. 2011;82(2):175–183. doi: 10.1016/j.bcp.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Kondo A, Blanchard JS, Englard S. Purification and properties of calf liver gamma-butyrobetaine hydroxylase. Arch Biochem Biophys. 1981;212(2):338–346. doi: 10.1016/0003-9861(81)90374-X. [DOI] [PubMed] [Google Scholar]
- Leung IK, Krojer TJ, Kochan GT, Henry L, von Delft F, Claridge TD, Oppermann U, McDonough MA, Schofield CJ. Structural and mechanistic studies on γ-butyrobetaine hydroxylase. Chem Biol. 2010;17(12):1316–1324. doi: 10.1016/j.chembiol.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Williams J, Phillips AL, Gaskin P, Hedden P. Function and substrate specificity of the gibberellin 3beta-hydroxylase encoded by the Arabidopsis GA4 gene. Plant Physiol. 1998;117:559–563. doi: 10.1104/pp.117.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M. A mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), 'green revolution' rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci USA. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc Natl Acad Sci USA. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JAD, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 1999;19:65–73. doi: 10.1046/j.1365-313X.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- Lester DR, Phillips A, Hedden P, Andersson I. Purification and kinetic studies of recombinant gibberellin dioxygenases. BMC Plant Biol. 2005;5:19–19. doi: 10.1186/1471-2229-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004;136:3660–3669. doi: 10.1104/pp.104.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Sekimoto H, von Numers C, Phillips AL, Hedden P, Kamiya Y. Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol Plantarum. 1997;99:111–118. doi: 10.1111/j.1399-3054.1997.tb03438.x. [DOI] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45(5):804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Appleford NE, Evans DJ, Lenton JR, Gaskin P, Croker SJ, Devos KM, Phillips AL, Hedden P. Function and transcript analysis of gibberellin-biosynthetic enzymes in wheat. Planta. 2006;223:568–582. doi: 10.1007/s00425-005-0104-0. [DOI] [PubMed] [Google Scholar]
- Bou-Torrent J, Martínez-García JF, García-Martínez JL, Prat S. Gibberellin A1 metabolism contributes to the control of photoperiod-mediated tuberization in potato. PLoS One. 2011;6(9):e24458. doi: 10.1371/journal.pone.0024458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman B, Navarro C, Bijsterbosch G, Lange T, Prat S, Visser RGF, Bachem CWB. StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. Plant J. 2007. [DOI] [PubMed]
- Carrera E, Jackson SD, Prat S. Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol. 1999;119:765–774. doi: 10.1104/pp.119.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Hedden P, Graebe JE. Expression cloning of a gibberellin 20-oxidase, a multifunctional enzyme involved in gibberellin biosynthesis. Proc Natl Acad Sci USA. 1994;91:8552–8556. doi: 10.1073/pnas.91.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T, Kappler J, Fischer A, Frisse A, Padeffke T, Schmidtke S, Lange MJ. Gibberellin biosynthesis in developing pumpkin seedlings. Plant Physiol. 2005;139:213–223. doi: 10.1104/pp.105.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis IS, Ward DA, Thomas SG, Phillips AL, Davey MR, Power JB, Lowe KC, Croker SJ, Lewis MJ, Magness SL, Hedden P. Induction of dwarfism in transgenic Solanum dulcamara by over-expression of a gibberellin 20-oxidase cDNA from pumpkin. Plant J. 2000;23:329–338. doi: 10.1046/j.1365-313x.2000.00784.x. [DOI] [PubMed] [Google Scholar]
- Lange T, Robatzek S, Frisse A. Cloning and expression of a gibberellin 2 beta,3 beta-hydroxylase cDNA from pumpkin endosperm. Plant Cell. 1997;9:1459–1467. doi: 10.1105/tpc.9.8.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc Natl Acad Sci USA. 1997;94:6553–6558. doi: 10.1073/pnas.94.12.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Li L, Gage DA, Zeevaart JA. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Zeevaart JA. Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach. Plant Physiol. 2002;130:2085–2094. doi: 10.1104/pp.008581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Zeevaart JA. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 2005;138:243–254. doi: 10.1104/pp.104.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol. 1997;33:1073–1084. doi: 10.1023/A:1005715722193. [DOI] [PubMed] [Google Scholar]
- Park SH, Nakajima M, Sakane M, Xu ZJ, Tomioka K, Yamaguchi I. Gibberellin 2-oxidases from seedlings of adzuki bean (Vigna angularis) show high gibberellin-binding activity in the presence of 2-oxoglutarate and Co2+ Biosci Biotechnol Biochem. 2005;69:1498–1507. doi: 10.1271/bbb.69.1498. [DOI] [PubMed] [Google Scholar]
- Serrani JC, Sanjuán R, Ruiz-Rivero O, Fos M, García-Martínez JL. Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 2007;145(1):246–257. doi: 10.1104/pp.107.098335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers M, Kaneta T, Kawaide H, Yamaguchi S, Yang YY, Imai R, Sekimoto H, Kamiya Y. Regulation of gibberellin biosynthesis genes during flower and early fruit development of tomato. Plant J. 1999;17(3):241–250. doi: 10.1046/j.1365-313X.1999.00366.x. [DOI] [PubMed] [Google Scholar]
- Martí E, Carrera E, Ruiz-Rivero O, García-Martínez JL. Hormonal regulation of tomato gibberellin 20-oxidase1 expressed in Arabidopsis. J Plant Physiol. 2010;167(14):1188–1196. doi: 10.1016/j.jplph.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Zhang JH, Zhang YY, Wang TT, Chen RG, Li HX, Ye ZB. Isolation and expression of GA 2-oxidase2 in tomato. DNA Seq. 2007;18:472–477. doi: 10.1080/10425170701462183. [DOI] [PubMed] [Google Scholar]
- Yang YY, Rebers M, Toyomasu T, Kawaide H, Kaneta T, Kamiya Y. Cloning of two cDNAs encoding gibberellin 3beta-hydroxylase (Accession No. AB010991, AB010992) of tomato (Solanum lycopersicum L.) seedlings (PGR98-200) Plant Physiol. 1998;118:1534–1534. [Google Scholar]
- Ward DA, Macmillan J, Gong F, Phillips AL, Hedden P. Gibberellin 3-oxidases in developing embryos of the southern wild cucumber, Marah macrocarpus. Phytochemistry. 2010;71:2010–2018. doi: 10.1016/j.phytochem.2010.09.015. [DOI] [PubMed] [Google Scholar]
- MacMillan J, Ward DA, Phillips AL, Sanchez-Beltran MJ, Gaskin P, Lange T, Hedden P. Gibberellin biosynthesis from gibberellin A12-aldehyde in endosperm and embryos of Marah macrocarpus. Plant Physiol. 1997;113:1369–1377. doi: 10.1104/pp.113.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Sawada Y, Suzuki M, Kenmoku H, Kawaide H, Mitsuhashi W, Sassa T, Inoue Y, Kamiya Y, Toyomasu T. Deactivation of gibberellin by 2-oxidation during germination of photoblastic lettuce seeds. Biosci Biotechnol Biochem. 2003;67:1551–1558. doi: 10.1271/bbb.67.1551. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Mitsuhashi W, Kamiya Y. Gibberellin biosynthetic enzyme. Submitted (AUG-1999) to the EMBL/GenBank/DDBJ databases. Strain: Grand Rapids EMBL Nos. BAB12438.1 (Ls20ox3); BAB12439.1 (Ls3h3)
- Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL. Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol. 2007;14:240–242. doi: 10.1038/nsmb1200. [DOI] [PubMed] [Google Scholar]
- Tanabe W, Suzuki S, Muto Y, Inoue M, Kigawa T, Terada T, Shirouzu M, Yokoyama S. Solution structure of the ARID domain of Jarid1b protein. J. p. . To be Published.
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Yue WW, Gileadi C, Krojer T, Pike ACW, Vondelft F, Ng S, Carpenter L, Arrowsmith C, Weigelt J, Edwards A, Bountra C, Oppermann U. Crystal Structure of Human Jmjd2C Catalytic Domain Journal. p. . To be Published.
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, Roberts TM, Chang HY, Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(Suppl 7163):689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Che KH, Yue WW, Krojer T, Muniz JRC, Ng SS, Tumber A, Daniel M, Burgess-Brown N, Savitsky P, Von Delft F, Ugochukwu E, Filippakopoulos P, Arrowsmith C, Weigelt J, Edwards A, Bountra C, Oppermann U. Crystal Structure of the Human Jmjd3 Jumonji Domain Journal. p. . To be Published.
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Tanabe W, Suzuki S, Muto Y, Inoue M, Kigawa T, Terada T, Shirouzu M, Yokoyama S. Solution structure of the ARID domain of JARID1D protein Journal. p. . To be Published.
- Chen Z, Zhang G. Crystal structure of the catalytic core domain of jmjd2d Journal. p. . To be Published.
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PAC, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WW, Hozjan V, Ge W, Loenarz C, Cooper CD, Schofield CJ, Kavanagh KL, Oppermann U, McDonough MA. Crystal structure of the PHF8 Jumonji domain, an N(epsilon)-methyl lysine demethylase. FEBS Lett. 2010;584:825–830. doi: 10.1016/j.febslet.2009.12.055. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432–437. doi: 10.1101/gad.1864410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol. 2007;14:341–343. doi: 10.1038/nsmb1216. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- Mills JL, Lee D, Kohan E, Sahdev S, Acton TB, Xiao R, Everett JK, Montelione GT, Szyperski T. Northeast Structural Genomics Consortium Target FR824D Journal. p. . To be Published.
- Lorbeck MT, Singh N, Zervos A, Dhatta M, Lapchenko M, Yang C, Elefant F. The histone demethylase Dmel\Kdm4A controls genes required for life span and male-specific sex determination in Drosophila. Gene. 2010;450(1–2):8–17. doi: 10.1016/j.gene.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carré C, Vaquero A, de Olano N, Azorín F. Characterization of Drosophila melanogaster JmjC + N histone demethylases. Nucleic Acids Res. 2008;36(9):2852–2863. doi: 10.1093/nar/gkn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho YW, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29(8):1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Hsia DA, Tepper CG, Pochampalli MR, Hsia EY, Izumiya C, Huerta SB, Wright ME, Chen HW, Kung HJ, Izumiya Y. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc Natl Acad Sci USA. 2010;107:9671–9676. doi: 10.1073/pnas.1000401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Kim JY, Choe NW, Cho IH, Kim JR, Kim DW, Seol JE, Lee SE, Kook H, Nam KI, Kook H, Bhak YY, Seo SB. Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res. 2010;38(19):6389–6403. doi: 10.1093/nar/gkq491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaleva II, Prudchenko IA, Ivanov VT, Voitenkov VB. JmjC-domain-containing histone demethylases of the JMJD1B type as putative precursors of endogenous DSIP. Peptides. 2011;32(4):826–831. doi: 10.1016/j.peptides.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Koehler C, Bishop S, Dowler EF, Schmieder P, Diehl A, Oschkinat H, Ball LJ. Backbone and sidechain 1 H, 13 C and 15 N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein. Biomol Nmr Assign. 2008;2:9–11. doi: 10.1007/s12104-007-9071-7. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Ortiz-Tello PA, Brunzelle JS, Trievel RC. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat Struct Mol Biol. 2007;14(8):689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Chen Z, Zang J, Whetstine J, Hong X, Davrazou F, Kutateladze TG, Simpson M, Mao Q, Pan C-H, Dai S, Hagman J, Hansen K, Shi Y, Zhang G. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Jin C, Li J, Green CD, Yu X, Tang X, Han D, Xian B, Wang D, Huang X, Cao X, Yan Z, Hou L, Liu J, Shukeir N, Khaitovich P, Chen CD, Zhang H, Jenuwein T, Han JD. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;2:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10(6):980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol. 2008;28(3):1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol. 2010;30(10):2485–2497. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011;155(2):906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers (Basel) 2011;3(1):1383–1404. doi: 10.3390/cancers3011383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122(5):1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, Rose NR, Offer J, Scheinost JC, Borowski T, Sundstrom M, Schofield CJ, Oppermann U. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448(7149):87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- McNeill LA, Hewitson KS, Gleadle JM, Horsfall LE, Oldham NJ, Maxwell PH, Pugh CW, Ratcliffe PJ, Schofield CJ. The use of dioxygen by HIF prolyl hydroxylase (PHD1) Bioorg Med Chem Lett. 2002;12:1547–1550. doi: 10.1016/S0960-894X(02)00219-6. [DOI] [PubMed] [Google Scholar]
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- Tian YM, Mole DR, Ratcliffe PJ, Gleadle JM. Characterization of different isoforms of the HIF prolyl hydroxylase PHD1 generated by alternative initiation. Biochem J. 2006;397:179–186. doi: 10.1042/BJ20051996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappalardi MB, McNulty DE, Martin JD, Fisher KE, Jiang Y, Burns MC, Zhao H, Ho T, Sweitzer S, Schwartz B, Annan RS, Copeland RA, Tummino PJ, Luo L. Biochemical characterization of human HIF hydroxylases using HIF protein substrates that contain all three hydroxylation sites. Biochem J. 2011;436:363–369. doi: 10.1042/BJ20101201. [DOI] [PubMed] [Google Scholar]
- van der Wel H, Ercan A, West CM. The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-alpha class of animal prolyl 4-hydroxylases. J Biol Chem. 2005;280:14645–14655. doi: 10.1074/jbc.M500600200. [DOI] [PubMed] [Google Scholar]
- Elkins JM, Hewitson KS, McNeill LA, Seibel JF, Schlemminger I, Pugh CW, Ratcliffe PJ, Schofield CJ. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J Biol Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann CE, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible factor 1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc Natl Acad Sci USA. 2002;99:15351–15356. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough MA, Li V, Flashman E, Chowdhury R, Mohr C, Lienard BMR, Zondlo J, Oldham NJ, Clifton IJ, Lewis J, McNeill LA, Kurzeja RJ, Hewitson KS, Yang E, Jordan S, Syed RS, Schofield CJ. Cellular oxygen sensing: crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2) Proc Natl Acad Sci USA. 2006;103:9814–9819. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim HL, Kim KH, do Kim J, Lee SJ, Yoon JY, Yoon HJ, Lee HY, Park SB, Kim SJ, Lee JY, Suh SW. Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex. Nucleic Acids Res. 2010;38(6):2099–2110. doi: 10.1093/nar/gkp1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Adachi N, Koyama H, Matsushita M. OGFOD1, a member of the 2-oxoglutarate and iron dependent dioxygenase family, functions in ischemic signaling. FEBS Lett. 2010;584(15):3340–3347. doi: 10.1016/j.febslet.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Hughes BT, Espenshade PJ. Oxygen-regulated degradation of fission yeast SREBP by Ofd1, a prolyl hydroxylase family member. EMBO J. 2008;27(10):1491–1501. doi: 10.1038/emboj.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Hirsilä M, Günzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279(11):9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- Barth S, Edlich F, Berchner-Pfannschmidt U, Gneuss S, Jahreis G, Hasgall PA, Fandrey J, Wenger RH, Camenisch G. Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J Biol Chem. 2009;284(34):23046–23058. doi: 10.1074/jbc.M109.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda J, Okabe S, Hashimoto T, Yamada Y. Molecular cloning of hyoscyamine 6 beta-hydroxylase, a 2-oxoglutarate-dependent dioxygenase, from cultured roots of Hyoscyamus niger. J Biol Chem. 1991;266:9460–9464. [PubMed] [Google Scholar]
- Vazquez-Flota F, De Carolis E, Alarco A-M, De Luca V. Molecular cloning and characterization of desacetoxyvindoline-4-hydroxylase, a 2-oxoglutarate dependent-dioxygenase involved in the biosynthesis of vindoline in Catharanthus roseus (L.) G. Don. Plant Mol Biol. 1997;34:935–948. doi: 10.1023/A:1005894001516. [DOI] [PubMed] [Google Scholar]
- De Carolis E, De Luca V. Purification, characterization, and kinetic analysis of a 2-oxoglutarate-dependent dioxygenase involved in vindoline biosynthesis from Catharanthus roseus. J Biol Chem. 1993;268:5504–5511. [PubMed] [Google Scholar]
- Cardillo AB, Talou JR, Giulietti AM. Expression of Brugmansia candida Hyoscyamine 6beta-Hydroxylase gene in Saccharomyces cerevisiae and its potential use as biocatalyst. Microb Cell Fact. 2008;7:17. doi: 10.1186/1475-2859-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Zhu P, Cheng KD, Meng C, He HX. Molecular cloning, expression and characterization of hyoscyamine 6beta-hydroxylase from hairy roots of Anisodus tanguticus. Planta Med. 2005;71:249–253. doi: 10.1055/s-2005-837825. [DOI] [PubMed] [Google Scholar]
- Pramod KK, Singh S, Jayabaskaran C. Biochemical and structural characterization of recombinant hyoscyamine 6β-hydroxylase from Datura metel L. Plant Physiol Biochem. 2010;48(12):966–970. doi: 10.1016/j.plaphy.2010.09.003. [DOI] [PubMed] [Google Scholar]
- El Jaber-Vazdekis N, González C, Ravelo AG, Zárate R. Cloning, characterization and analysis of expression profiles of a cDNA encoding a hyoscyamine 6beta-hydroxylase (H6H) from Atropa baetica Willk. Plant Physiol Biochem. 2009;47(1):20–25. doi: 10.1016/j.plaphy.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Montero-Morán GM, Li M, Rendòn-Huerta E, Jourdan F, Lowe DJ, Stumpff-Kane AW, Feig M, Scazzocchio C, Hausinger RP. Purification and characterization of the FeII- and alpha-ketoglutarate-dependent xanthine hydroxylase from Aspergillus nidulans. Biochemistry. 2007;46(18):5293–5304. doi: 10.1021/bi700065h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cultrone A, Scazzocchio C, Rochet M, Montero-Morán G, Drevet C, Fernández-Martín R. Convergent evolution of hydroxylation mechanisms in the fungal kingdom: molybdenum cofactor-independent hydroxylation of xanthine via alpha-ketoglutarate-dependent dioxygenases. Mol Microbiol. 2005;57(1):276–290. doi: 10.1111/j.1365-2958.2005.04686.x. [DOI] [PubMed] [Google Scholar]
- Vainio S, Genest P-A, ter Riet B, van Luenen HGAM, Borst P. Evidence that J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base J. Mol Biochem Parasitol. 2009;164:157–161. doi: 10.1016/j.molbiopara.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Yu Z, Genest P-A, ter Riet B, Sweeney K, DiPaolo C, Kieft R, Christodoulou E, Perrakis A, Simmons JM, Hausinger RP, van Luenen HGAM, Rigden DJ, Sabatini R, Borst P. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35:2107–2115. doi: 10.1093/nar/gkm049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliffe LJ, Kieft R, Southern T, Birkeland SR, Marshall M, Sweeney K, Sabatini R. JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 2009;37:1452–1462. doi: 10.1093/nar/gkn1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidigh JW, Darwanto A, Williams AA, Wall NR, Sowers LC. Cloning and characterization of Rhodotorula glutinis thymine hydroxylase. Chem. 2009;22:885–893. doi: 10.1021/tx8004482. [DOI] [PubMed] [Google Scholar]
- Hsu CA, Saewert MD, Polsinelli LF, Abbott MT. Uracil's uncoupling of the decarboxylation of alpha-ketoglutarate in the thymine 7-hydroxylase reaction of Neurospora crassa. J Biol Chem. 1981;256(12):6098–6101. [PubMed] [Google Scholar]
- Holme E. A kinetic study of thymine 7-hydroxylase from neurospora crassa. Biochemistry. 1975;14(22):4999–5003. doi: 10.1021/bi00693a033. [DOI] [PubMed] [Google Scholar]
- McDonough MA, Kavanagh KL, Butler D, Searls T, Oppermann U, Schofield CJ. Structure of human phytanoyl-CoA 2-hydroxylase identifies molecular mechanisms of Refsum disease. J Biol Chem. 2005;280(49):41101–41110. doi: 10.1074/jbc.M507528200. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kochan GT, Ng SS, Kavanagh KL, Oppermann U, Schofield CJ, McDonough MA. Crystal structure of PHYHD1A, a 2OG oxygenase related to phytanoyl-CoA hydroxylase. Biochem Biophys Res Commun. 2011;408(4):553–558. doi: 10.1016/j.bbrc.2011.04.059. [DOI] [PubMed] [Google Scholar]
- Jansen GA, Ofman R, Denis S, Ferdinandusse S, Hogenhout EM, Jakobs C, Wanders RJA. Phytanoyl-CoA hydroxylase from rat liver: protein purification and cDNA cloning with implications for the subcellular localization of phytanic acid alpha-oxidation. J Lipid Res. 1999;40:2244–2254. [PubMed] [Google Scholar]
- Iwano M, Ueno M, Miyazaki M, Harada T, Nagai Y, Hirano M, Dohi Y, Akai Y, Kurioka H, Dohi K. Molecular cloning and expression of a novel peptide (LN1) gene: reduced expression in the renal cortex of lupus nephritis in MRL/lpr mouse. Biochem Biophys Res Commun. 1996;229:355–360. doi: 10.1006/bbrc.1996.1805. [DOI] [PubMed] [Google Scholar]
- p. . Arabidopsis thaliana [At2g01490/F2I9.11] 283 aa Accession AAM51599 GI:21436043 AY116965.1 (mRNA)
- Kershaw NJ, Mukherji M, MacKinnon CH, Claridge TD, Odell B, Wierzbicki AS, Lloyd MD, Schofield CJ. Studies on phytanoyl-CoA 2-hydroxylase and synthesis of phytanoyl-coenzyme A. Bioorg Med Chem Lett. 2001;11(18):2545–2548. doi: 10.1016/S0960-894X(01)00494-2. [DOI] [PubMed] [Google Scholar]
- You Z, Omura S, Ikeda H, Cane DE, Jogl G. Crystal structure of the non-heme iron dioxygenase PtlH in pentalenolactone biosynthesis. J Biol Chem. 2007;282:36552–36560. doi: 10.1074/jbc.M706358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller I, Kahnert A, Pape T, Sheldrick GM, Meyer-Klaucke W, Dierks T, Kertesz M, Usón I. Crystal structure of the alkylsulfatase AtsK: insights into the catalytic mechanism of the Fe(II) alpha-ketoglutarate-dependent dioxygenase superfamily. Biochemistry. 2004;43(11):3075–3088. doi: 10.1021/bi035752v. [DOI] [PubMed] [Google Scholar]
- Kahnert A, Kertesz MA. Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J Biol Chem. 2000;275:31661–31667. doi: 10.1074/jbc.M005820200. [DOI] [PubMed] [Google Scholar]
- Elkins JM, Ryle MJ, Clifton IJ, Dunning Hotopp JC, Lloyd JS, Burzlaff NI, Baldwin JE, Hausinger RP, Roach PL. X-ray crystal structure of Escherichia coli taurine/alpha-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry. 2002;41(16):5185–5192. doi: 10.1021/bi016014e. [DOI] [PubMed] [Google Scholar]
- Eichhorn E, van der Ploeg JR, Kertesz MA, Leisinger T. Characterization of alpha-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J Biol Chem. 1997;272:23031–23036. doi: 10.1074/jbc.272.37.23031. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Auchtung TA, Hausinger RP. Cloning and characterization of a sulfonate/alpha-ketoglutarate dioxygenase from Saccharomyces cerevisiae. J Bacteriol. 1999;181:5876–5879. doi: 10.1128/jb.181.18.5876-5879.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori F, Hausinger RP. Alcaligenes eutrophus JMP134 "2,4-dichlorophenoxyacetate monooxygenase" is an alpha-ketoglutarate- dependent dioxygenase. J Bacteriol. 1993;175(7):2083–2086. doi: 10.1128/jb.175.7.2083-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Smith SR, Saari EA, McCracken J, Hausinger RP. Site-directed mutagenesis of 2,4-dichlorophenoxyacetic acid/alpha-ketoglutarate dioxygenase. Identification of residues involved in metallocenter formation and substrate binding. J Biol Chem. 2000;275:12400–12409. doi: 10.1074/jbc.275.17.12400. [DOI] [PubMed] [Google Scholar]
- Müller RH, Kleinsteuber S, Babel W. Physiological and genetic characteristics of two bacterial strains utilizing phenoxypropionate and phenoxyacetate herbicides. Microbiol Res. 2001;156(2):121–131. doi: 10.1078/0944-5013-00089. [DOI] [PubMed] [Google Scholar]
- Westendorf A, Benndorf D, Kleinsteuber S, Mueller RH, Harms H. The S-enantiomer specific enzymes from Rhodoferax sp. P230 and Sphingobium herbicidovorans MH involved in the cleavage of 2-phenoxypropionate (dichlorprop and mecoprop) and phenoxyacetate herbicides (2,4-D and MCPA), kinetic characteristics and protein data. p. . Direct submission 2006, EMBL/GenBank/DDBJ database.
- Schleinitz KM, Kleinsteuber S, Vallaeys T, Babel W. Localization and characterization of two novel genes encoding stereospecific dioxygenases catalyzing 2(2,4-dichlorophenoxy)propionate cleavage in Delftia acidovorans MC1. Appl Environ Microbiol. 2004;70:5357–5365. doi: 10.1128/AEM.70.9.5357-5365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Uniprot and PDB ids of sequences used in this work.
Table S2. Comparative domain analysis of template sequences.
Table S3. Pair wise structural alignment of selected proteins.
Table S4. Analysis of HMM profiles with highlighted active site residues. (PDF 29 kb)
Table S5. Link to HMM profile database “profile-DB\aKG-profile-database.hmm”.