Controlling the chemical composition of supported phospholipid bilayers (SLBs) on the micrometer and submicrometer scale has been a widely pursued goal in bioanalytical chemistry. For example, Jackson and Groves1 applied scanning probe lithography to completely remove lipid membranes in pre-patterned 1 × 1 μm chromium arrays and then backfilled these regions with new lipid components. Orth and co-workers demonstrated ~1 μm scale SLB patterning using an elegant polymer lift-off method.2 Lenhert et al.3 developed a dippen nanolithography method for patterning bilayer/multi-bilayer structures down to the 100 nm scale.
These patterned membranes can be employed to address fundamental biophysical questions about cell membrane behavior.4–6 For instance, Mossman and colleagues5 showed that the radial location of T-cell receptors can be finely tuned in immunological synapses by using micropatterned SLBs. Wu et al.6 demonstrated that micropatterned bilayers are useful for the visualization of membrane compartmentalization during receptor-mediated signaling. The fine patterning of lipid membranes may also serve as the basis for a variety of biosensor technologies.7 One example is the formation of submicrometer-sized SLBs inside Au nanoholes (~110 nm), which have been employed for label-free biorecognition in conjunction with localized surface plasmon resonance.8,9
Despite these successes, it is generally agreed that supported membrane patterning below the 100 nm scale would add an important new dimension to biophysical and bioanalytical studies. There should, however, be an ultimate size limit to free-standing SLB formation because these supramolecular architectures pay the cost of an edge energy in order to fuse to solid supports.10,11 Herein, we used atomic force microscopy (AFM) based nanoshaving to control the formation of SLBs down to the sub-100 nm level. Nanoshaving employs an AFM tip to selectively remove a pre-existing thin film from a substrate.12–17 The shaved region can be subsequently backfilled with new materials such as an SLB. Our results revealed that lines of phosphatidylcholine bilayers possessing widths as thin as 55 nm can be patterned on borosilicate supports, but not lines with 36 nm widths.
A three-step process was conducted to create lines of SLBs as shown schematically in Figure 1. First, a bovine serum albumin (BSA) monolayer was formed on a planar borosilicate substrate by incubation with a phosphate buffered saline (PBS) solution containing 10 mg/mL BSA.18 Excess protein molecules were washed away with purified water and the BSA monolayer was subsequently dried under streaming nitrogen. In a second step, immobilized protein molecules were selectively removed with an ultrasharp AFM tip in air in contact mode to create vacant lines with varying widths. The force applied to the tip was ~300 nN, which was sufficient to remove the protein without damaging the underlying surface. It should be noted that the Si tip was moved laterally in 2 nm steps in a process controlled by patterning software. The nanoshaving speed was 40 μm/s. The width of the patterned lines was directly measured by AFM (see Supporting Information). For convenience, 1 in 20 proteins was fluorescently tagged so that vacant regions could be observed by epifluorescence microscopy.
Figure 1.
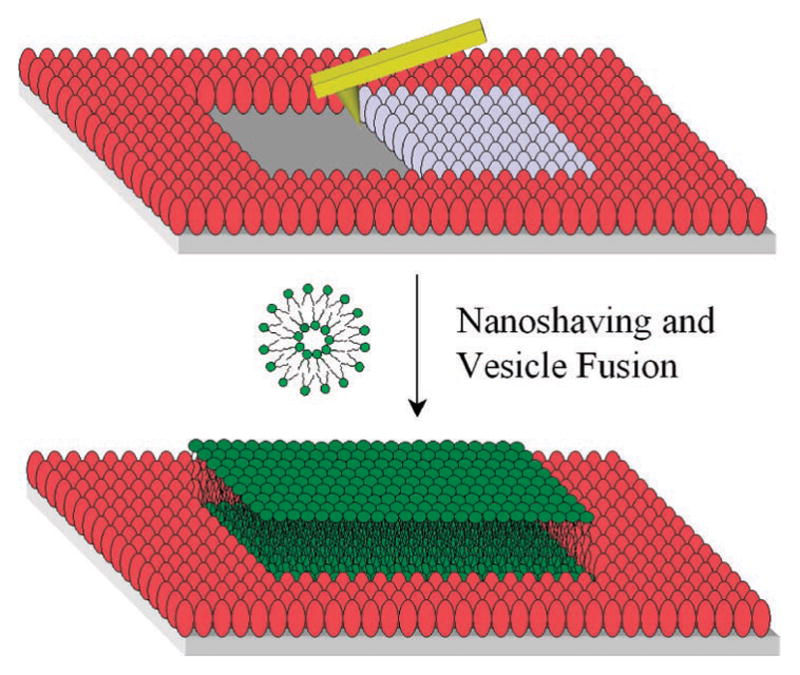
Schematic diagram of AFM-based nanoshaving lithography for nanoscale SLB formation. The red and gray ellipsoids represent adsorbed BSA molecules. The gray ones are being removed by the AFM tip. A subsequently deposited lipid bilayer is shown in green.
Backfilling was performed with a 0.5 mg/mL vesicle solution composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) with 2.0 mol % of a dye-labeled lipid (NBD-PE).19 Small unilamellar lipid vesicles were incubated over the nanopatterned BSA substrate for 10 min. After rinsing with PBS solution, fluorescence microscopy clearly showed uniform fluorescence from 55 nm wide lines down to the diffraction limit (Figure 2a). This is consistent with the presence of lipid material within the shaved regions. The mobility of the bilayer in this narrow region was confirmed by one-dimensional fluorescence recovery after photo-bleaching (FRAP)20 measurements. Figure 2 panels b and c show the bleached spot immediately after it was made and 120 s later, respectively. The diffusion constant was ~2.5 × 10−8 cm2/sec with a mobile fraction of ~0.97. The calculations employed for measuring diffusion constants and mobile fractions with 1-D geometries are discussed in the Supporting Information section.
Figure 2.
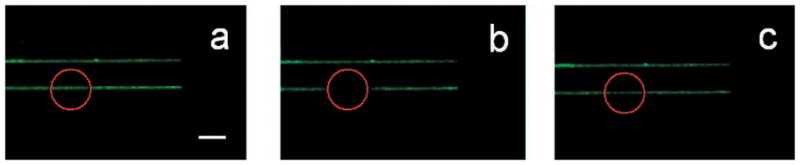
FRAP images of 2.0 mol % NBD-PE/POPC bilayer lines as a function of time that are ~55 nm in width. The bleached bilayer spot is shown (a) before bleaching, (b) immediately after bleaching, and (c) 120 s later. The red circle denotes the location of the bleached spot. Only the bottom line was bleached and the upper one was used as a reference. The scale bar is 3 μm.
Next, a series of parallel lines ranging from 15 to 600 nm were formed under an identical set of nanoshaving conditions. After nanoshaving, the Texas Red-labeled BSA monolayer was imaged by epifluorescence microscopy (Figure 3a). Close-up AFM images for the identical protein line widths are provided in the Supporting Information section. At this point, POPC vesicles containing 2.0 mol % NBD-PE were introduced over the sample. Epifluorescence images of the nascently formed lipid bilayers from this process are shown in Figure 3b. The smallest bilayer lines created by this method were 55 nm. On the other hand, the lipid material completely and consistently washed away from the surface when the line width was 36 nm or below. This finding was not changed by modulating the size of the vesicles or through the use of osmotic pressure to rupture them (see Supporting Information).
Figure 3.
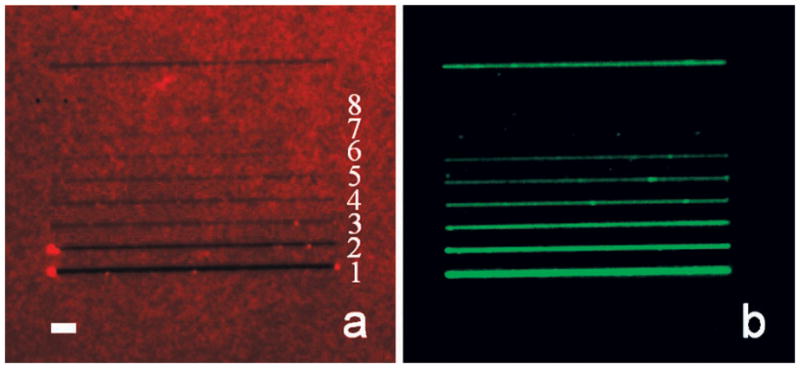
Epifluorescence images of (a) a nanoshaved BSA monolayer and (b) SLB lines. The top line, which is ~200 nm in width, was used as a reference marker. The widths of shaved lines in (a) from 1 to 8 are ~600, ~300, ~142, ~103, ~78, ~55, ~36, and ~15 nm, respectively, as measured by AFM. The length of each line is 40 μm. The scale bar is 3 μm. Note, the vacant lines in (a) become increasingly difficult to observe by epifluorescence microscopy as the line width narrows. Green fluorescence emanating from these regions, however, should be trivial to observe even for the thinnest lines under the conditions of this experiment, if a bilayer is indeed present.
Although sub-100 nm bilayer lines could be created, there is a clear size limit for POPC/glass bilayers. All free-standing SLB patches formed on solid supports (e.g., borosilicate glass) have an edge energy associated with them. Indeed, the bilayer is, presumably, highly curved along its perimeter to avoid exposing the hydrophobic lipid tails directly to water.10,21 The concept of edge energy per unit length (γ, J/m)10 describes the curvature cost over a given distance. The reason micrometer-scale lipid bilayers readily fuse to planar supports is because the adhesion energy more than compensates for such line tension.11 By contrast, the edge energy can be greater than the adhesion energy for nanoscale bilayers.
In the present case, bilayers are w μm wide and l μm long. Therefore, the edge energy, Ee, and surface adhesion energy, Es, should be:
| (1) |
| (2) |
where W is the surface adhesion energy per unit area. Neglecting any interactions between the bilayer edge and the surrounding BSA molecules, the surface adhesion energy (eq 2) must be greater than or equal to the edge energy (eq 1) in order for a stable SLB to form:
| (3) |
since the length of the bilayer lines (e.g., 40 μm) is 2 to 3 orders of magnitude larger than their widths, eq 3 can be reduced and rearranged to:
| (4) |
where the equality describes the minimum size for an SLB. The adhesion energy per unit area (W) of phosphocholine bilayers on glass substrates has been estimated to be ~2.0 × 10−4 J/m2 in several reports.9,22–25 Moreover, the typical edge energy per unit length (γ) of a phosphocholine bilayer is known to be ~1 × 10−11 J/m.26–30 This leads to an estimated lower width limit of ~100 nm. Such a value is in reasonably good agreement with our finding of 55 nm. However, our number implies that W may be somewhat larger and/or that γ might be somewhat smaller. A slightly lower value of γ in the present case, for example, might stem from a slightly favorable interaction between the bilayer edge and neighboring BSA molecules.
The size limitations found herein are almost certainly specific to the lipid composition of the membrane. Modulating the composition should lead to changes in γ. For example, supported bilayers composed of a mixture of long-chain phospholipids with a small concentration of 1,2-dihexanoyl-sn-glycero-3-phosphocholine (DHPC) would be expected to have a lower edge energy and thus allow narrower line widths to be achieved. In fact, such compositions are often used to make lipid bicelles in bulk solution.31 These pancake-like bilayer structures probably reduce edge energy by having a high concentration of DHPC along the edge. In fact, nanoshaving experiments with POPC bilayers containing 6.0 mol % DHPC showed that ~36 nm wide lines could be made, but not ~ 15 nm (see Supporting Information).
Supplementary Material
Acknowledgments
We thank the NIH (Grant R01 GM070622) and the ARO (Grant W911NF-05-1-0494) for funding. Discussions with Dr. Xin Chen, Yanjie Zhang, and Tinglu Yang were also appreciated.
Footnotes
Supporting Information Available: Epifluorescence images of BSA monolayers, experimental details of SLB formation, vesicle size and lipid composition effects, diffusion constant measurements, and AFM measurements. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jackson BL, Groves JT. J Am Chem Soc. 2004;126:13878–13879. doi: 10.1021/ja046040a. [DOI] [PubMed] [Google Scholar]
- 2.Orth RN, Kameoka J, Zipfel WR, Ilic B, Webb WW, Clark TG, Craighead HG. Biophys J. 2003;85:3066–3073. doi: 10.1016/S0006-3495(03)74725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenhert S, Sun P, Wang YH, Fuchs H, Mirkin CA. Small. 2007;3:71–75. doi: 10.1002/smll.200600431. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka M, Sackmann E. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 5.Mossman KD, Campi G, Groves JT, Dustin ML. Science. 2005;310:1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 6.Wu M, Holowka D, Craighead HG, Baird B. Proc Natl Acad Sci. 2004;101:13798–13803. doi: 10.1073/pnas.0403835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellana ET, Cremer PS. Surf Sci Rep. 2006;61:429–444. doi: 10.1016/j.surfrep.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlin A, Zach M, Rindzevicius T, Kall M, Sutherland DS, Höök F. J Am Chem Soc. 2005;127:5043–5048. doi: 10.1021/ja043672o. [DOI] [PubMed] [Google Scholar]
- 9.Jonsson MP, Jönsson P, Dahlin AB, Höök F. Nano Lett. 2007;7:3462–3468. doi: 10.1021/nl072006t. [DOI] [PubMed] [Google Scholar]
- 10.Litster JD. Phys Lett A. 1975;A 53:193–194. [Google Scholar]
- 11.Lipowsky R, Seifert U. Mol Cryst Liq Cryst. 1991;202:17–25. [Google Scholar]
- 12.Xu S, Liu GY. Langmuir. 1997;13:127–129. [Google Scholar]
- 13.Wadu-Mesthrige K, Amro NA, Garno JC, Xu S, Liu GY. Biophys J. 2001;80:1891–1899. doi: 10.1016/s0006-3495(01)76158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang CH, Stevens BD, Carlier PR, Calter MA, Ducker WA. J Am Chem Soc. 2002;124:12114–12115. doi: 10.1021/ja017686v. [DOI] [PubMed] [Google Scholar]
- 15.Liu MZ, Amro NA, Chow CS, Liu GY. Nano Lett. 2002;2:863–867. [Google Scholar]
- 16.Schonherr H, Rozkiewicz DI, Vancso GJ. Langmuir. 2004;20:7308–7312. doi: 10.1021/la0498915. [DOI] [PubMed] [Google Scholar]
- 17.Tinazli A, Piehler J, Beuttler M, Guckenberger R, Tampe R. Nat Nanotechnol. 2007;2:220–225. doi: 10.1038/nnano.2007.63. [DOI] [PubMed] [Google Scholar]
- 18.Wertz CF, Santore MM. Langmuir. 2001;17:3006–3016. [Google Scholar]
- 19.1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl), ammonium salt (NBD-PE).
- 20.Mullineaux CW, Tobin MJ, Jones GR. Nature. 1997;390:421–424. [Google Scholar]
- 21.Hamai C, Cremer PS, Musser SM. Biophys J. 2007;92:1988–1999. doi: 10.1529/biophysj.106.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radler J, Strey H, Sackmann E. Langmuir. 1995;11:4539–4548. [Google Scholar]
- 23.Schonherr H, Johnson JM, Lenz P, Frank CW, Boxer SG. Langmuir. 2004;20:11600–11606. doi: 10.1021/la049302v. [DOI] [PubMed] [Google Scholar]
- 24.Cremer PS, Boxer SG. J Phys Chem B. 1999;103:2554–2559. [Google Scholar]
- 25.Nissen J, Gritsch S, Wiegand G, Radler JO. Eur Phys J B. 1999;10:335–344. [Google Scholar]
- 26.Jiang FY, Bouret Y, Kindt JT. Biophys J. 2004;87:182–192. doi: 10.1529/biophysj.103.031054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhelev DV, Needham D. Biochim Biophys Acta. 1993;1147:89–104. doi: 10.1016/0005-2736(93)90319-u. [DOI] [PubMed] [Google Scholar]
- 28.Taupin C, Dvolaitzky M, Sauterey C. Biochemistry. 1975;14:4771–4775. doi: 10.1021/bi00692a032. [DOI] [PubMed] [Google Scholar]
- 29.Genco I, Gliozzi A, Relini A, Robello M, Scalas E. Biochim Biophys Acta. 1993;1149:10–18. doi: 10.1016/0005-2736(93)90019-v. [DOI] [PubMed] [Google Scholar]
- 30.Chernomordik LV, Kozlov MM, Melikyan GB, Abidor IG, Markin VS, Chizmadzhev YA. Biochim Biophys Acta. 1985;812:643–655. [Google Scholar]
- 31.Sanders CR, Schwonek JP. Biochemistry. 1992;31:8898–8905. doi: 10.1021/bi00152a029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


