Abstract
The mammalian nervous system is the most complex organ of any living organism. How this complexity is generated during neural development is just beginning to be elucidated. This article discusses the signaling, transcriptional, and epigenetic mechanisms that are involved in neural development. The first part focuses on molecules that control neuronal numbers through regulation of the timing of onset of neurogenesis, the timing of the neuronal-to-glial switch, and the rate of progenitor proliferation. The second part focuses on molecules that control neuronal diversity by generating spatially or temporally distinct populations of neuronal progenitors. Most of the studies discussed in this article are focused on the developing mammalian cerebral cortex, because this is one of the main model systems for neural developmental studies and many of the mechanisms identified in this tissue also operate elsewhere in the developing brain and spinal cord.
In the mammalian cortex, various molecules control neuronal numbers and diversity through spatial and temporal patterning mechanisms. These mechanisms may also operate elsewhere in the developing brain and spinal cord.
1. INTRODUCTION
The primordium of the central nervous system (CNS), the neural plate, originates from the ectoderm of gastrulating vertebrate embryos. It is made of a single sheet of neuroepithelial (NE) cells, which undergo rapid symmetric divisions that result in planar expansion of the neural plate and in generation of the neural tube. At mid-gestation, between embryonic day 9 (E9) and E10 in the mouse, the first neurons of the CNS are born, heralding an important developmental transition in the development of the neural progenitor cells of the brain. Coincident with the acquisition of neurogenic potential, the progenitors acquire the identity of radial glial (RG) cells and begin to express glial markers such as GLAST (Glutamate-aspartate transporter) and BLBP (Brain lipid-binding protein). In addition, the tight junctions that couple early NE progenitors are replaced by apically located adherens junctions (Gotz and Huttner 2005; Kriegstein and Alvarez-Buylla 2009). RG stem cells persist as the principal progenitor type during development of the embryonic and postnatal CNS. Importantly, RG cells are thought to be the primary progenitors of most neurons throughout the CNS and also to give rise, via lineage-restricted intermediate precursors, to the two main macroglial cell types, astrocytes and oligodendrocytes (Kriegstein and Alvarez-Buylla 2009). In the nervous system, there is a distinct progression of lineage differentiation whereby RG cells first give rise to neurons and later to astrocytes and oligodendrocytes.
Following the transition to the RG fate, some progenitor cells begin to divide asymmetrically to generate another radial glial cell and a more differentiated daughter cell, which migrates away from the apical progenitor domain and commences neuronal differentiation. In the forebrain, the cell lineages that produce neurons have become significantly more complex, with the insertion of fate-restricted progenitor stages between stem cells and postmitotic cells. These intermediate cells have been called basal or abventricular progenitors (BPs). They are generated in the developing telencephalon by asymmetric division of radial glial stem cells (Haubensak et al. 2004; Miyata et al. 2004; Noctor et al. 2004). BPs are not attached to the ventricular surface and divide away from it in the subventricular zone (SVZ). To maintain their self-renewing asymmetric divisions, RG stem cells must localize the cell polarity determinants of the Par family, including Par3 and Par6 and their regulator, the small Rho GTPase Cdc42, to their ventricular end feet. Disruption of the Par complex results in premature generation of BPs (Cappello et al. 2006; Costa et al. 2008; Bultje et al. 2009). In the dorsal telencephalon (which gives rise to the cerebral cortex), BPs divide symmetrically only once or a few times and are thought to generate most cortical projection neurons, including early-born neurons in deep cortical layers and late-born neurons in superficial cortical layers (Farkas and Huttner 2008). The number of divisions of BPs in the ventral telencephalon (which gives rise to the basal ganglia and interneurons of the cortex) and the extent of their contribution to the generation of basal ganglia neurons and cortical interneurons are less well characterized.
As neurogenesis progresses, there is an ever-increasing propensity for progenitors to undergo a symmetric terminal division, in which both daughters differentiate. Consequently the expansion of the progenitor pool gradually slows and then stops. Many studies over the last decade have shed light on specific signaling, transcriptional, and epigenetic mechanisms that are integrated to ensure a timely and coordinate switch in progenitor properties. This is discussed in the first section of this article, which examines the molecular mechanisms that instruct the acquisition of radial glial features, the proliferation of neural progenitors, the promotion of neurogenesis, and the subsequent termination of neurogenesis and concomitant switch to gliogenesis.
In most regions of the CNS, different types of neurons are generated at different times during neurogenesis. In the cerebral cortex, for example, neurons located in deep layers, which mostly project to subcortical targets such as the thalamus and spinal cord, are generated before neurons in more superficial layers, which project to other parts of the cortex. Premature transition from proliferative symmetric divisions to neurogenic asymmetric divisions, which is thought to underlie several developmental brain abnormalities such as microcephaly (Manzini and Walsh 2011; Rubenstein 2011), therefore reduces brain size but also alters the balance between different neuronal populations. Conversely, protraction of the phase of progenitor expansion over evolutionary time is thought to contribute greatly to an increased number of late-born cortical neurons connecting different cortical areas in humans compared with lower primates (Rakic 1995). The second section of this article discusses the mechanisms that promote the generation of different types of neurons by progenitors in different brain regions and by the same progenitors over time. Most of the examples discussed refer to studies performed in the cerebral cortex, but many of the principal mechanisms also operate in other CNS regions.
2. CONTROLLING NEURONAL NUMBER IN THE EMBRYONIC BRAIN
2.1. Onset and Progression of Neurogenesis
2.1.1. Signaling Mechanisms Regulating the Onset and Maintenance of Neurogenesis
The onset of neurogenesis and the transition from NE to RG coincides with the onset of Notch signaling in the dorsal telencephalon, as detected by the expression of the major Notch ligand Delta-like 1 (Dll1) and the downstream transcription factors Hes1 and Hes5 (Hatakeyama et al. 2004). In support of the idea that Notch signaling promotes the NE-to-RG transition, Gaiano and Fishell (Gaiano et al. 2000) showed that premature Notch pathway activation strongly induces RG markers and cellular phenotype. Notch signaling has also been reported to inhibit the generation of BPs by RG cells (Mizutani et al. 2007; Ohata et al. 2011). It is not clear what initiates Notch signaling in the cortex, although the induction of Dll1 coincides with the appearance of the pro-neural proteins Ngn2 and Ascl1, which are major transcriptional regulators of neurogenesis (see below) and have been shown to directly regulate Dll1 (Castro et al. 2006). Notch can instruct acquisition of RG identity, but strong perturbation of the pathway via deletion of Hes1, Hes5, or of the Notch effector transcription factor RBPJ did not block the appearance of RG, although Notch-deficient RGs commenced neurogenesis prematurely and lost their apico–basal polarity (Hatakeyama et al. 2004; Imayoshi et al. 2010). Thus Notch is important, if not essential, for the instigation of RG development and to maintain neurogenic RG in an undifferentiated state (Fig. 1B).
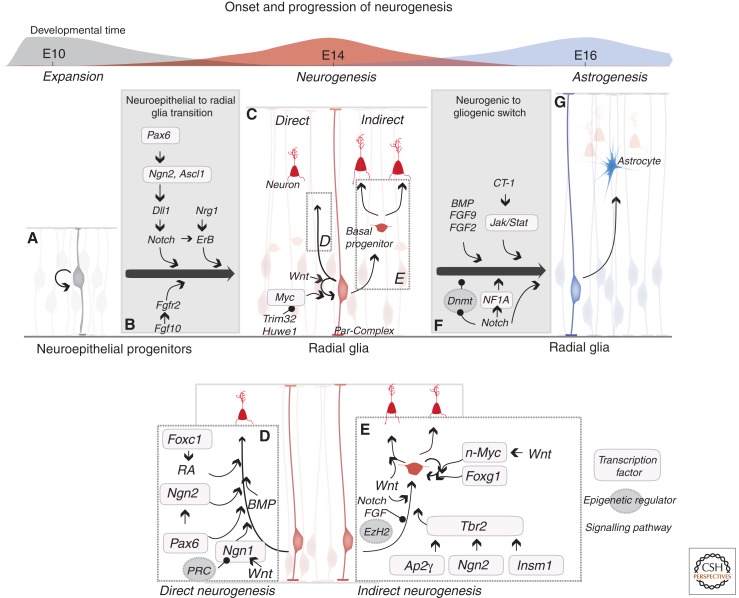
Figure 1.
Molecular pathways regulating the onset, progression, and termination of neurogenesis in the rodent cerebral cortex. The onset of neurogenesis is concomitant with the transformation of neuroepithelial stem cells (A) into radial glial (RG) stem cells (C). Several signaling pathways, including the Dll1/Notch, Nrg1/ErB, and Fgf10/Fgfr2 pathways have been implicated in this transformation (see text). RG cells then generate neurons directly (D) or via basal progenitors (E). Several transcription factors (Ap2γ, Ngn2, Insm1, Tbr2) have been shown to promote the generation of basal progenitors from RGs, whereas the Notch and FGF pathways and the epigenetic regulator Ezh2 inhibit this step. Whether AP2γ, Ngn2, and Insm1 act primarily by inducing Tbr2 expression or also via Tbr2-independent mechanisms is unclear. Other transcription factors and signaling molecules promote the self-renewal of RGs (Wnt, Myc) and the proliferation of basal progenitors (Foxg1, Wnt/n-Myc). Whether the same factors and pathways that promote the direct generation of neurons by RGs (Pax6, Ngn1/2, RA) also drive the generation of neurons by basal progenitors (data not shown) is unclear. The termination of neurogenesis results from the terminal differentiation of RGs into astrocytes (G). Multiple signaling pathways (Jack/Stat, Notch, BMP, FGF) synergize to elicit the neurogenic-to-gliogenic switch (see text). It is noteworthy that the same pathways frequently operate in different temporal contexts to exert contrasting cellular effects.
Consistent with the specification of RG cells continuing in the absence of Notch signaling, other pathways are clearly implicated in this process. For example Neuregulin 1 (Nrg1) is expressed in the developing cortex and signals through its receptors ErB2 and ErB4 to promote RG identity and to suppress the differentiation of RG into astrocytes (Schmid et al. 2003; Sardi et al. 2006). Interestingly, ErB2 seems to be a target of the Notch pathway, suggesting that it might account for some of the Notch pathway’s RG-promoting activity (Fig. 1B) (Schmid et al. 2003).
Similar to the effect of enforced early activation of the Notch pathway, expression of a constitutively active form of a receptor for fibroblast growth factors, Fgf receptor 2 (Fgfr2), also promotes precocious acquisition of RG cell identity (Yoon et al. 2004). A recent study has shed light on which of the many fibroblast growth factors (Fgf) ligands is likely to be most relevant in the NE-to-RG transition. Fgf10 expression appears in the cortical ventricular zone (VZ) at E9.5, and its overexpression promotes expression of RG markers, whereas Fgf10 mutant mouse embryos show an extended period of NE expansion and delayed neurogenesis (Fig. 1B) (Sahara and O’Leary 2009). FGF signaling is also required to slow down the progression from RG to BPs (Fig. 1E) (Kang et al. 2009).
In addition to this role in the maturation of cortical progenitors, FGF ligands have been shown to promote the proliferation of cortical progenitors and inhibit neurogenesis, by regulating the duration of the cell cycle. Cell cycle length and, specifically, the duration of the G1 phase of the cycle increase markedly as neurogenesis progresses in the mammalian brain. The cell cycle is also longer for progenitors undergoing neurogenic divisions, that is, producing postmitotic neurons, than for neighboring cells dividing to produce more progenitors (Calegari et al. 2005). Experimental manipulations of cortical progenitors have shown that cell cycle lengthening is, indeed, sufficient by itself to increase the fraction of progenitor divisions that produce neurons and to promote the generation and expansion of BPs (Lukaszewicz et al. 2005; Lange et al. 2009; Pilaz et al. 2009). FGF2 maintains the proliferation of progenitors at the onset of neurogenesis (Raballo et al. 2000) by controlling the duration of their divisions. FGF2 up-regulates expression of cyclin D1 and down-regulates expression of the cyclin-dependent kinase (cdk) inhibitor p27(kip1), thereby shortening the G1 phase of the cycle and decreasing the proportion of neurogenic divisions (Lukaszewicz et al. 2002). Another important mitogen for neural progenitors, Insulin-like growth factor-1 (IGF-1), also promotes the division of neural progenitors by inducing the expression of cyclin D1 as well as cyclin D3 and E while simultaneously reducing the expression of the cdk inhibitors p27(KIP1) and p57(KIP2) (Mairet-Coello et al. 2009). A related molecule, IGF-2, is secreted in the cortico–spinal fluid by the choroid plexus and provides a mitogenic signal to cortical progenitor cells lining the ventricular cavity (Lehtinen et al. 2011).
Until recently, it was not clear whether the acquisition of RG properties was inextricably linked to the ability to undertake asymmetric, neurogenic divisions. In mice mutant for Foxc1, a transcription factor expressed in the cortical meninges, where it is required for the production of retinoic acid (RA), there is a dramatic increase in the number of RG cells and a severe block in neuronal differentiation (Fig. 1D) (Siegenthaler et al. 2009). Thus, in the absence of RA signaling, RG are specified, apparently normally, yet they go through repeated rounds of symmetric, expanding divisions without producing neuronal progeny. Treatment of RA-deficient mice with dietary RA was able to prevent the lateral expansion of the cortex and rescued the production of differentiated neurons. The downstream transducers of RA signaling that promote neurogenesis are not known, although the pro-neural factors, Neurogenin1 and 2, might be involved (Ribes et al. 2008; Lee et al. 2009).
Another extracellular signal that may not be required for the specification of RG cells but is an important regulator of neuronal production in the developing cortex is the Wnt activity. Some gain- and loss-of-function studies have implicated Wnt in promoting proliferation and self-renewal of RG progenitors (Fig. 1C) (Chenn and Walsh 2002; Machon et al. 2003; Woodhead et al. 2006; Zhou et al. 2006), while other studies have shown that at subsequent stages, Wnt signaling promotes the maturation of RG cells into BPs and proliferation of the latter, through regulation of N-myc and the pro-neural gene Neurogenin1 (Viti et al. 2003; Hirabayashi et al. 2004; Kuwahara et al. 2010). Other work has also suggested that Wnt can act instructively to promote neuronal differentiation of BPs (Fig. 1E) (Munji et al. 2011). Thus the effects of Wnt signaling in cortical development are complex and tightly regulated in space and time. It should be noted that several of these studies involved the manipulation of β-catenin, which is a major transducer of canonical Wnt signaling but also has non-Wnt-associated functions, for example, in cadherin cell–cell adhesion complexes. Thus, the observed phenotypes may not be due solely to changes in Wnt pathway activity.
Another important pathway, with similarly elusive and complex inputs into the regulation of cortical neurogenesis, is the BMP (bone morphogenetic protein) pathway. Treatment of early (mouse E12–E13) cortical progenitors with BMP in culture induces neurogenesis (Li et al. 1998; Mabie et al. 1999), and deletion of BMP receptors in vivo causes localized reduction of neurogenesis in caudal regions of the CNS (Fig. 1D) (Wine-Lee et al. 2004). Later in cortical development (after E14), BMPs block neurogenesis and, instead, promote astrocyte differentiation (Fig. 1F) (Gross et al. 1996; Nakashima et al. 2001).
The difficulty in pinning down the effects of these signaling pathways in the regulation of cortical neurogenesis illustrates an important theme that applies to embryonic development generally. Frequently, the same signaling pathway operating in two neighboring cells, or in the same cell at different times, can cause completely different cellular effects, presumably due to the varying availability of effector molecules and epigenetic states of the target genes of signaling pathways. There is also extensive and complex cross talk between signaling pathways such that they modulate each other’s effects. These mechanisms allow a relatively small number of signaling pathways to evoke the complex spectrum of cellular behaviors required to build a functional brain.
2.1.2. Transcriptional Mechanisms Regulating Cortical Neurogenesis
Cell-intrinsic genetic controls and cell-extrinsic signaling mechanisms ultimately influence cell fate via the precise modulation of gene expression. A great deal of effort has been made in recent years in understanding the transcriptional control of neurogenesis in the cerebral cortex by sequence-specific transcription factors. Here we focus on some of the key transcription factors that have clearly demonstrated roles in the regulation of embryonic neurogenesis (Fig. 1B,D,E).
A large number of transcription factors have been implicated in the proliferation of neural progenitors, and thus ultimately in the control of neuronal numbers in the developing brain. Defects in progenitor divisions and brain growth have been found in mice mutant for the paired homeobox factor Pax6 (Arai et al. 2005), the homeobox proteins Lhx2 (Porter et al. 1997) and Arx (Friocourt et al. 2008), the winged-helix protein Foxg1 (Hanashima et al. 2002), and the nuclear receptor Tlx (Roy et al. 2004). The target genes that are regulated and the types of progenitor cells that are induced to proliferate by these factors were usually not examined, except in a few cases. Pax6, in particular, has been proposed to promote proliferation early during cortical development through induction of multiple target genes, including transcription factors (e.g., Hmga2), signaling molecules (e.g., Fabp7), and cell cycle regulators (e.g., Cdk4) (Arai et al. 2005; Sansom et al. 2009).
The basic-helix–loop–helix (bHLH) pro-neural transcription factors are crucial players in the regulation of neurogenesis, and this appears to be a highly conserved function (for review, see (Bertrand et al. 2002; Ross et al. 2003). Three of these pro-neural factors—Neurogenin (Ngn)1, Ngn2, and, at a lower level of expression, Ascl1 (also called Mash1)—are expressed in radial glial cells of the developing cortex. These factors start to be expressed around the time of the NE-to-RG transition but do not appear to be absolutely required for the acquisition of radial glia features, although in embryos mutant for both Ngn2 and Ascl1, radial glia are disorganized and differentiate into astrocytes prematurely (Nieto et al. 2001). Cortical neurogenesis is greatly impaired in these pro-neural-deficient embryos in vivo and in vitro (Nieto et al. 2001). Consistent with this requirement for pro-neural factors for neurogenesis, they are also sufficient to induce a full program of neurogenesis. If overexpressed in vivo or in vitro in neural progenitors, Ngn2 or Ascl1 can induce rapid and full neuronal differentiation (Mizuguchi et al. 2001; Nakada et al. 2004; Berninger et al. 2007). Even more impressively, pro-neural factors can respecify cells from other lineages into cortical projection neurons. For example, Ngn2 can drive postnatal astroglia to transdifferentiate into cortical projection neurons (Heinrich et al. 2010), whereas Ascl1 expression can achieve the remarkable feat of reprogramming a fibroblast into a neuron, although the efficiency of this latter step is much improved if Ascl1 is overexpressed in combination with cofactors such as Brn2 and Myt1l (Vierbuchen et al. 2010).
A recent study of Ascl1 targets provides compelling evidence that Ascl1 directly activates a wide array of genes required for neuronal differentiation in the ventral telencephalon (Castro et al. 2011). A genome-wide survey of Ngn2 targets in the mouse forebrain has not yet been performed, although Ngn2 has been shown to directly activate specific genes required for differentiation and migration of cortical projection neurons (Ge et al. 2006; Heng et al. 2008; Ochiai et al. 2009). Thus, rather than inducing a cascade of downstream transcription factors that, in turn, induce the program of neurogenesis, pro-neural bHLH factors are directly involved in activating the effectors of neuronal differentiation.
Pax6 is required for the progression of cortical neurogenesis. Pax6 is expressed in both the early neuroepithelial progenitors and the neurogenic radial glia in many regions of the CNS, and when it is mutated, cortical neurogenesis is impaired in vitro (Heins et al. 2002). Cortical neurogenesis defects in vivo have proven to be stage dependent and difficult to analyze because of cell-autonomous loss of dorsal and acquisition of ventral telencephalic features in Pax6 mutant progenitors (Quinn et al. 2007). Nevertheless, like Ngn2, Pax6 can instructively promote neurogenesis in cortical progenitors and in astrocytes (Heins et al. 2002). Pax6 directly induces Ngn2 expression in the cortex and the spinal cord (Scardigli et al. 2003), and consequently drives neurogenesis indirectly. Nevertheless, Pax6 can reprogram astrocytes into neurons without inducing Ngn2 (Heins et al. 2002), and there is evidence that it directly regulates many genes implicated in the progression of neurogenesis in the cortex (Sansom et al. 2009). Thus, Pax6 appears to act both upstream of and in parallel to pro-neural bHLH factors in promoting neurogenesis.
Several transcription factors, including Ngn2, Insm1, and AP2γ, have been implicated in the crucial step of generation of basal progenitors from RG stem cells (Fig. 1E). All three factors share the capacity to induce Tbr2, a transcription factor specifically expressed in the subventricular zone of the cortex and required for the generation of BPs (Miyata et al. 2004; Arnold et al. 2008; Farkas et al. 2008; Sessa et al. 2008; Pinto et al. 2009). Other factors act subsequently to promote the divisions of BPs, including Foxg1 in the cortex and Ascl1 in the ventral telencephalon, which acts by activating multiple cell cycle regulators including E2f1 and cyclin-dependent kinases (Siegenthaler et al. 2008; Castro et al. 2011). How transcription factors interact with signaling pathways that regulate the generation and expansion of the different populations of neural progenitors remains largely unknown.
Recent studies have highlighted the importance of another layer of control of the genetic program of neurogenesis at the posttranslational level (Fig. 1C). TRIM32 is a TRIM-NHL protein expressed in radial glia that is asymmetrically inherited by daughter cells during mitosis (Schwamborn et al. 2009). The TRIM32-inheriting daughter is more prone to differentiate into a neuron because of at least two activities of TRIM32. Firstly, it acts as a ubiquitin ligase that promotes the destruction of the pro-proliferative transcription factor c-Myc; and secondly, it binds to the Argonaute complex and enhances the activity of microRNAs including Let-7, which can, in turn, promote neuronal differentiation (Schwamborn et al. 2009). The HECT domain E3 ubiquitin ligase Huwe1 also promotes neurogenesis in the cortex, in this case, by promoting the degradation of n-Myc, which can no longer activate the Notch ligand Dll3 and promote self-renewing rather than differentiative divisions of RG (Zhao et al. 2009). The E3 ubiquitin ligase TRIM11 acts in the opposite direction and suppresses neurogenesis by promoting the degradation of Pax6 (Tuoc and Stoykova, 2008).
2.1.3. Epigenetic Mechanisms
Recently, compelling evidence has emerged to show that epigenetic modifications of chromatin provide another crucial level of control of gene expression and underpin the complex choreography of embryonic neurogenesis in the cerebral cortex and beyond (Fig. 1D–F) (for review, see Hirabayashi and Gotoh 2010). Much of the progress in this area has been achieved by mutating enzymes that catalyze specific epigenetic modifications. For example, the Polycomb complex catalyzes the tri-methylation of lysine 27 on the tail of histone H3 (H3K27me3), a modification mainly associated with transcriptional repression. When Polycomb complex members Ring1b or EZH2 were ablated during cortical neurogenesis, the rate of neurogenesis was increased, and the length of the neurogenic period was extended with a corresponding delay in the onset of astrogliogenesis (Hirabayashi et al. 2004). At least one of the important targets for Polycomb-mediated antagonism of neurogenesis is the promoter of the pro-neural factor Ngn1, which gradually accumulates the H3K27me3 mark during extended culture of cortical progenitors, in parallel with their reduced ability to produce neuronal progeny (Hirabayashi et al. 2004). Consistently, Polycomb-deficient cortices showed up-regulation of Ngn1 in vivo (Fig. 1D). Two other studies have provided evidence that Polycomb complex activity must be antagonized to sustain cortical neurogenesis. Jepsen et al. (2007) showed that the retinoic acid pathway, which is an important determinant of the onset of neurogenesis (Fig. 1B), directly induces Jmjd3, an H3K27me3 demethylase, which when forcibly expressed can activate genes associated with neurogenesis in cortical progenitors. In a similar vein, Lim et al. (2009) described the activity of the Trithorax complex member, Mll1, in antagonizing H3K27me3 deposition on the promoter of the neurogenic transcription factor Dlx2, which can then sustain neurogenesis.
In addition to histone methylation, direct methylation of chromatin also has a role in sustaining embryonic neurogenesis. DNA methyltransferase 1 (Dnmt1) is an enzyme that maintains methylation of CpG dinucleotides, a DNA modification mainly associated with repressed genes. When Dnmt1 is mutated in the developing CNS, neurogenesis is terminated prematurely (Fan et al. 2005).
2.2. The Neuronal-to-Glial Switch: Timing and Control Mechanisms
Around E18.5 in mouse cortical development, another major developmental transition occurs. RG progenitors stop producing neurons, and the first astrocytes appear (Fig. 1F,G). During this process, most RG release their apical attachment to the ventricle and move upward, away from the ventricular surface. They lose their radial processes and gradually take on a multipolar, astrocytic morphology. It is still not clear whether further cell division occurs to amplify the population of astrocytes in the cerebral cortex during peri- and postnatal development of the brain, although this does seem likely (Kriegstein and Alvarez-Buylla 2009). Apart from small populations of RG that are maintained and retain the ability to produce neurons in defined regions throughout life (namely, the subventricular zone of the lateral ventricles and the dentate gyrus of the hippocampus), very few neurons are produced in the rodent or primate cerebral cortex beyond the neurogenic-to-astrogenic transition (Kriegstein and Alvarez-Buylla 2009). Owing to this relatively abrupt transition, the timing of the end of neurogenesis must be very carefully controlled to ensure that all of the neurons required for the function of the adult brain are born before gliogenesis commences.
Most prominent among the pro-astrocytic signals is the Jak/Stat pathway (Fig. 1F). Although Jak/Stat signaling is active at a low level in progenitors during neurogenesis and may even be required for radial glial identity (Kamakura et al. 2004), at the end of this period pathway activity increases greatly (He et al. 2005). The sudden increase in Jak/Stat signaling seems to be caused by several mechanisms. Firstly, several promoter elements on glial and Stat pathway genes that are responsive to Stat transcription factors (the main transducers of Jak/Stat signaling) are specifically methylated during the neurogenic period, which precludes Stat binding (Takizawa et al. 2001; Fan et al. 2005). This methylation is removed at the end of neurogenesis in part by Notch signaling (see below), and the promoters are derepressed. The second important factor is the reduced expression of the pro-neural factors Ngn1 and Ngn2. Ngn1 (and probably also Ngn2, although this has been less studied) is a potent antagonist of the Jak/Stat pathway and seems to achieve this by sequestering the important transcriptional coactivators CREB binding protein (CBP), p300, and Smad1 away from Stat3, which cannot then activate astroglial genes such as GFAP (Sun et al. 2001; He et al. 2005). Thirdly, the cytokine Cardiotrophin-1 (CT-1) starts to be expressed at high levels by differentiating neurons, and this molecule appears to be the main activating ligand for the Jak/Stat pathway in vivo (Fig. 1F) (Barnabé-Heider et al. 2005; Miller and Gauthier 2007). The end result of these events is an increase in Stat activity, which becomes even more amplified because the Stat factors start to positively autoregulate their own transcription (He et al. 2005).
While the Jak/Stat pathway activity becomes quantitatively amplified at the neurogenesis-to-gliogenesis transition, the output of Notch signaling appears to undergo a dramatic qualitative shift. As discussed above, early in corticogenesis, Notch mainly promotes the RG progenitor state (Fig. 1B), but later in cortical development, Notch activation instructively promotes astrocyte differentiation and blocks neuronal differentiation both in vitro and in vivo (Fig. 1F) (Chambers et al. 2001; Grandbarbe et al. 2003). In further support of an instructive rather than a permissive role in gliogenesis, Ge et al. (2002) showed that the key transducers of Notch signaling—the intracellular domain of the Notch receptor itself and the RBPJ transcription factor (also known as CSL or CBF1)—form a transcriptional activation complex on the GFAP promoter. Furthermore, cultured neural progenitors derived from RBPJ mutant embryonic stem cells show a significant delay in astrocyte development in vitro. Recently an elegant study by Namihira et al. (2009) showed that the main source of Notch signaling during corticogenesis is likely to be from young neurons and BPs, which express the Notch ligand Dll1. Namihira and colleagues went on to show that Notch signaling in cortical progenitors directly activates Nuclear Factor IA (NFIA), a transcription factor already shown to be necessary and sufficient to promote glial specification in the spinal cord (Deneen et al. 2006; Namihira et al. 2009). The combined actions of Notch and NFIA then cause demethylation of the promoter of GFAP (and presumably of other astrocytic genes) via the displacement of DNA methyltransferase enzymes, rendering it responsive to further Jak/Stat- and Notch-mediated activation (Namihira et al. 2009). Interestingly, there is good evidence that Hes proteins, which are the main targets of Notch/RBPJ signaling in the developing CNS, physically interact with Jak and Stat factors and that these interactions augment Jak/Stat pathway activation (Kamakura et al. 2004). Thus, the Notch and Jak/Stat pathways converge to provide a strong pro-astroglial signal.
Like Notch, the BMP pathway also imparts different effects on early and late corticogenesis. Early BMP signaling can promote neurogenesis (Fig. 1D), whereas late expression drives astroglial development (Fig. 1F). When BMP4 was overexpressed by cortical neurons from E16, RG progenitors were more likely to differentiate into astrocytes (Gomes et al. 2003). This in vivo result is consistent with earlier in vitro work showing that BMP treatment of cortical progenitors could induce astrocyte differentiation (Nakashima et al. 1999). Just like the cooperation between Notch and Jak/Stat signaling, there is pro-astrocytic synergy between BMP and LIF (another activator of Jak/Stat signaling) mediated by the formation of a complex between Stat3, Smad1, and the coactivator p300, which together strongly activate the GFAP promoter (Nakashima et al. 1999). Another important effect of BMP signaling in late cortical development is the induction of negative helix–loop–helix factors of the Id family, which inhibit the pro-neural transcription factors Ngn1/2 and Ascl1 (Nakashima et al. 2001).
Yet another signaling pathway that seems to promote glial differentiation late in development but not early, is the Fgf pathway. In vitro Fgf2 is required for Jak/Stat-mediated activation of astroglial genes (Song and Ghosh 2004), again showing the codependence of multiple pathways in the transition from neurogenesis to gliogenesis (Fig. 1F). In an in vivo context, the most relevant Fgf ligand in this context might be Fgf9, which is induced by the transcription factor Sip1 in neurons of the cortical plate and signals back to ventricular zone progenitors to promote glial differentiation and inhibit neurogenesis (Seuntjens et al. 2009).
Therefore, as the rate of neurogenesis reaches its peak, the levels of CT-1, Fgf9, BMP, and Notch ligand from neurons and neuronal precursors are greatly amplified. This quantitative effect, coupled with direct cross talk and collaboration between the pathways, ultimately results in the rather sudden cessation of neurogenesis and commencement of gliogenesis. By this means, it is the production of neurons themselves that provides the timing mechanism necessary to signal the end of neurogenesis. Although it has been argued on the basis of elegant clonal assays that neural progenitor cells themselves encode cell-intrinsic timers that ensure the progression from neurogenesis to gliogenesis (Shen et al. 2006), in these experiments, the progenitor cell is always in contact with its own progeny and therefore amenable to local signals from those daughter cells. As such, a cell-extrinsic accumulation of signals that alter the outcome of progenitor divisions is also consistent with the results of clonal cultures.
3. GENERATING NEURONAL DIVERSITY IN THE EMBRYONIC BRAIN
3.1. Spatial Mechanisms of Neuronal Fate Specification
In the developing telencephalon, several signaling centers secrete diffusible signaling molecules that form overlapping gradients and act as morphogens (Hoch et al. 2009). Sonic hedgehog (Shh) is secreted from the ventral midline of the forebrain, starting well before the closure of the neural tube at E7.5. Members of the FGF family, including FGF8, 15, and 17, are secreted anteriorly by the midline of the telencephalon or commissural plate. Multiple members of the Wnt and BMP families are secreted from medial and caudal aspects of the cortex (Hoch et al. 2009). These extracellular factors induce in progenitors the regionalized expression of homeodomain and helix–loop–helix transcription factors, which is subsequently refined by cross-repressive interactions, thereby subdividing the telencephalic vesicles into a dorsal or pallial and a ventral or subpallial territory (Campbell 2003). These patterning transcription factors, in turn, induce another group of transcription factors (sometimes called lineage determinants) that define progenitor identities and contribute to the selection of a neuronal fate and the specification of defined neuronal phenotypes (Fig. 2) (Guillemot 2005; Hoch et al. 2009). The same basic principles that underlie the specification of dorsal and ventral identities in the telencephalon also apply to the spatial patterning of other developing neural tissues (Zhuang and Sockanathan 2006) and also to the subdivision of the cerebral cortex into distinct architectonic and functional areas later during development (O’Leary et al. 2007).
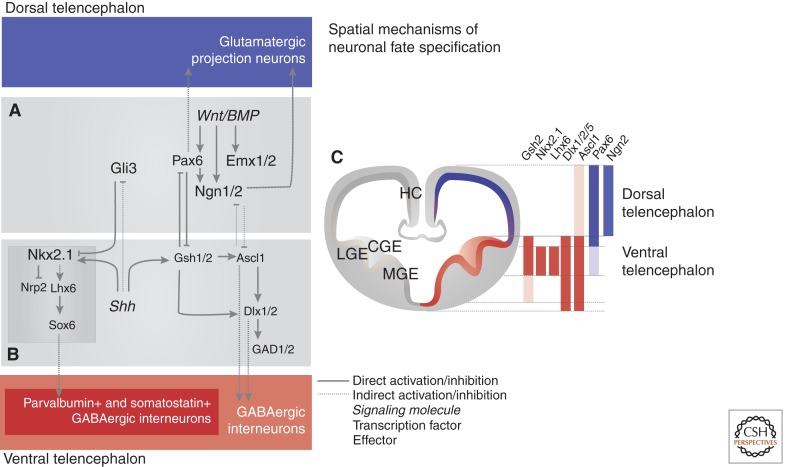
Figure 2.
Spatial mechanisms of neuronal fate specification. (A) BMP and Wnt signals, diffusing from the dorsal midline of the telencephalon, induce expression of important transcriptional regulators of dorsal telencephalic fates (Emx1/2, Pax6, Ngn1/2). Pax6 and Ngn1/2 promote neurogenesis and specify the glutamatergic projection neuron identity of neurons born in the cerebral cortex. (B) The morphogen Shh secreted by the ventral midline of the telencephalon, represses the activity of Gli3 and induces the expression of transcriptional determinants of ventral telencephalic cell fates, including Nkx2.1 and Gsh1/2. Gsh1/2 induce expression of the pro-neural gene Ascl1, which together with its targets Dlx1/2 promotes neurogenesis and contributes to the specification of GABAergic neurons, including basal ganglial neurons and cortical interneurons, in the ventral telencephalon. Nkx2.1, through induction of Lhx6 and Sox6, further specifies subpopulations of cortical interneurons defined by the expression of the calcium-binding protein parvalbumin and the peptide somatostatin. (C) Dorsally or ventrally restricted expression of transcriptional determinants of neuronal fates. The sharp boundary of their expression domains is established by cross-repression (shown in A and B).
Shh establishes ventral identities in the telencephalon by opposing the dorsalizing activity of the transcriptional repressor Gli3 (Rallu et al. 2002). Shh induces the expression of the patterning homeodomain proteins Gsh1 and Gsh2 throughout the ventral telencephalon and Nkx2.1 in the medial part of the ventral telencephalon (Rallu et al. 2002). Nkx2.1 and Gsh1/2 act independently as transcriptional effectors of Shh signaling by inducing different lineage determinants, including induction of the LIM homeobox protein Lhx6 by Nkx2.1, and of Ascl1 and the homeodomain proteins Dlx1 and Dlx2 by Gsh1/2 (Toresson et al. 2000; Du et al. 2008; Wang et al. 2009). Ascl1 and Dlx1/2, in turn, initiate the neuronal differentiation of ventral telencephalic progenitors (Casarosa et al. 1999; Yun et al. 2002; Long et al. 2009).
In the dorsal telencephalon, Wnt induces the expression of Pax6 and the pro-neural proteins Ngn1, whereas Pax6 induces expression of the related protein Ngn2, as mentioned above (Gunhaga et al. 2003; Scardigli et al. 2003; Hirabayashi et al. 2004). Ngn1/2 and Pax6 are the main transcriptional activators of neurogenesis in the cerebral cortex (Nieto et al. 2001; Heins et al. 2002; Schuurmans et al. 2004).
The dorsal and ventral transcriptional cascades are mutually repressive, thus stabilizing the fates of progenitor domains responsible for the production of cortical and basal ganglia neurons, respectively. For example, in the dorsal telencephalon, Gli3 represses Shh target genes such as Gsh2 and Nkx2.1; Pax6 represses Gsh2; and Ngn1/2 repress Ascl1 (Fig. 2) (Fode et al. 2000; Toresson et al. 2000; Rallu et al. 2002). Deletion of dorsal transcription factors, including Gli3, Pax6, and Ngn1/Ngn2, thus leads to expansion of ventral determinants into pallial territory, and vice versa.
The neurogenic factors that are induced by the dorsal and ventral transcription factor cascades, and particularly the pro-neural factors Ngn1 and Ngn2 dorsally and Ascl1 ventrally, have two distinct roles in the generation of neurons from telencephalic progenitors (Bertrand et al. 2002). Firstly, they activate a generic program of neurogenesis, which arrests the divisions of progenitor cells, selects the neuronal fate, suppresses the alternative astroglial fate, initiates the migration of the new neurons to their appropriate locations, and initiates the growth of axon and dendrites and the terminal differentiation of the neurons. Secondly, these factors specify the regional identity of the new neurons, and particularly their mode of neurotransmission and general morphology. Ngn1/2 specify the glutamatergic and pyramidal phenotype of cortical projection neurons, whereas Ascl1 specifically promotes the GABAergic and multipolar properties of basal ganglia neurons and cortical interneurons (Fode et al. 2000; Parras et al. 2002; Hand et al. 2005). It should be noted, however, that different transcription factors are involved in specification of the same neurotransmitter phenotypes, GABAergic and glutamatergic, in other regions of the nervous system (Cheng et al. 2005).
Although pro-neural factors play a prominent role in neurogenesis, other transcription factors that participate in the transcriptional cascades in dorsal and ventral telencephalon also control specific aspects of the neuronal phenotype (Fig. 2B). For example, in the ventral telencephalon, Lhx6 and its target the Sox protein Sox6 promote the specification and differentiation of parvalbumin- and somatostatin-containing cortical interneurons (Gelman and Marin 2010), whereas the LIM protein Lhx7 specifies the cholinergic neurotransmission phenotype of a subset of striatal interneurons (Fragkouli et al. 2009), and the homedomain protein Nkx2.1 promotes the migration of interneurons to the striatum (Nobrega-Pereira et al. 2008).
3.2. Temporal Mechanisms of Neuronal Fate Specification
An additional and equally important mechanism for the generation of neuronal diversity is temporal patterning, that is, the sequential production by the same progenitors of different types of neurons at different times during neurogenesis. It has long been known that a strong correlation exists between the location of neurons in different layers of the cerebral cortex or the retina and the time of their birth. For example, neurons located in the deepest layer of the cortex are generated first, and neurons located in each of the layers above are generated at progressively later times during cortical development (McConnell 1995). Cell transplantations and lineage tracing using viruses or dye injections have shown that these sequential phases of neurogenesis represent changes in the output of individual retinal and cortical progenitors, rather than the generation of different neurons by different types of progenitors (McConnell 1995). The sequential generation of different types of neurons by the same progenitors is a widespread strategy to diversify neuronal populations that also operates in other CNS regions, such as the vertebrate dorsal spinal cord (Muller et al. 2002), and in other distantly related species, such as Drosophila (Isshiki et al. 2001; Maurange et al. 2008).
The molecular mechanisms that confer temporal identities to progenitors and control the temporal order of neuronal specification have been partially elucidated in Drosophila (Isshiki et al. 2001; Maurange et al. 2008), but they remain largely unknown in vertebrates. Transplantation of cortical progenitors into cortices of a different developmental stage (heterochronic transplantations) (McConnell 1995) as well as in vitro progenitor cultures (Shen et al. 2006; Eiraku et al. 2008; Gaspard et al. 2008) suggest that the generation of layer-specific cortical neurons in a precise temporal order is largely controlled by cell-intrinsic mechanisms. Dissociated cortical progenitors in culture maintain their capacity to sequentially generate different types of neurons (Shen et al. 2006). Even more surprising, embryonic stem cell cultures that acquire a cortical fate also generate neurons of different laminar identities in the same temporal order as occurs during cortical development in vivo (Eiraku et al. 2008; Gaspard et al. 2008). However, in these experiments, the progenitor cell is not isolated from its progeny; thus, even in a single clone, the influence of extrinsic signals emanating from postmitotic daughter cells cannot be formally excluded.
Progress has been made recently in identifying transcription factors involved in the sequential generation of specific types of cortical neuron during different stages of neurogenesis (Fig. 3). For example, the zinc finger protein Fezf2 and its target gene Ctip2 play a central role in the specification of neurons located in layer V of the cortex and projecting out of the cortex (Arlotta et al. 2005; Molyneaux et al. 2005). Repression of Ctip2 has been shown to be required for the specification of both earlier-born and later-born neurons: Tbr1 represses Ctip2 to promote generation of layer VI neurons early, whereas the homeobox protein Satb2 represses Ctip2 later to promote layer II/III neurogeneses. Conversely, part of Fezf2’s function in promoting layer V neuron production is to block precocious expression of the Satb2 (Fig. 3) (Britanova et al. 2008; Chen et al. 2008; McKenna et al. 2011). Thus, a network of mutually cross-repressive cell-intrinsic regulators plays a central role in the temporal control of cortical laminar fate specification in the developing cortex.
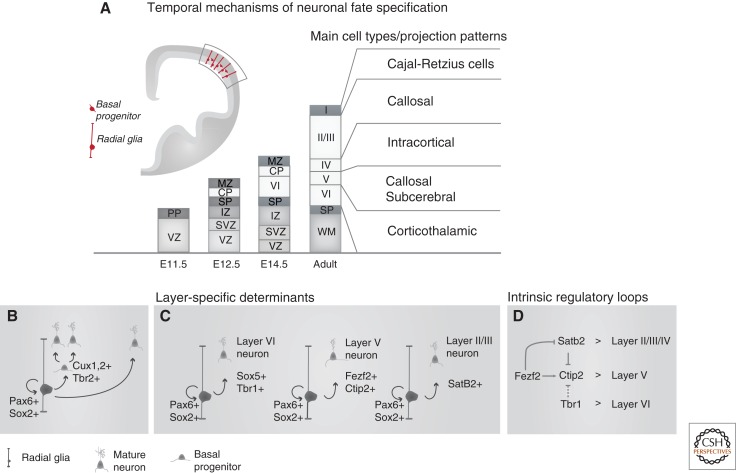
Figure 3.
Temporal mechanisms of neuronal fate specification. (A) Distinct neuronal subtypes are generated sequentially in an inside-first–outside-last order during neurogenesis, by progenitors located in the cortical ventricular zone (VZ) and subventricular zone (SVZ), resulting in the layered organization of the adult cerebral cortex. The intermediate zone (IZ), subplate (SP), and marginal zone (MZ) are present only transiently in the embryonic cortex. (B) Cortical projection (CP) neurons are generated either directly from radial glial stem cells or indirectly via intermediate basal progenitors (BPs). (C) The transcription factors that specify the identity of the distinct classes of projection neurons forming the different cortical layers (see Molyneaux et al. 2007). (D) Transcription factors that specify cortical neuron identities are involved in cross-repressive interactions whereby a layer-specific determinant represses directly (solid lines) or indirectly (dashed lines) the transcription factors involved in neuronal specification in neighboring layers. Abbreviations: VI, layer VI; V, layer V; IV, layer IV; I/II/III, layers I, II, and III.
4. CONCLUDING REMARKS
Although we have achieved a better understanding of the mechanisms that control neuronal identity in the rodent cerebral cortex and in other CNS regions (Dalla Torre di Sanguinetto et al. 2008), it remains unclear how subtype specification mechanisms are temporally coordinated to generate different types of neurons sequentially. Transcription factors that control the timing of generation of the different types of cortical neurons (Naka et al. 2008; Seuntjens et al. 2009) appear also to influence the switch from neurogenesis to gliogenesis. This finding suggests that a common machinery controls the temporal competence of cortical progenitors to produce first different types of neurons and then glial cells. The mechanistic basis of temporal patterning in the nervous system is a challenging issue in developmental neurobiology.
The current effort to develop therapies for neurological disorders is also renewing interest in how neurons of defined identities are generated. Several types of neurons, including spinal motor neurons and layer V cortical projection neurons, can now be differentiated in vitro from embryonic stem cells (Peljto and Wichterle 2011). New strategies are also devised to generate specific neuronal populations from non-neuronal cells through cellular reprogramming (Heinrich et al. 2010; Vierbuchen et al. 2010). Research into the basic mechanisms of neural development has contributed greatly to these recent successes. Further research in this field will continue to facilitate the design of rational strategies to generate clinically relevant neuronal types in vitro.
ACKNOWLEDGMENTS
B.M., D.D., and F.G. are supported by the Medical Research Council, UK. Research in F.G.’s laboratory is funded by grants from the Wellcome Trust and institutional funds from the Medical Research Council.
Footnotes
Editors: Patrick P.L. Tam, W. James Nelson, and Janet Rossant
Additional Perspectives on Mammalian Development available at www.cshperspectives.org
REFERENCES
- Arai Y, Funatsu N, Numayama-Tsuruta K, Nomura T, Nakamura S, Osumi N 2005. Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J Neurosci 25: 9752–9761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD 2005. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45: 207–221 [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnar Z, Robertson EJ, Groszer M 2008. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev 22: 2479–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabé-Heider F, Wasylnka JA, Fernandes KJL, Porsche C, Sendtner M, Kaplan DR, Miller FD 2005. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron 48: 253–265 [DOI] [PubMed] [Google Scholar]
- Berninger B, Guillemot F, Götz M 2007. Directing neurotransmitter identity of neurones derived from expanded adult neural stem cells. Eur J Neurosci 25: 2581–2590 [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F 2002. Proneural genes and the specification of neural cell types. Nat Rev Neurosci 3: 517–530 [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. 2008. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57: 378–392 [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH 2009. Mammalian Par3 regulates progenitor cell asymmetric division via Notch signaling in the developing neocortex. Neuron 63: 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calegari F, Haubensak W, Haffner C, Huttner WB 2005. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci 25: 6533–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K 2003. Dorsal–ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol 13: 50–56 [DOI] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, et al. 2006. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci 9: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F 1999. Mash1 regulates neurogenesis in the ventral telencephalon. Development 126: 525–534 [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, et al. 2006. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell 11: 831–844 [DOI] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, et al. 2011. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev 25: 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers CB, Peng Y, Nguyen H, Gaiano N, Fishell G, Nye JS 2001. Spatiotemporal selectivity of response to Notch1 signals in mammalian forebrain precursors. Development 128: 689–702 [DOI] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK 2008. The Fezf2–Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci 105: 11382–11387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q 2005. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci 8: 1510–1515 [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA 2002. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297: 365–369 [DOI] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Götz M 2008. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development 135: 11–22 [DOI] [PubMed] [Google Scholar]
- Dalla Torre di Sanguinetto SA, Dasen JS, Arber S 2008. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr Opin Neurobiol 18: 36–43 [DOI] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ 2006. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52: 953–968 [DOI] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA 2008. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development 135: 1559–1567 [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y 2008. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3: 519–532 [DOI] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H, et al. 2005. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 132: 3345–3356 [DOI] [PubMed] [Google Scholar]
- Farkas LM, Huttner WB 2008. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol 20: 707–715 [DOI] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Paabo S, Huttner WB 2008. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60: 40–55 [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F 2000. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev 14: 67–80 [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V 2009. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development 136: 3841–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G, Kanatani S, Tabata H, Yozu M, Takahashi T, Antypa M, Raguenes O, Chelly J, Ferec C, Nakajima K, et al. 2008. Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J Neurosci 28: 5794–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G 2000. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron 26: 395–404 [DOI] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, et al. 2008. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455: 351–357 [DOI] [PubMed] [Google Scholar]
- Ge W, Martinowich K, Wu X, He F, Miyamoto A, Fan G, Weinmaster G, Sun YE 2002. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J Neurosci Res 69: 848–860 [DOI] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI-T, Martinowich K, et al. 2006. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci 103: 1319–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Marin O 2010. Generation of interneuron diversity in the mouse cerebral cortex. Eur J Neurosci 31: 2136–2141 [DOI] [PubMed] [Google Scholar]
- Gomes WA, Mehler MF, Kessler JA 2003. Transgenic overexpression of BMP4 increases astroglial and decreases oligodendroglial lineage commitment. Dev Biol 255: 164–177 [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB 2005. The cell biology of neurogenesis. Nat Rev Mol Cell Biol 6: 777–788 [DOI] [PubMed] [Google Scholar]
- Grandbarbe L, Bouissac J, Rand M, Hrabé de Angelis M, Artavanis-Tsakonas S, Mohier E 2003. Delta–Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development 130: 1391–1402 [DOI] [PubMed] [Google Scholar]
- Gross RE, Mehler MF, Mabie PC, Zang Z, Santschi L, Kessler JA 1996. Bone morphogenetic proteins promote astroglial lineage commitment by mammalian subventricular zone progenitor cells. Neuron 17: 595–606 [DOI] [PubMed] [Google Scholar]
- Guillemot F 2005. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol 17: 639–647 [DOI] [PubMed] [Google Scholar]
- Gunhaga L, Marklund M, Sjodal M, Hsieh JC, Jessell TM, Edlund T 2003. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat Neurosci 6: 701–707 [DOI] [PubMed] [Google Scholar]
- Hanashima C, Shen L, Li SC, Lai E 2002. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci 22: 6526–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. 2005. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48: 45–62 [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R 2004. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131: 5539–5550 [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB 2004. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc Natl Acad Sci 101: 3196–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D, Guillemot F, Fan G, et al. 2005. A positive autoregulatory loop of Jak–STAT signaling controls the onset of astrogliogenesis. Nat Neurosci 8: 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Blum R, Gascón S, Masserdotti G, Tripathi P, Sánchez R, Tiedt S, Schroeder T, Götz M, Berninger B 2010. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol 8: e1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde Y-A, Götz M 2002. Glial cells generate neurons: The role of the transcription factor Pax6. Nat Neurosci 5: 308–315 [DOI] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, et al. 2008. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455: 114–118 [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y 2010. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci 11: 377–388 [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y 2004. The Wnt/β-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131: 2791–2801 [DOI] [PubMed] [Google Scholar]
- Hoch RV, Rubenstein JL, Pleasure S 2009. Genes and signaling events that establish regional patterning of the mammalian forebrain. Semin Cell Dev Biol 20: 378–386 [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R 2010. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci 30: 3489–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ 2001. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106: 511–521 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim H-J, Glass CK, Hermanson O, Rosenfeld MG 2007. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature 450: 415–419 [DOI] [PubMed] [Google Scholar]
- Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y 2004. Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat Cell Biol 6: 547–554 [DOI] [PubMed] [Google Scholar]
- Kang W, Wong LC, Shi SH, Hebert JM 2009. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J Neurosci 29: 14571–14580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A 2009. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci 32: 149–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A, Hirabayashi Y, Knoepfler PS, Taketo MM, Sakai J, Kodama T, Gotoh Y 2010. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development 137: 1035–1044 [DOI] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F 2009. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5: 320–331 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Lee JW, Lee SK 2009. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron 62: 641–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. 2011. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69: 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cogswell CA, LoTurco JJ 1998. Neuronal differentiation of precursors in the neocortical ventricular zone is triggered by BMP. J Neurosci 18: 8853–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang Y-C, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A 2009. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458: 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lee T 2011. Generating neuronal diversity in the Drosophila central nervous system. Dev Dyn 241: 457–468 [DOI] [PubMed] [Google Scholar]
- Long JE, Cobos I, Potter GB, Rubenstein JL 2009. Dlx1&2 and Mash1 transcription factors control MGE and CGE patterning and differentiation through parallel and overlapping pathways. Cereb Cortex 19: i96–i106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Savatier P, Cortay V, Kennedy H, Dehay C 2002. Contrasting effects of basic fibroblast growth factor and neurotrophin 3 on cell cycle kinetics of mouse cortical stem cells. J Neurosci 22: 6610–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, Kennedy H, Dehay C 2005. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47: 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabie PC, Mehler MF, Kessler JA 1999. Multiple roles of bone morphogenetic protein signaling in the regulation of cortical cell number and phenotype. J Neurosci 19: 7077–7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machon O, van den Bout CJ, Backman M, Kemler R, Krauss S 2003. Role of β-catenin in the developing cortical and hippocampal neuroepithelium. Neurosci 122: 129–143 [DOI] [PubMed] [Google Scholar]
- Mairet-Coello G, Tury A, DiCicco-Bloom E 2009. Insulin-like growth factor-1 promotes G1/S cell cycle progression through bidirectional regulation of cyclins and cyclin-dependent kinase inhibitors via the phosphatidylinositol 3-kinase/Akt pathway in developing rat cerebral cortex. J Neurosci 29: 775–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Walsh CA 2011. What disorders of cortical development tell us about the cortex: One plus one does not always make two. Curr Opin Genet Dev 21: 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurange C, Cheng L, Gould AP 2008. Temporal transcription factors and their targets schedule the end of neural proliferation in Drosophila. Cell 133: 891–902 [DOI] [PubMed] [Google Scholar]
- McConnell SK 1995. Strategies for the generation of neuronal diversity in the developing central nervous system. J Neurosci 15: 6987–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B 2011. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci 31: 549–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS 2007. Timing is everything: Making neurons versus glia in the developing cortex. Neuron 54: 357–369 [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M 2004. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131: 3133–3145 [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M 2001. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31: 757–771 [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N 2007. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449: 351–355 [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD 2005. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron 47: 817–831 [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD 2007. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci 8: 427–437 [DOI] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C 2002. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron 34: 551–562 [DOI] [PubMed] [Google Scholar]
- Munji RN, Choe Y, Li G, Siegenthaler JA, Pleasure SJ 2011. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J Neurosci 31: 1676–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, Nakamura S, Shimazaki T, Okano H 2008. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci 11: 1014–1023 [DOI] [PubMed] [Google Scholar]
- Nakada Y, Hunsaker TL, Henke RM, Johnson JE 2004. Distinct domains within Mash1 and Math1 are required for function in neuronal differentiation versus neuronal cell-type specification. Development 131: 1319–1330 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T 1999. Synergistic signaling in fetal brain by STAT3–Smad1 complex bridged by p300. Science 284: 479–482 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takizawa T, Ochiai W, Yanagisawa M, Hisatsune T, Nakafuku M, Miyazono K, Kishimoto T, Kageyama R, Taga T 2001. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc Natl Acad Sci 98: 5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K 2009. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell 16: 245–255 [DOI] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F 2001. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29: 401–413 [DOI] [PubMed] [Google Scholar]
- Nobrega-Pereira S, Kessaris N, Du T, Kimura S, Anderson SA, Marin O 2008. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron 59: 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci 7: 136–144 [DOI] [PubMed] [Google Scholar]
- Ochiai W, Nakatani S, Takahara T, Kainuma M, Masaoka M, Minobe S, Namihira M, Nakashima K, Sakakibara A, Ogawa M, et al. 2009. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci 40: 225–233 [DOI] [PubMed] [Google Scholar]
- Ohata S, Aoki R, Kinoshita S, Yamaguchi M, Tsuruoka-Kinoshita S, Tanaka H, Wada H, Watabe S, Tsuboi T, Masai I, Okamoto H 2011. Dual roles of Notch in regulation of apically restricted mitosis and apicobasal polarity of neuroepithelial cells. Neuron 69: 215–230 [DOI] [PubMed] [Google Scholar]
- O’Leary DD, Chou SJ, Sahara S 2007. Area patterning of the mammalian cortex. Neuron 56: 252–269 [DOI] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F 2002. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev 16: 324–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peljto M, Wichterle H 2011. Programming embryonic stem cells to neuronal subtypes. Curr Opin Neurobiol 21: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker M-W, Tarakhovsky A, Livesey FJ 2010. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci 107: 15957–15962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, et al. 2009. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci 106: 21924–21929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto L, Drechsel D, Schmid MT, Ninkovic J, Irmler M, Brill MS, Restani L, Gianfranceschi L, Cerri C, Weber SN, et al. 2009. AP2γ regulates basal progenitor fate in a region- and layer-specific manner in the developing cortex. Nat Neurosci 12: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, et al. 1997. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 124: 2935–2944 [DOI] [PubMed] [Google Scholar]
- Quinn JC, Molinek M, Martynoga BS, Zaki PA, Faedo A, Bulfone A, Hevner RF, West JD, Price DJ 2007. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev Biol 302: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raballo R, Rhee J, Lyn-Cook R, Leckman JF, Schwartz ML, Vaccarino FM 2000. Basic fibroblast growth factor (Fgf2) is necessary for cell proliferation and neurogenesis in the developing cerebral cortex. J Neurosci 20: 5012–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P 1995. A small step for the cell, a giant leap for mankind: A hypothesis of neocortical expansion during evolution. Trends Neurosci 18: 383–388 [DOI] [PubMed] [Google Scholar]
- Rallu M, Machold R, Gaiano N, Corbin JG, McMahon AP, Fishell G 2002. Dorsoventral patterning is established in the telencephalon of mutants lacking both Gli3 and Hedgehog signaling. Development 129: 4963–4974 [DOI] [PubMed] [Google Scholar]
- Ribes V, Stutzmann F, Bianchetti L, Guillemot F, Dolle P, Le Roux I 2008. Combinatorial signalling controls Neurogenin2 expression at the onset of spinal neurogenesis. Dev Biol 321: 470–481 [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD 2003. Basic helix–loop–helix factors in cortical development. Neuron 39: 13–25 [DOI] [PubMed] [Google Scholar]
- Roy K, Kuznicki K, Wu Q, Sun Z, Bock D, Schutz G, Vranich N, Monaghan AP 2004. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci 24: 8333–8345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL 2011. Development of the cerebral cortex: Implications for neurodevelopmental disorders. J Child Psychol Psychiatry 52: 339–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, O’Leary DDM 2009. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron 63: 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ 2009. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet 5: e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi SP, Murtie J, Koirala S, Patten BA, Corfas G 2006. Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127: 185–197 [DOI] [PubMed] [Google Scholar]
- Scardigli R, Bäumer N, Gruss P, Guillemot F, Le Roux I 2003. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130: 3269–3281 [DOI] [PubMed] [Google Scholar]
- Schmid RS, McGrath B, Berechid BE, Boyles B, Marchionni M, Sestan N, Anton ES 2003. Neuregulin 1–erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex. Proc Natl Acad Sci 100: 4251–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, et al. 2004. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J 23: 2892–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA 2009. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136: 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V 2008. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60: 56–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V 2009. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci 12: 1373–1380 [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S 2006. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci 9: 743–751 [DOI] [PubMed] [Google Scholar]
- Siegenthaler JA, Tremper-Wells BA, Miller MW 2008. Foxg1 haploinsufficiency reduces the population of cortical intermediate progenitor cells: Effect of increased p21 expression. Cereb Cortex 18: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, et al. 2009. Retinoic acid from the meninges regulates cortical neuron generation. Cell 139: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR, Ghosh A 2004. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci 7: 229–235 [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME 2001. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104: 365–376 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell 1: 749–758 [DOI] [PubMed] [Google Scholar]
- Toresson H, Potter SS, Campbell K 2000. Genetic control of dorsal–ventral identity in the telencephalon: Opposing roles for Pax6 and Gsh2. Development 127: 4361–4371 [DOI] [PubMed] [Google Scholar]
- Tuoc TC, Stoykova A 2008. Trim11 modulates the function of neurogenic transcription factor Pax6 through ubiquitin–proteosome system. Genes Dev 22: 1972–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M 2010. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463: 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viti J, Gulacsi A, Lillien L 2003. Wnt regulation of progenitor maturation in the cortex depends on Shh or fibroblast growth factor 2. J Neurosci 23: 5919–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Waclaw RR, Allen ZJ II, Guillemot F, Campbell K 2009. Ascl1 is a required downstream effector of Gsx gene function in the embryonic mouse telencephalon. Neural Dev 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wine-Lee L, Ahn KJ, Richardson RD, Mishina Y, Lyons KM, Crenshaw EB III 2004. Signaling through BMP type 1 receptors is required for development of interneuron cell types in the dorsal spinal cord. Development 131: 5393–5403 [DOI] [PubMed] [Google Scholar]
- Woodhead GJ, Mutch CA, Olson EC, Chenn A 2006. Cell-autonomous β-catenin signaling regulates cortical precursor proliferation. J Neurosci 26: 12620–12630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N 2004. Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J Neurosci 24: 9497–9506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL 2002. Modulation of the Notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development 129: 5029–5040 [DOI] [PubMed] [Google Scholar]
- Zhao X, D’Arca D, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, et al. 2009. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell 17: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C-J, Borello U, Rubenstein JLR, Pleasure SJ 2006. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience 142: 1119–1131 [DOI] [PubMed] [Google Scholar]
- Zhuang B, Sockanathan S 2006. Dorsal–ventral patterning: A view from the top. Curr Opin Neurobiol 16: 20–24 [DOI] [PubMed] [Google Scholar]