Abstract
A protein’s function is intimately linked to its correct subcellular location, yet the machinery required for protein synthesis is predominately cytosolic. How proteins are trafficked through the confines of the cell and integrated into the appropriate cellular compartments has puzzled and intrigued researchers for decades. Indeed, studies exploring this premise revealed elaborate cellular protein translocation and sorting systems, which ensure that all proteins are shuttled to the appropriate cellular destination, where they fulfill their specific functions. This holds true for mitochondria, where sophisticated molecular machines serve to recognize incoming precursor proteins and integrate them into the functional framework of the organelle. We summarize the recent progress in our understanding of mitochondrial protein sorting and the machineries and mechanisms that mediate and regulate this highly dynamic cellular process essential for survival of virtually all eukaryotic cells.
About 99% of mitochondrial proteins are encoded by the nuclear genome and synthesized on cytosolic ribosomes. Mechanisms that target and integrate the proteins into mitochondria may be the most intricate in eukaryotic cells.
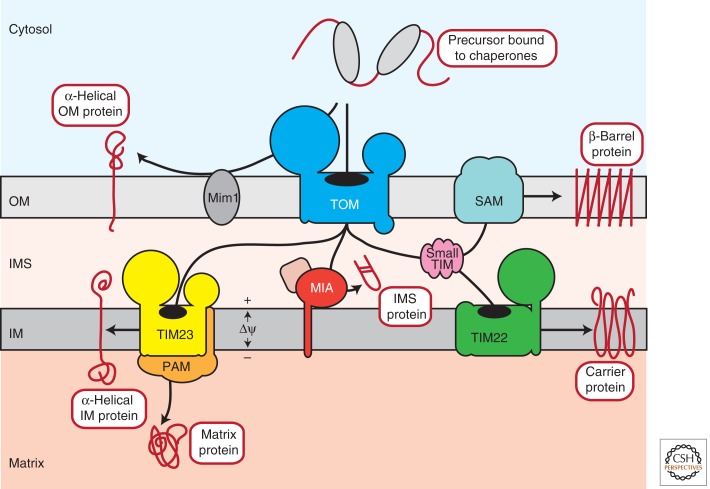
Mitochondria are multifunctional double-membrane-bound organelles that arose from a bacterial endosymbiont during the evolution of eukaryotic cells. Known as the powerhouses of the cell, mitochondria harbor the oxidative phosphorylation machinery for ATP synthesis, but also a large number of biosynthetic pathways. Moreover, they are intimately involved in complex cellular processes, like calcium homeostasis and programmed cell death. As a relic of their evolutionary origin, mitochondria contain their own genetic material and machineries to manufacture their own RNAs and proteins. However, the small circular mitochondrial genome encodes only a few proteins (8 and 13 polypeptides in yeast and humans, respectively). All remaining mitochondrial proteins (approximately 99%) are encoded by the nuclear genome and synthesized on cytosolic ribosomes in their precursor forms. To acquire their mature, functional state these precursor proteins need to be efficiently targeted and imported into mitochondria and sorted to the correct submitochondrial compartment: outer membrane, intermembrane space (IMS), inner membrane, and matrix. The inner mitochondrial membrane is further subdivided into the inner boundary membrane, which is closely opposed to the outer membrane, and large tubular invaginations, termed cristae membranes. Within the four mitochondrial compartments, sophisticated translocation, sorting, and assembly machineries serve to establish incoming precursors in a functional state within the context of their new environment. Advances in the last decade, particularly because of the application of proteomic approaches, have significantly extended the number of components and machineries known to be involved in mitochondrial protein import (Sickmann et al. 2003; Prokisch et al. 2004; Reinders et al. 2006; Pagliarini et al. 2008). These and previous discoveries have provided us with the current framework, which suggests the presence of at least six distinct translocation and assembly machineries within mitochondria (Fig. 1). In this article, we will summarize our current understanding of the machineries for mitochondrial protein import and describe the different molecular mechanisms that execute this essential task.
Figure 1.
Overview of mitochondrial protein sorting pathways. Cytosolic chaperones deliver precursor proteins to the organelle in a translocation-competent state. Some α-helical proteins are inserted into the outer membrane with the help of Mim1. Virtually all other precursors initially traverse the outer membrane via the TOM complex and are subsequently routed to downstream sorting pathways. Biogenesis of outer membrane β-barrel proteins requires the small TIM chaperones of the IMS and the SAM complex. Cysteine-containing IMS proteins are imported via the MIA pathway. Metabolite carriers of the inner mitochondrial membrane are transferred by the small TIM chaperones to the TIM22 complex, which mediates their membrane integration. Presequence-containing precursors are directly taken over from the TOM complex by the TIM23 machinery that either inserts these proteins into the membrane or translocates them into the matrix in cooperation with the import motor PAM. OM, outer membrane; IMS, intermembrane space; IM, inner membrane, Δψ, membrane potential across the inner mitochondrial membrane.
MITOCHONDRIAL PRECURSOR PROTEINS: SYNTHESIS AND TARGETING
It is widely accepted that the vast majority of mitochondrial precursor proteins are imported in a posttranslational manner. To this end, precursor proteins must be kept in an unfolded or loosely folded conformation to allow their passage through tightly gated membrane pores. This is achieved by the binding of cytosolic factors to nascent precursors that stabilize them in a translocation-competent form and guide them to dedicated receptors on the mitochondrial surface (Fig. 1). The best characterized machineries that escort mitochondrial precursor proteins through the cytosol are the Hsp90/p23 and Hsc70/Hsp40 chaperone systems (Young et al. 2003; Bhangoo et al. 2007; Zara et al. 2009). Additional cytosolic proteins, like the arylhydrocarbon receptor-interacting protein (AIP), have been implicated in this process (Yano et al. 2003). However, there is evidence that at least some precursor proteins, like fumarase and Sod2, are imported cotranslationally into mitochondria in vivo (Luk et al. 2005; Yogev et al. 2007). Interestingly, approximately one-half of the mRNAs coding for mitochondrial proteins were reported to be localized to the mitochondrial surface (Marc et al. 2002; Garcia et al. 2007). Puf3, a member of the Pumilio/FBF family of RNA-binding proteins, has been shown to be involved in the targeting of mRNAs to mitochondria (Garcia-Rodriguez et al. 2007; Saint-Georges et al. 2008). A seemingly cotranslational import reaction may in some cases result from the fact that the initiation of precursor translocation is simply faster than the termination of translation for mitochondrial-associated mRNAs. From a physiological perspective, mRNA targeting to mitochondria may allow specific posttranscriptional processes to closely cooperate with the mitochondrial import machinery permitting rapid responses to metabolic changes and/or cellular stimuli (Devaux et al. 2010).
The final destination of a protein in the cell is determined by specific sequence elements within the precursor form that are referred to as signal sequences or targeting signals. These signals are recognized by companion receptors on the surface of the organelles. Mitochondrial targeting signals are categorized in two major classes: (i) “classical” amino-terminal cleavable presequences, and (ii) noncleavable internal import signals (Vögtle et al. 2009). Amino-terminal presequences are the most frequently found import signals and form positively charged amphipathic α-helices, which facilitate a chain of interactions between the precursors and defined import components by gradually increasing affinities (Moczko et al. 1997; Komiya et al. 1998; Marom et al. 2011). Presequences direct precursor proteins to the inner membrane or the matrix and are cleaved on import by the mitochondrial processing peptidase (MPP) (Taylor et al. 2001). Some precursors are further processed at their amino-termini by other proteases, like Oct1 or Icp55 (Naamati et al. 2009; Vögtle et al. 2009, 2011). Other precursor proteins, like cytochrome c1 or cytochrome b2, are initially inserted into the inner membrane by means of a hydrophobic stop-transfer signal downstream of the presequence and subsequently released into the IMS by inner membrane peptidase (IMP) cleavage (Glick et al. 1992; Nunnari et al. 1993). The presence of multiple internal targeting signals was first described for inner membrane proteins of the mitochondrial metabolite carrier family. These import signals largely overlap with the hydrophobic transmembrane segments of the mature carrier proteins (Wiedemann et al. 2001; Brandner et al. 2005). Only recently, membrane proteins that adopt a β-barrel conformation, like Tom40 or porin, were found to contain a specific import signal in the last β-strand of the precursors (β-signal) directing them into the outer mitochondrial membrane (Fig. 2A) (Kutik et al. 2008a). These rather short, evolutionary conserved β-signals are characterized by a bulky polar residue (predominantly lysine or glutamine), followed by an invariant glycine and two large hydrophobic residues. Shortly after the identification of the β-signal, a consensus sequence was identified that targets precursor proteins to the mitochondrial IMS (Milenkovic et al. 2009; Sideris et al. 2009). This mitochondrial IMS sorting signal (MISS) consists of a conserved leucine and valine residue together with a cysteine that forms a transient disulfide bond with the IMS receptor Mia40 (Fig. 3; see below).
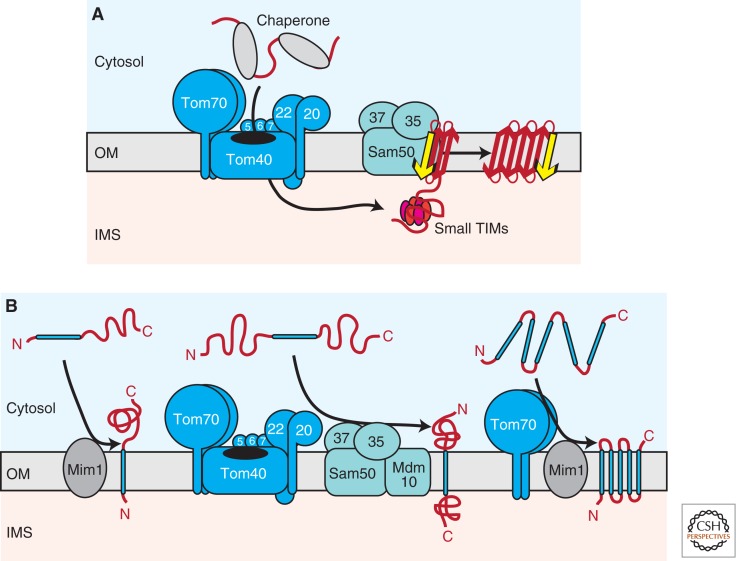
Figure 2.
Biogenesis of outer membrane proteins. (A) Several mitochondrial outer membrane proteins are characterized by a β-barrel topology. On passage of the TOM complex, β-barrel precursors are guided through the aqueous IMS by hexameric small TIM complexes to the SAM complex. A specific import signal (β-signal), which is located within the last β-strand of the precursor, is recognized by the Sam35 receptor. Sam50 forms a central cavity in the SAM complex, where membrane insertion and folding of precursors takes place. (B) Multiple pathways mediate the insertion of α-helical proteins into the outer membrane. Signal-anchored precursors require Mim1 for membrane integration (left). The biogenesis of Tom22, which contains a single central transmembrane segment, depends on both the TOM and SAM–Mdm10 complexes (middle). Polytopic α-helical outer membrane proteins are recognized by the Tom70 receptor and transferred to Mim1 for membrane insertion (right). OM, outer membrane; IMS, intermembrane space; red arrow, β-strand; yellow arrow, β-signal; blue bar, transmembrane α-helix C, carboxy; N, amino.
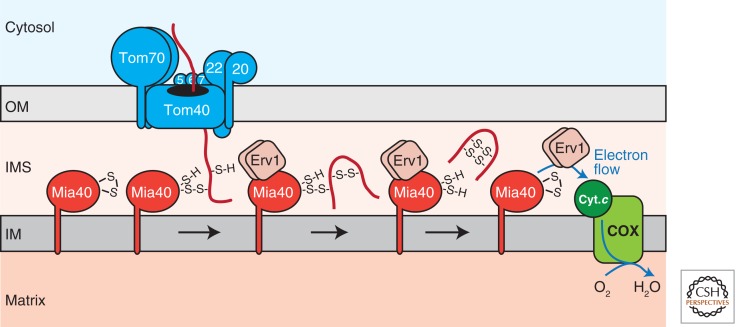
Figure 3.
Import of cysteine-containing precursor proteins into the IMS. The Mia40 receptor forms transient intermolecular disulfide bonds with precursors emerging from the TOM complex. Mia40 functions not only as a receptor, but also as an oxidoreductase by catalyzing the formation of intramolecular disulfide bonds within the precursor together with the sulfhydryl oxidase Erv1. A ternary complex of Mia40, Erv1, and a bound precursor protein allows for the incorporation of multiple disulfide bonds. Erv1 re-oxidizes Mia40 for another round of import and transfers electrons via cytochrome c (Cyt.c) and cytochrome c oxidase (COX) to molecular oxygen. OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
THE TOM COMPLEX: A COMMON JUNCTION IN DISTINCT BIOGENESIS PATHWAYS
Independent of the type of import signal, the general translocase of the outer membrane (TOM complex) is the first line of contact at mitochondria for nearly all precursor proteins (Fig. 1). The central subunit of the TOM complex is the pore-forming β-barrel protein Tom40 (Hill et al. 1998; Künkele et al. 1998; Ahting et al. 1999; Becker et al. 2005). The primary receptors Tom20 and Tom70 are involved in precursor recognition; precursors with amino-terminal targeting signals generally bind to Tom20, whereas those with internal targeting signals preferentially interact with Tom70 (Brix et al. 1997; Abe et al. 2000; Wu and Sha 2006; Saitoh et al. 2007). Tom22 is crucial for the stability of the TOM complex and acts as a central receptor mediating the transfer of precursors from the primary receptor sites to the Tom40 channel (van Wilpe et al. 1999; Shiota et al. 2011). Moreover, the IMS domain of Tom22 contains a trans binding site for amino-terminal presequences (Moczko et al. 1997; Komiya et al. 1998). The small TOM proteins Tom5, Tom6, and Tom7 are involved in the biogenesis, stabilization, and dynamics of the TOM machinery (Dekker et al. 1998; Model et al. 2001; Dukanovic et al. 2009; Becker et al. 2010, 2011a).
In a new twist, it has recently been reported that the function of the TOM complex is extensively regulated by cytosolic protein kinases (Schmidt et al. 2011). It was found that casein kinase 2 phosphorylates the central receptor Tom22 to facilitate its import and interaction with Tom20. Consequently, mutational inactivation of casein kinase 2 leads to defects in the TOM machinery. Additionally, phosphorylation of the Tom70 receptor by protein kinase A was shown to negatively regulate the import of metabolite carrier proteins in response to metabolic changes in the cell (Schmidt et al. 2011). These findings support the notion that the mitochondrial import machinery is not just a static entity, but rather is integrated into and regulated by cellular signaling networks that govern organelle biogenesis.
Once at the TOM complex, the majority of precursors are translocated across the outer membrane into the IMS, where they are transferred over to specialized protein sorting machineries of other submitochondrial compartments (Fig. 1). The mechanisms that allow for the initial translocation of several different classes of precursors through the TOM complex, and subsequent selective sorting into distinct downstream protein biogenesis pathways remain poorly understood.
DIVERSE PATHWAYS FOR TRAFFICKING TO THE MITOCHONDRIAL OUTER MEMBRANE
From a topological perspective, the outer membrane of mitochondria is an intriguing environment because it contains both β-barrel proteins and proteins that span the lipid bilayer with single or multiple α-helical transmembrane domains. Thus, the integrity of this subcompartment is upheld by the coexistence of multiple biogenesis pathways, which are likely acting in concert with each other. The presence of β-barrel proteins in the outer membranes of mitochondria and chloroplasts reflects the bacterial origin of these organelles (Dolezal et al. 2006). The basic mechanisms of β-barrel biogenesis appear to be conserved in evolution. BamA (Omp85, YaeT), the central component of the bacterial β-barrel assembly machinery (Knowles et al. 2009), is homologous to Sam50, the essential pore-forming component of the outer membrane sorting and assembly machinery (SAM complex; also termed TOB complex), which mediates the membrane insertion and folding of β-barrel proteins in mitochondria (Kozjak et al. 2003; Paschen et al. 2003; Gentle et al. 2004). Remarkably, the SAM complex is able to insert recombinantly expressed bacterial β-barrel proteins into the outer mitochondrial membrane (Walther et al. 2009). The SAM complex contains two additional peripheral membrane proteins, Sam35, which is essential for cell growth and the nonessential Sam37 (Wiedemann et al. 2003; Ishikawa et al. 2004; Milenkovic et al. 2004; Waizenegger et al. 2004). Mitochondrial β-barrel precursors are initially translocated across the outer membrane via the TOM complex (Fig. 2A). As β-barrel precursors emerge at the trans side of the TOM complex they are met by members of the small TIM chaperone family (Tim9/Tim10; Tim8/Tim13), which likely trap the β-barrel precursors in the IMS and shield the unfolded proteins from this aqueous environment on their way to the SAM machinery (Hoppins and Nargang 2004; Wiedemann et al. 2004). The molecular mechanism of precursor transfer from the TOM complex to the TIM chaperones and further to the SAM complex awaits to be elucidated. Either a concerted hand-over mechanism or the existence of soluble IMS intermediates appears conceivable. There is good evidence that on passage through the IMS, the aforementioned β-signal directs insertion of the precursor into the SAM complex by binding to the receptor Sam35 (Fig. 2A) (Kutik et al. 2008a). Complex formation of Sam35 and Sam50 with the β-signal is believed to initiate the entry of the precursor into a hydrophilic and proteinaceous membrane environment, which then accommodates β-barrel folding and insertion into the lipid phase (Kutik et al. 2008a). The physical nature of this membrane-embedded protein folding chamber is still under debate. Current models suggest that it may be formed mainly by a Sam50 oligomer (Paschen et al. 2003; Kutik et al. 2008a). Finally, both Sam37 and the amino-terminal IMS domain of Sam50 are involved in the release of the β-barrel precursor into the outer membrane (Chan and Lithgow 2008; Stroud et al. 2011a).
Several other outer membrane proteins participate in the assembly of β-barrel proteins. Mdm10 and Mim1 were found to interact with the SAM complex and are differentially required for the assembly of TOM complex components (Meisinger et al. 2004; Waizenegger et al. 2005; Becker et al. 2008). Mdm10 is also found in the endoplasmic reticulum-mitochondria encounter structure (ERMES), also known as mitochondrial distribution and morphology (MDM) complex, which contains four additional proteins: Mdm12, Mmm1, Mdm34, and the Miro GTPase Gem1 (Boldogh et al. 2003; Kornmann et al. 2011; Stroud et al. 2011b). ERMES was shown to tether the mitochondrial outer membrane to the endoplasmic reticulum (Kornmann et al. 2009). Anchoring of ERMES to the ER is mediated by the amino-terminal transmembrane segment of Mmm1 (Kornmann et al. 2009; Stroud et al. 2011b). ERMES was proposed to facilitate the exchange of phospholipids and calcium between the ER and mitochondria (Kornmann et al. 2009; Kornmann and Walter 2010; Gebert et al. 2011a). Interestingly, inactivation of ERMES proteins also affects β-barrel protein assembly (Meisinger et al. 2007; Wideman et al. 2010). The suggested role in lipid transport provides a possible explanation for the severe mitochondrial morphology defects observed with the loss of ERMES proteins (Boldogh et al. 2003; Meisinger et al. 2004, 2007). However, the true function of ERMES in the biogenesis of β-barrel proteins has not yet been clarified. The available data raise the intriguing possibility that lipid transfer from the ER to mitochondria and the biogenesis of mitochondrial β-barrel proteins may be coupled processes. It seems likely that the dually localized Mdm10 protein links the different roles of ERMES. Interestingly, the small TOM protein Tom7 was shown to regulate the dynamic interaction of Mdm10 with the SAM and ERMES complexes (Meisinger et al. 2006; Yamano et al. 2010; Becker et al. 2011a).
For proteins anchored to the outer membrane via α-helical regions, transmembrane domains are located at either the amino- (signal anchor) or carboxyl-terminus (tail anchor). Other α-helical outer membrane proteins span the lipid bilayer multiple times. There is a striking variety in the mechanisms adopted for their insertion (Fig. 2B). Membrane integration of signal-anchored proteins, such as Tom20 and Tom70, is facilitated by the outer membrane protein Mim1 (Becker et al. 2008; Hulett et al. 2008; Popov-Čeleketić et al. 2008a). However, a Mim1 counterpart for tail-anchored proteins has not yet been discovered, but rather the lipid composition of the outer membrane appears be crucial for specific insertion of these proteins (Setoguchi et al. 2006; Kemper et al. 2008). The precursor of Tom22, which contains one central transmembrane α-helix, requires both the TOM complex and the SAM–Mdm10 complex for its biogenesis (Fig. 2B) (Stojanovski et al. 2007; Thornton et al. 2010). This finding revealed that the substrate spectrum of the SAM complex is not limited to β-barrel precursors, but also includes specific α-helical outer membrane proteins. Finally, multi-spanning proteins, like the membrane fusion protein Ugo1, are assisted by the Tom70 receptor (but no other TOM components) and the Mim1 protein for their insertion into the outer membrane (Fig. 2B) (Otera et al. 2007; Becker et al. 2011b; Papic et al. 2011).
REDOX-DRIVEN PROTEIN IMPORT INTO THE MITOCHONDRIAL INTERMEMBRANE SPACE
Many IMS proteins are small in size (<20 kDa) and contain conserved cysteine residues in Cx3C or Cx9C motives that are involved in binding of cofactors and metals or the formation of disulfide bonds. The best characterized proteins of this kind are the small TIM chaperones that form hetero-oligomeric complexes with each subunit containing two disulfide bonds (Webb et al. 2006). Import of these proteins into mitochondria is mediated by the mitochondrial IMS import and assembly (MIA) machinery (Fig. 3). The MIA pathway involves two essential conserved core components, the import receptor Mia40 and the sulfhydryl oxidase Erv1 (ALR in humans) (Chacinska et al. 2004; Naoé et al. 2004; Mesecke et al. 2005). Mia40 is firmly attached to the inner mitochondrial membrane by an amino-terminal membrane anchor in yeast, whereas human Mia40 is a soluble IMS protein (Chacinska et al. 2008; Banci et al. 2009). Structural analysis has revealed that client proteins associate with Mia40 via a hydrophobic cleft on the surface and a redox-active CPC motif of Mia40 (Banci et al. 2009; Kawano et al. 2009). Polypeptide segments of both, Mia40 itself and Erv1/ALR, have been suggested to occupy the hydrophobic substrate binding site of Mia40 in the absence of a precursor (Banci et al. 2009; Kawano et al. 2009). Docking of substrate to Mia40 facilitates folding of the precursor and generation of a transient precursor-receptor disulfide intermediate with a cysteine residue in the precursor’s import signal (Fig. 3) (Grumbt et al. 2007; Milenkovic et al. 2007, 2009; Sideris and Tokatlidis 2007; Sideris et al. 2009; Banci et al. 2010). Thus, Mia40 behaves as an oxidoreductase donating disulfide bonds to incoming precursors thereby trapping them in the IMS. This reaction involves the removal of electrons from the precursor and reduction of cysteine residues in Mia40. Analagous to other disulfide-generating systems, the sulfhydryl oxidase Erv1 reoxidizes Mia40 for further rounds of the reaction cycle (Fig. 3) (Mesecke et al. 2005; Stojanovski et al. 2008; Banci et al. 2011). Erv1 acts as a homo-dimer, and an intersubunit electron exchange mechanism is involved in the redox reaction (Bien et al. 2010). A role for the small redox-active peptide glutathione in proofreading of Mia40/Erv1-generated disulfide bonds has been suggested (Bien et al. 2010). For its own reoxidation, Erv1 transfers electrons via cytochrome c to the respiratory chain and, hence, molecular oxygen (Fig. 3) (Bihlmaier et al. 2007; Dabir et al. 2007). Interestingly, Erv1 was found in a ternary complex with Mia40 and a precursor protein. This direct association may facilitate the acquisition of multiple disulfides through a channeling mechanism, which does not require continual association and dissociation of the components (Stojanovski et al. 2008; Banci et al. 2011).
A recent study by von der Malsburg et al. (2011) revealed that mitofilin/Fcj1, an inner membrane protein required for the maintenance of inner membrane architecture, is involved in the MIA pathway. Mitofilin is part of the mitochondrial inner membrane organizing system (MINOS; also termed MICOS or MitOS), a large inner membrane protein complex that is crucial for the morphology of cristae membranes and involved in multiple contact sites between inner and outer mitochondrial membranes (Harner et al. 2011; Hoppins et al. 2011; von der Malsburg et al. 2011). Mitofilin associates with both the TOM machinery in the outer membrane and Mia40, thereby recruiting Mia40 into the proximity of incoming client proteins, as soon as they emerge from the Tom40 channel.
Insertion of Metabolite Carriers into the Inner Membrane via the TIM22 Complex
The large family of mitochondrial inner membrane metabolite carriers, such as the ADP/ATP carriers (AAC), functions in the transfer of metabolites between the mitochondrial matrix and IMS. Metabolite carriers as well as a few other integral membrane proteins with internal import signals, like the Tim23 protein, are inserted into the inner membrane by the translocase of the inner membrane 22 (TIM22 complex; carrier translocase) (Fig. 4). The TIM22 machinery consists of four membrane-integral components, Tim22, Tim54, Tim18, and Sdh3, and the peripherally associated IMS chaperones Tim9, Tim10, and Tim12 (Sirrenberg et al. 1996, 1998; Kerscher et al. 1997, 2000; Koehler et al. 1998a,b, 2000; Gebert et al. 2011b). Tim22 forms a channel in the inner membrane for precursor insertion (Rehling et al. 2003), whereas Tim54 exposes a large domain to the IMS and is believed to provide a binding site for the Tim9–Tim10–Tim12 complex (Fig. 4) (Gebert et al. 2008; Wagner et al. 2008). Tim18 facilitates the assembly of Tim54 with Tim22 (Wagner et al. 2008). A recent study reported on the surprising finding that the integral inner membrane protein Sdh3, which has been known as a subunit of respiratory chain complex II (succinate dehydrogenase, SDH complex), is also a genuine subunit of the TIM22 complex. Sdh3 forms a subcomplex with Tim18, which is homologous to Sdh4, the binding partner of Sdh3 in the SDH complex (Gebert et al. 2011b). This finding is interesting from the evolutionary perspective, as it exemplifies how certain protein modules have been used to build different membrane protein machineries in mitochondria.
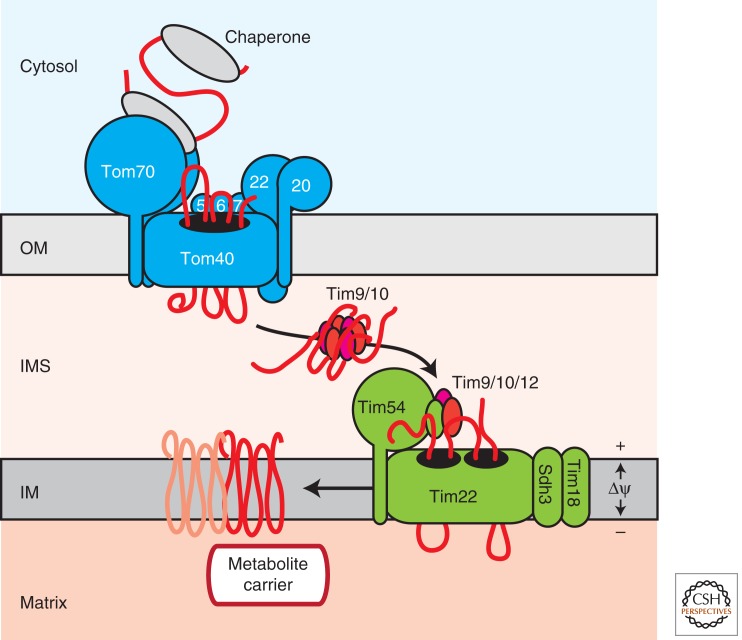
Figure 4.
Insertion of metabolite carriers into the inner mitochondrial membrane. The Tom70 receptor takes over metabolite carrier precursors from cytosolic chaperones. The precursors pass the outer membrane in a loop conformation. In the IMS, the hydrophobic transmembrane segments of carriers are shielded by the Tim9/Tim10 chaperone complex, which delivers the precursor to the TIM22 machinery in the inner membrane. Tim54 and the adaptor protein Tim12 transfer the precursor to the protein-conducting channel formed by Tim22. Tim18, and Sdh3 are required for assembly and stability of the translocase. The membrane potential across the inner mitochondrial membrane (Δψ) drives the insertion of metabolite carriers, which finally adopt their functional, dimeric state. OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
Cytosolic chaperones deliver carrier precursors to the Tom70 receptor in an ATP-dependent process (Wiedemann et al. 2001; Young et al. 2003; Bhangoo et al. 2007; Zara et al. 2009). They pass the TOM complex in a loop-like conformation and are bound in the IMS by the small TIM chaperones (Fig. 4). The hexameric Tim9/Tim10 and Tim8/Tim13 complexes transport these hydrophobic substrates across the aqueous IMS and deliver them to the inner membrane (Curran et al. 2002; Webb et al. 2006; Baker et al. 2009). Biochemical data suggest that the Tim9/Tim10 and Tim8/Tim13 complexes bind to the hydrophobic segments of the model substrates AAC and Tim23, respectively, concomitant with their chaperone-like role (Curran et al. 2002; Vergnolle et al. 2005; Davis et al. 2007). The crystal structure of the Tim9/Tim10 complex revealed an α-propeller topology with helical blades pointing away from a central opening that is lined by polar residues—a similar domain arrangement as for the chaperones Skp and prefoldin (Webb et al. 2006). With the help of the small TIM protein Tim12, the precursor-loaded chaperone complex docks to the membrane-embedded core of the TIM22 machinery, where the precursor is integrated into the inner membrane again by a looping mechanism (Fig. 4) (Rehling et al. 2003; Gebert et al. 2008). Membrane-inserted metabolite carriers subsequently assemble into their mature dimeric form. The only required energy source for the insertion process is the membrane potential (Δψ) across the inner mitochondrial membrane.
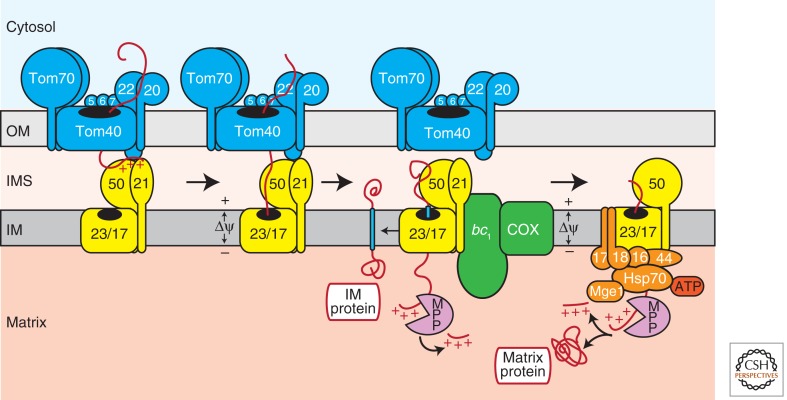
INNER MEMBRANE INSERTION AND MATRIX TRANSLOCATION BY THE TIM23 MACHINERY
Precursor proteins with amino-terminal presequences (preproteins) are handed over from the TOM complex in the outer membrane to the translocase of the inner membrane 23 (TIM23 complex; presequence translocase). This process does not involve soluble IMS stages, but leads to the formation of two-membrane-spanning transport intermediates (TOM–TIM23 supercomplexes), in which the amino-terminal portion of the preprotein has been inserted into the TIM23 complex, whereas the carboxy-terminal domain is still inside the TOM complex (Dekker et al. 1997; Chacinska et al. 2003). The essential membrane-embedded core of the TIM23 complex (TIM23CORE) is composed of three subunits (Fig. 5): the channel-forming Tim23 protein (Meinecke et al. 2006; van der Laan et al. 2007; Alder et al. 2008); Tim17, which is involved in gating of the Tim23 channel and membrane insertion of preproteins (Chacinska et al. 2005; Meier et al. 2005) and Tim50, which induces the closing of the Tim23 protein-conducting channel in the absence of preproteins (Meinecke et al. 2006). Tim50 exposes a large domain to the IMS, which is in close proximity to the outer membrane TOM complex, and together with the amino-terminal IMS domain of Tim23 constitutes the receptor domain of the TIM23 machinery (Gevorkyan-Airapetov et al. 2009; Mokranjac et al. 2009; Tamura et al. 2009a; Marom et al. 2011; Qian et al. 2011; Shiota et al. 2011). Two recent studies have indicated that Tim50 might have two distinct binding sites for preproteins in the conserved core domain and in the carboxy-terminal domain, respectively (Qian et al. 2011; Schulz et al. 2011). The Tim21 protein connects with TIM23CORE and promotes preprotein transfer from the TOM to the TIM23 complex, probably by directly interacting with the IMS domain of Tom22 (Fig. 5) (Chacinska et al. 2005; Albrecht et al. 2006).
Figure 5.
Import of presequence-containing precursor proteins (preproteins) into the inner membrane and matrix. Amino-terminal positively charged presequences are preferentially recognized by the Tom20 receptor on the mitochondrial surface. On the trans side of the protein-conducting Tom40 channel, presequences are bound by the IMS domain of Tom22. Membrane potential (Δψ)-dependent transfer of preproteins to the TIM23 machinery in the inner membrane involves Tim50 and Tim21. Preproteins with a hydrophobic stop-transfer signal (blue bar) are directly released from the protein-conducting channel formed by Tim23 into the inner membrane. This Δψ-dependent process is supported by the Tim21-dependent coupling of respiratory chain supercomplexes to TIM23. Stepwise translocation of soluble preproteins into the matrix requires the dynamic coupling of the ATP-driven mtHsp70-containing import motor, which leads to a loss of Tim21 from the TIM23 complex. Finally, the matrix processing peptidase (MPP) proteolytically removes presequences from both inner membrane and matrix preproteins. OM, outer membrane; IMS, intermembrane space; IM, inner membrane; bc1, cytochrome bc1 complex (complex III); COX, cytochrome c oxidase (complex IV).
The TIM23CORE-Tim21 complex has been termed TIM23SORT, as this complex is sufficient to integrate preproteins with a hydrophobic stop-transfer signal adjacent to the amino-terminal presequence into the inner membrane in a Δψ-dependent manner (van der Laan et al. 2007). Remarkably, TIM23SORT is dynamically coupled to respiratory chain supercomplexes consisting of complex III (cytochrome bc1 complex) and complex IV (cytochrome c oxidase), most likely via a direct interaction between Tim21 and the complex III subunit Qcr6 (Fig. 5) (van der Laan et al. 2006, 2007; Wiedemann et al. 2007; Saddar and Stuart 2008). Association of respiratory chain complexes to TIM23 was shown to stimulate the Δψ-dependent step of preprotein membrane insertion, which appears to be particularly important under conditions, where the overall Δψ across the inner membrane becomes limiting (van der Laan et al. 2006).
For complete import of precursors into the mitochondrial matrix the energy derived from Δψ in not sufficient. Therefore, the TIM23 complex has to be coupled to the ATP-driven presequence translocase-associated import motor (PAM) for matrix translocation (Fig. 5). The central subunit of PAM is the mitochondrial heat shock protein 70 (mtHsp70) that converts the energy from repeated cycles of ATP hydrolysis into a matrix-directed vectorial movement of the preprotein (Neupert and Herrmann 2007; Mapa et al. 2010). This activity of mtHsp70 is regulated by the multiple membrane-bound co-chaperones of PAM: The J-protein Pam18 stimulates the ATPase activity of mtHsp70 (D’Silva et al. 2003; Mokranjac et al. 2003; Truscott et al. 2003). The J-like protein Pam16 associates with Pam18 in the J-complex, recruits Pam18 to TIM23CORE, and regulates its ATPase-stimulating function (Frazier et al. 2004; Kozany et al. 2004; Li et al. 2004; Mokranjac et al. 2006; D’Silva et al. 2008, Pais et al. 2011). Tim44 is crucial for the binding of both, mtHsp70 and J-complex to TIM23CORE (D’Silva et al. 2004; Hutu et al. 2008; Schiller et al. 2008). The matrix-localized nucleotide exchange factor Mge1 is required for the completion of the mtHsp70 reaction cycle (Slutsky-Leiderman et al. 2007). Pam17 is thought to trigger the dynamic assembly of PAM (van der Laan et al. 2005; Hutu et al. 2008). The stepwise recruitment of PAM components goes along with a release of Tim21 from TIM23CORE (Chacinska et al. 2005; van der Laan et al. 2005; Popov-Čeleketić et al. 2008b). Thus, the majority of TIM23CORE complexes in mitochondria are either associated with Tim21 or PAM (Fig. 5) (Chacinska et al. 2005, 2010; van der Laan et al. 2005, 2006).
The picture of the TIM23 complex emerging from these findings is that of a highly dynamic molecular machine that switches between different functional and structural states depending on the signal information within an incoming preprotein. This view is best exemplified by the recently unraveled biogenesis pathway of the mitochondrial ATP-binding cassette (ABC) transporter Mdl1, a polytopic inner membrane protein with an amino-terminal presequence (Bohnert et al. 2010). Earlier studies suggested that polytopic inner membrane proteins are first fully translocated across the membrane in a PAM-dependent manner and afterwards integrated into the bilayer from the matrix side by the oxidase assembly (OXA) complex, a membrane protein insertase of bacterial origin that also mediates the biogenesis of mitochondrially encoded respiratory chain subunits (Neupert and Herrmann 2007). However, membrane insertion of Mdl1 was found to be a modular process, in which certain transmembrane segment are laterally released from the TIM23 complex independently of PAM, whereas other transmembrane segments are routed to the inner membrane via the matrix and the OXA machinery (Bohnert et al. 2010). Such a complex import mechanism requires multiple functional switches and modular rearrangements within the TIM23 complex that must be tightly controlled by specific import signals and partner proteins of TIM23.
CONCLUSIONS AND PERSPECTIVES
Protein trafficking to and within mitochondria is possibly the most intricate in the context of a eukaryotic cell (the same would apply to chloroplasts). Indeed, it is fascinating how nature has answered the difficult question of protein sorting within mitochondria by providing elaborate transport systems to ensure that precursor proteins acquire their mature and functional state. Future perspectives for the field include addressing the role of phospholipids in protein transport processes. Recent studies have indicated that cardiolipin is important for the stability and functionality of not only respiratory chain complexes, but also protein translocation devices, like the TIM23/PAM machinery and the TOM and SAM complexes (Kutik et al. 2008b; Gebert et al. 2009; Tamura et al. 2009b). Certainly, high-resolution structures of the membrane-embedded core domains of translocation complexes will be invaluable for learning more about the organization and operation of these machineries. More information is also required on how the import systems are regulated and how the effectiveness of import is promoted through specific coupling of translocation machineries. Recent insights into the role of cytosolic kinases for the assembly and activity of the TOM complex and the cooperation of the TOM machinery with downstream protein sorting pathways have highlighted the central importance of this aspect (Chacinska et al. 2010; Schmidt et al. 2011; von der Malsburg et al. 2011). Despite many efforts, we still do not fully understand how specific import signals in precursor proteins are recognized by receptor domains and protein-conducting channels and how these signals are transmitted to induce large-scale rearrangements and functional switches in the mitochondrial protein import systems. Finally, an exciting link between protein import and machineries implicated in the regulation of mitochondrial morphology has emerged, which needs to be further explored (Meisinger et al. 2007; Kornmann et al. 2010; von der Malsburg et al. 2011).
In this article, we have highlighted our current understanding of mitochondrial import based on research that mainly utilized fungal models. It is undeniable that this field will continue to rely on such models; however, the crucial importance of understanding these processes in mammalian cells should be emphasized. Despite a sound knowledge of the protein import machinery in mammalian cells, there has so far been little focus on how problems in mitochondrial protein import can contribute to human disease, although evidence supporting such connections is mounting. For instance, Mohr-Tranebjaerg syndrome (MTS) is caused by mutations in the small TIM chaperone deafness dystonia polypeptide 1 (DDP1/TIMM8/Tim8) (Koehler et al. 1999). The underlying severity of the disease may be attributable to impaired biogenesis of the TIM23 complex as a result of dysfunctional Tim8. Defects in Pam18 (DNAJC19) have been linked to dilated cardiomyopathy with ataxia (DCMA) (Davey et al. 2006; Sinha et al. 2010). TIMM17A mRNA expression was recently shown to be significantly higher in breast cancer compared with normal or benign breast tissue (Salhab et al. 2010). Moreover, TIM17 was identified as a multicopy suppressor of mtDNA instability in specific mutant backgrounds in yeast, which may be relevant for certain forms of mtDNA depletion syndrome (Iacovino et al. 2009). Hsp60, a central player in protein folding and assembly of mitochondria, has been implicated in spastic paraplegia-13 (SPG13), an autosomally dominant form of pure hereditary spastic paraplegia (Fontaine et al. 2000; Hansen et al. 2002). These observations reiterate the significance of a functional mitochondrial protein biogenesis network for health and disease and the importance of gaining a greater understanding of the import machinery in higher eukaryotes.
ACKNOWLEDGMENTS
This work is supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, Excellence Initiative of the German Federal and State Governments (EXC 294), Gottfried Wilhelm Leibniz Program, the Fonds der Chemischen Industrie, the Australian Research Council, and the Clive and Vera Ramaciotti Foundation.
Footnotes
Editors: Douglas C. Wallace and Richard J. Youle
Additional Perspectives on Mitochondria available at www.cshperspectives.org
REFERENCES
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D 2000. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100: 551–560 [DOI] [PubMed] [Google Scholar]
- Ahting U, Thun C, Hegerl R, Typke D, Nargang FE, Neupert W, Nussberger S 1999. The TOM core complex: The general protein import pore of the outer membrane of mitochondria. J Cell Biol 147: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht R, Rehling P, Chacinska A, Brix J, Cadamuro SA, Volkmer R, Guiard B, Pfanner N, Zeth K 2006. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep 7: 1233–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder NN, Jensen RE, Johnson AE 2008. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 134: 439–450 [DOI] [PubMed] [Google Scholar]
- Baker MJ, Webb CT, Stroud DA, Palmer CS, Frazier AE, Guiard B, Chacinska A, Gulbis JM, Ryan MT 2009. Structural and functional requirements for activity of the Tim9-Tim10 complex in mitochondrial protein import. Mol Biol Cell 20: 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K 2009. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol 16: 198–206 [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Cenacchi L, Ciofi-Baffoni S, Felli IC, Gallo A, Gonnelli L, Luchinat E, Sideris D, et al. 2010. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc Natl Acad Sci 107: 20190–20195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Calderone V, Cefaro C, Ciofi-Baffoni S, Gallo A, Kallergi E, Lionaki E, Pozidis C, Tokatlidis K 2011. Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc Natl Acad Sci 108: 4811–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker L, Bannwarth M, Meisinger C, Hill K, Model K, Krimmer T, Casadio R, Truscott KN, Schulz GE, Pfanner N, et al. 2005. Preprotein translocase of the outer mitochondrial membrane: Reconstituted Tom40 forms a characteristic TOM pore. J Mol Biol 353: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Becker T, Pfannschmidt S, Guiard B, Stojanovski D, Milenkovic D, Kutik S, Pfanner N, Meisinger C, Wiedemann N 2008. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J Biol Chem 283: 120–127 [DOI] [PubMed] [Google Scholar]
- Becker T, Guiard B, Thornton N, Zufall N, Stroud DA, Wiedemann N, Pfanner N 2010. Assembly of the mitochondrial protein import channel: Role of Tom5 in two-stage interaction of Tom40 with the SAM complex. Mol Biol Cell 21: 3106–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Thornton N, Stroud D, Meisinger C, Wiedemann N, Pfanner N 2011a. Biogenesis of mitochondria: Dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J Mol Biol 405: 113–124 [DOI] [PubMed] [Google Scholar]
- Becker T, Wenz LS, Krüger V, Lehmann W, Müller JM, Goroncy L, Zufall N, Lithgow T, Guiard B, Chacinska A, et al. 2011b. The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J Cell Biol 194: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhangoo MK, Tzankov S, Fan AC, Dejgaard K, Thomas DY, Young JC 2007. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol Biol Cell 18: 3414–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J 2010. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol Cell 37: 516–528 [DOI] [PubMed] [Google Scholar]
- Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM 2007. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol 179: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert M, Rehling P, Guiard B, Herrmann JM, Pfanner N, van der Laan M 2010. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol 20: 1227–1232 [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell 14: 4618–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K, Rehling P, Truscott KN 2005. The carboxyl-terminal third of the dicarboxylate carrier is crucial for productive association with the inner membrane twin-pore translocase. J Biol Chem 280: 6215–6221 [DOI] [PubMed] [Google Scholar]
- Brix J, Dietmeier K, Pfanner N 1997. Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22, and Tom70. J Biol Chem 272: 20730–20735 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Rehling P, Guiard B, Frazier AE, Schulze-Specking A, Pfanner N, Voos W, Meisinger C 2003. Mitochondrial translocation contact sites: Separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J 22: 5370–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuán Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N 2004. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 23: 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, et al. 2005. Mitochondrial presequence translocase: Switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Guiard B, Müller JM, Schulze-Specking A, Gabriel K, Kutik S, Pfanner N 2008. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J Biol Chem 283: 29723–29729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, van der Laan M, Mehnert CS, Guiard B, Mick DU, Hutu DP, Truscott KN, Wiedemann N, Meisinger C, Pfanner N, et al. 2010. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol 30: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Lithgow T 2008. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell 19: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran SP, Leuenberger D, Oppliger W, Koehler CM 2002. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J 21: 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM 2007. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J 26: 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey KM, Parboosingh JS, McLeod DR, Chan A, Casey R, Ferreira P, Snyder FF, Bridge PJ, Bernier FP 2006. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet 43: 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Alder NN, Jensen RE, Johnson AE 2007. The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochondrial protein import. Mol Biol Cell 18: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Martin F, Maarse AC, Bömer U, Müller H, Guiard B, Meijer M, Rassow J, Pfanner N 1997. The Tim core complex defines the number of mitochondrial translocation sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J 16: 5408–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Ryan MT, Brix J, Müller H, Hönlinger A, Pfanner N 1998. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol 18: 6515–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux F, Lelandais G, Garcia M, Goussard S, Jacq C 2010. Posttranscriptional control of mitochondrial biogenesis: Spatio-temporal regulation of the protein import process. FEBS Lett 584: 4273–4279 [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313: 314–318 [DOI] [PubMed] [Google Scholar]
- D’Silva PD, Schilke B, Walter W, Andrew A, Craig EA 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci 100: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Silva P, Liu Q, Walter W, Craig EA 2004. Regulated interactions of mtHsp70 with Tim44 at the translocon in the mitochondrial inner membrane. Nat Struct Mol Biol 11: 1084–1091 [DOI] [PubMed] [Google Scholar]
- D’Silva PR, Schilke B, Hayashi M, Craig EA 2008. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol Biol Cell 19: 424–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukanovic J, Dimmer KS, Bonnefoy N, Krumpe K, Rapaport D 2009. Genetic and functional interactions between the mitochondrial outer membrane proteins Tom6 and Sam37. Mol Cell Biol 29: 5975–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine B, Davoine CS, Durr A, Paternotte C, Feki I, Weissenbach J, Hazan J, Brice A 2000. A new locus for autosomal dominant pure spastic paraplegia, on chromosome 2q24-q34. Am J Hum Genet 66: 702–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, et al. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat Struct Mol Biol 11: 226–233 [DOI] [PubMed] [Google Scholar]
- Garcia M, Darzacq X, Delaveau T, Jourdren L, Singer RH, Jacq C 2007. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol Biol Cell 18: 362–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rodriguez LJ, Gay AC, Pon LA 2007. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol 176: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Chacinska A, Wagner K, Guiard B, Koehler CM, Rehling P, Pfanner N, Wiedemann N 2008. Assembly of the three small Tim proteins precedes docking to the mitochondrial carrier translocase. EMBO Rep 9: 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, et al. 2009. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: Implications for Barth syndrome. Curr Biol 19: 2133–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Ryan MT, Pfanner N, Wiedemann N, Stojanovski D 2011a. Mitochondrial protein import machineries and lipids: A functional connection. Biochim Biophys Acta 1808: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Gebert N, Gebert M, Oeljeklaus S, von der Malsburg K, Stroud DA, Kulawiak B, Wirth C, Zahedi RP, Dolezal P, Wiese S, et al. 2011b. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell 44: 811–818 [DOI] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevorkyan-Airapetov L, Zohary K, Popov-Čeleketić D, Mapa K, Hell K, Neupert W, Azem A, Mokranjac D 2009. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J Biol Chem 284: 4865–4872 [DOI] [PubMed] [Google Scholar]
- Glick BS, Brandt A, Cunningham K, Müller S, Hallberg RL, Schatz G 1992. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69: 809–822 [DOI] [PubMed] [Google Scholar]
- Grumbt B, Stroobant V, Terziyska N, Israel L, Hell K 2007. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J Biol Chem 282: 37461–37470 [DOI] [PubMed] [Google Scholar]
- Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, et al. 2002. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet 70: 1328–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Körner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W 2011. The mitochondrial contact site complex, a determinant of mitochondrial ultrastructure. EMBO J 30: 4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Model K, Ryan MT, Dietmeier K, Martin F, Wagner R, Pfanner N 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395: 516–521 [DOI] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE 2004. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem 279: 12396–12405 [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J 2011. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol 195: 323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett JM, Lueder F, Chan NC, Perry AJ, Wolynec P, Likic VA, Gooley PR, Lithgow T 2008. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J Mol Biol 376: 694–704 [DOI] [PubMed] [Google Scholar]
- Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, van der Laan M 2008. Mitochondrial protein import motor: Differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol Biol Cell 19: 2642–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovino M, Granycome C, Sembongi H, Bokori-Brown M, Butow RA, Holt IJ, Bateman JM 2009. The conserved translocase Tim17 prevents mitochondrial DNA loss. Hum Mol Genet 18: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol 166: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Yamano K, Naoé M, Momose T, Terao K, Nishikawa S, Watanabe N, Endo T 2009. Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc Natl Acad Sci 106: 14403–14407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper C, Habib SJ, Engl G, Heckmeyer P, Dimmer KS, Rapaport D 2008. Integration of tail-anchored proteins into the mitochondrial outer membrane does not require any known import components. J Cell Sci 121: 1990–1998 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Holder J, Srinivasan M, Leung RS, Jensen RE 1997. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J Cell Biol 139: 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Sepuri NB, Jensen RE 2000. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol Biol Cell 11: 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR 2009. Membrane protein architecture: the role of the BAM complex in outer membrane protein assembly. Nat Rev Microbiol 7: 206–214 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G 1998a. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science 279: 369–373 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Merchant S, Oppliger W, Schmid K, Jarosch E, Dolfini L, Junne T, Schatz G, Tokatlidis K 1998b. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J 17: 6477–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Leuenberger D, Merchant S, Renold A, Junne T, Schatz G 1999. Human deafness dystonia syndrome is a mitochondrial disease. Proc Natl Acad Sci 96: 2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler CM, Murphy MP, Bally NA, Leuenberger D, Oppliger W, Dolfini L, Junne T, Schatz G, Or E 2000. Tim18p, a new subunit of the TIM22 complex that mediates insertion of imported proteins into the yeast mitochondrial inner membrane. Mol Cell Biol 20: 1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya T, Rospert S, Koehler C, Looser R, Schatz G, Mihara K 1998. Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: Evidence for the “acid chain” hypothesis. EMBO J 17: 3886–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Walter P 2010. ERMES-mediated ER-mitochondria contacts: Molecular hubs for the regulation of mitochondrial biology. J Cell Sci 123: 1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325: 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Osman C, Walter P 2011. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci 108: 14151–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat Struct Mol Biol 11: 234–241 [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278: 48520–48523 [DOI] [PubMed] [Google Scholar]
- Künkele KP, Heins S, Dembowski M, Nargang FE, Benz R, Thieffry M, Walz J, Lill R, Nussberger S, Neupert W 1998. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Kutik S, Stojanovski D, Becker L, Becker T, Meinecke M, Krüger V, Prinz C, Meisinger C, Guiard B, Wagner R, et al. 2008a. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 132: 1011–1024 [DOI] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, et al. 2008b. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol 183: 1213–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W 2004. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J Biol Chem 279: 38047–38054 [DOI] [PubMed] [Google Scholar]
- Luk E, Yang M, Jensen LT, Bourbonnais Y, Culotta VC 2005. Manganese activation of superoxide dismutase 2 in the mitochondria of Saccharomyces cerevisiae. J Biol Chem 280: 22715–22720 [DOI] [PubMed] [Google Scholar]
- Mapa K, Sikor M, Kudryavtsev, Waegemann K, Kalinin S, Seidel CA, Neupert W, Lamb DC, Mokranjac D 2010. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell 38: 89–100 [DOI] [PubMed] [Google Scholar]
- Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C 2002. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 3: 159–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom M, Dayan D, Demishtein-Zohary K, Mokranjac D, Neupert W, Azem A 2011. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 complex. J Biol Chem 286: 43809–43815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Neupert W, Herrmann JM 2005. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J Biol Chem 280: 7777–7785 [DOI] [PubMed] [Google Scholar]
- Meinecke M, Wagner R, Kovermann P, Guiard B, Mick DU, Hutu DP, Voos W, Truscott KN, Chacinska A, Pfanner N, et al. 2006. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312: 1523–1526 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Rissler M, Chacinska A, Sanjuán Szklarz LK, Milenkovic D, Kozjak V, Schönfisch B, Lohaus C, Meyer HE, Yaffe MP, et al. 2004. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell 7: 61–71 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Wiedemann N, Rissler M, Strub A, Milenkovic D, Schönfisch B, Müller H, Kozjak V, Pfanner N 2006. Mitochondrial protein sorting: Differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem 281: 22819–22826 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfannschmidt S, Rissler M, Milenkovic D, Becker T, Stojanovski D, Youngman MJ, Jensen RE, Chacinska A, Guiard B, et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochondria. EMBO J 26: 2229–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM 2005. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121: 1059–1069 [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, Meisinger C 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem 279: 22781–22785 [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Gabriel K, Guiard B, Schulze-Specking A, Pfanner N, Chacinska A 2007. Biogenesis of the essential Tim9-Tim10 chaperone complex of mitochondria: Site-specific recognition of cysteine residues by the intermembrane space receptor Mia40. J Biol Chem 282: 22472–22480 [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Ramming T, Müller JM, Wenz LS, Gebert N, Schulze-Specking A, Stojanovski D, Rospert S, Chacinska A 2009. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell 20: 2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M, Bömer U, Kübrich M, Zufall N, Hönlinger A, Pfanner N 1997. The intermembrane space domain of mitchondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol Cell Biol 17: 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol 8: 361–370 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J 22: 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M 2006. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J 25: 4675–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Popov-Čeleketić D, Mapa K, Gevorkyan-Airapetov L, Zohary K, Hell K, Azem A, Neupert W 2009. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol Biol Cell 20: 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naamati A, Regey-Rudzki N, Galperin S, Lill R, Pines O 2009. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J Biol Chem 284: 30200–30208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoé M, Ohwa Y, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T 2004. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem 279: 47815–47821 [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM 2007. Translocation of proteins into mitochondria. Annu Rev Biochem 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Nunnari J, Fox TD, Walter P 1993. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262: 1997–2004 [DOI] [PubMed] [Google Scholar]
- Otera H, Taira Y, Horie C, Suzuki Y, Suzuki H, Setoguchi K, Kato H, Oka T, Mihara K 2007. A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments. J Cell Biol 179: 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais JE, Schilke B, Craig EA 2011. Re-evaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor. Mol Biol Cell 22: 4740–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papic D, Krumpe K, Dukanovic J, Dimmer KS, Rapaport D 2011. Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway. J Cell Biol 194: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Popov-Čeleketić J, Waizenegger T, Rapaport D 2008a. Mim1 functions in an oligomeric form to facilitate the integration of Tom20 into the mitochondrial outer membrane. J Mol Biol 376: 671–680 [DOI] [PubMed] [Google Scholar]
- Popov-Čeleketić D, Mapa K, Neupert W, Mokranjac D 2008b. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J 27: 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokisch H, Scharfe C, Camp DG 2nd, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, et al. 2004. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol 2: e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Gebert M, Höpker J, Yan M, Li J, Wiedemann N, van der Laan M, Pfanner N, Sha B 2011. Structural basis for the function of Tim50 in the mitochondrial presequence translocase. J Mol Biol 411: 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N 2003. Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5: 1543–1554 [DOI] [PubMed] [Google Scholar]
- Saddar S, Dienhart MK, Stuart RA 2008. F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J Biol Chem 282: 6677–6686 [DOI] [PubMed] [Google Scholar]
- Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C 2008. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE 4: e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Igura M, Obita T, Ose T, Kojima R, Maenaka K, Endo T, Kohda D 2007. Tom20 recognizes mitochondrial presequences through dynamic equilibrium among multiple bound states. EMBO J 26: 4777–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salhab M, Patani N, Jiang W, Mokbel K 2010. High TIMM17A expression is associated with adverse pathological and clinical outcomes in human breast cancer. Breast Cancer 19: 153–160 [DOI] [PubMed] [Google Scholar]
- Schiller D, Cheng YC, Liu Q, Walter W, Craig EA 2008. Residues of Tim44 involved in both association with the translocon of the inner mitochondrial membrane and regulation of mitochondrial Hsp70 tethering. Mol Cell Biol 28: 4424–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, et al. 2011. Regulation of mitochondrial protein import by cytosolic kinases. Cell 144: 227–239 [DOI] [PubMed] [Google Scholar]
- Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P 2011. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J Cell Biol 195: 643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoguchi K, Otera H, Mihara K 2006. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J 25: 5635–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T 2011. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci 108: 15179–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris DP, Tokatlidis K 2007. Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space. Mol Microbiol 65: 1360–1373 [DOI] [PubMed] [Google Scholar]
- Sideris DP, Petrakis N, Katrakili N, Mikropoulou D, Gallo A, Ciofi-Baffoni S, Banci L, Bertini I, Tokatlidis K 2009. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol 187: 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Joshi N, Chittoor B, Samji P, D’Silva P 2010. Role of magmas in protein transport and human mitochondria biogenesis. Hum Mol Genet 19: 1248–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M 1996. Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384: 582–585 [DOI] [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Folsch H, Stuart RA, Neupert W, Brunner M 1998. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature 391: 912–915 [DOI] [PubMed] [Google Scholar]
- Slutsky-Leiderman O, Marom M, Iosefson O, Levy R, Maoz S, Azem A 2007. The interplay between components of the mitochondrial protein translocation motor studied using purified components. J Biol Chem 282: 33935–33942 [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Guiard B, Kozjak-Pavlovic V, Pfanner N, Meisinger C 2007. Alternative function for the mitochondrial SAM complex in biogenesis of α-helical TOM proteins. J Cell Biol 179: 881–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Milenkovic D, Müller JM, Gabriel K, Schulze-Specking A, Baker MJ, Ryan MT, Guiard B, Pfanner N, Chacinska A 2008. Mitochondrial protein import: Precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J Cell Biol 183: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Becker T, Qiu J, Stojanovski D, Pfannschmidt S, Wirth C, Hunte C, Guiard B, Meisinger C, Pfanner N, et al. 2011a. Biogenesis of mitochondrial β-barrel proteins: The POTRA domain is involved in precursor release from the SAM complex. Mol Biol Cell 22: 2823–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Oeljeklaus S, Wiese S, Bohnert M, Lewandrowski U, Sickmann A, Guiard B, van der Laan M, Warscheid B, Wiedemann N 2011b. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol 413: 743–750 [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T 2009a. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol 184: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Endo T, Iijima M, Sesaki H 2009b. Ups1p and Ups2p antagonistically regulare cardiolipin metabolism in mitochondria. J Cell Biol 185: 1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J 2001. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9: 615–625 [DOI] [PubMed] [Google Scholar]
- Thornton N, Stroud DA, Milenkovic D, Guiard B, Pfanner N, Becker T 2010. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J Mol Biol 396: 540–549 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Voos W, Frazier AE, Lind M, Li Y, Geissler A, Dudek J, Müller H, Sickmann A, Meyer HE, et al. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol 163: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P 2005. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol Cell Biol 25: 7449–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N 2006. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol 16: 2271–2276 [DOI] [PubMed] [Google Scholar]
- van der Laan M, Meinecke M, Dudek J, Hutu DP, Lind M, Perschil I, Guiard B, Wagner R, Pfanner N, Rehling P 2007. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat Cell Biol 9: 1152–1159 [DOI] [PubMed] [Google Scholar]
- van Wilpe S, Ryan MT, Hill K, Maarse AC, Meisinger C, Brix J, Dekker PJ, Moczko M, Wagner R, Meijer M, et al. 1999. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401: 485–489 [DOI] [PubMed] [Google Scholar]
- Vergnolle MA, Baud C, Golovanov AP, Alcock F, Luciano P, Lian LY, Tokatlidis K 2005. Distinct domains of small Tims involved in subunit interaction and substrate recognition. J Mol Biol 351: 839–849 [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, et al. 2009. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139: 428–439 [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Prinz C, Kellermann J, Lottspeich F, Pfanner N, Meisinger C 2011. Mitochondrial protein turnover: Role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell 22: 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, Müller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, et al. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell 21: 694–707 [DOI] [PubMed] [Google Scholar]
- Wagner K, Gebert N, Guiard B, Brandner K, Truscott KN, Wiedemann N, Pfanner N, Rehling P 2008. The assembly pathway of the mitochondrial carrier translocase involves four preprotein translocases. Mol Cell Biol 28: 4251–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D 2004. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep 5: 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T, Schmitt S, Zivkovic J, Neupert W, Rapaport D 2005. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep 6: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther DM, Papic D, Bos MP, Tommassen J, Rapaport D 2009. Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc Natl Acad Sci 106: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM 2006. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed α-propeller. Mol Cell 21: 123–133 [DOI] [PubMed] [Google Scholar]
- Wideman JG, Go NE, Klein A, Redmond E, Lackey SW, Tao T, Kalbacher H, Rapaport D, Neupert W, Nargang FE 2010. Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol Biol Cell 21: 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Ryan MT 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J 20: 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424: 565–571 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N 2004. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: Intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem 279: 18188–18194 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol 179: 1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sha B 2006. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol 13: 589–593 [DOI] [PubMed] [Google Scholar]
- Yamano K, Tanaka-Yamano S, Endo T 2010. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J Biol Chem 285: 41222–41231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Terada K, Mori M 2003. AIP is a mitochondrial import mediator that binds to both import receptor Tom20 and preproteins. J Cell Biol 163: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O, Karniely S, Pines O 2007. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J Biol Chem 282: 29222–29229 [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50 [DOI] [PubMed] [Google Scholar]
- Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC 2009. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem J 419: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]