Abstract
When the cell-surface receptor Notch interacts with a ligand (e.g., Delta), its intracellular domain is cleaved and travels to the nucleus to regulate transcription. This influences cell division, fate, and death in metazoans.
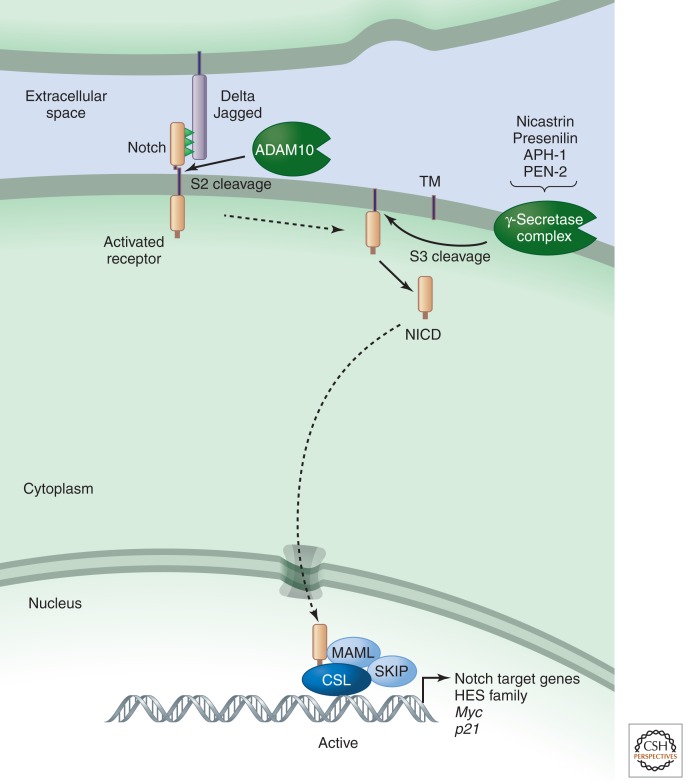
The Notch pathway regulates cell proliferation, cell fate, differentiation, and cell death in all metazoans. Notch itself is a cell-surface receptor that transduces short-range signals by interacting with transmembrane ligands such as Delta (termed Delta-like in humans) and Serrate (termed Jagged in humans) on neighboring cells (Fig. 1). Some soluble ligands have also been identified in Caenorhabditis elegans, but these bind to Notch together with transmembrane adaptors (Komatsu et al. 2008). Ligand binding leads to cleavage and release of the Notch intracellular domain (NICD), which then travels to the nucleus to regulate transcriptional complexes containing the DNA-binding protein CBF1/RBPjk/Su(H)/Lag1 (CSL).
Figure 1.
Notch signaling (simplified view).
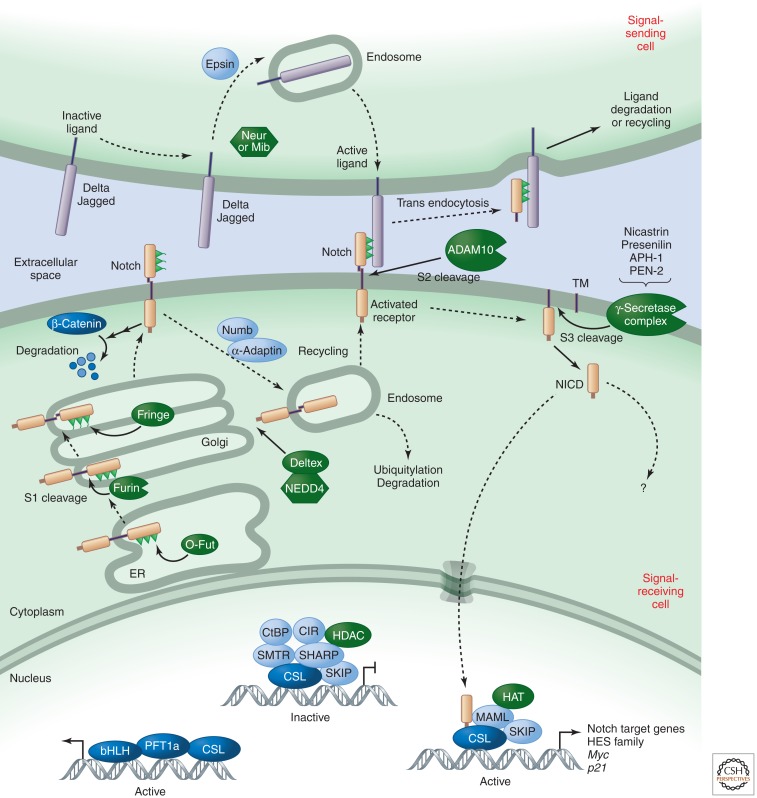
Following their synthesis, Notch receptors are cleaved by protein convertases during exocytosis at site 1 (S1), which regulates their trafficking and signaling activity (Logeat et al. 1998; Gordon et al. 2009). During passage through the Golgi, they can be glycosylated by glycosyltransferases such as Fringe, which determines the subsequent response to different subfamilies of ligands. These and other posttranslational modifications of the receptors and ligands tune the amplitude and timing of Notch activity to generate context-specific signals. Several proteins, including E3 ubiquitin ligases (e.g., Deltex and Nedd4), Numb, and α-adaptin, regulate the steady-state levels of the Notch receptor at the cell surface. In signal-sending cells, E3 ubiquitin ligases (Neur and MIB) similarly ubiquitylate the intracellular domain of the ligand to promote epsin-mediated endocytosis, which is associated with ligand activation (Fig. 2).
Figure 2.
Notch signaling.
Following ligand binding, signaling is initiated when endocytosis of ligand–receptor complexes induces unfolding of a juxtamembrane negative control region (NRR) unique to Notch proteins. Unfolding of the NRR allows access by the protease ADAM10 (also known as KUZ), which removes the Notch extracellular domain by cleaving at site 2 (S2); γ-secretase then cleaves Notch within its transmembrane domain at site 3 (S3) to release various forms of the NICD. Those that have valine or methionine residues at the amino terminus escape the N-end-rule degradation pathway (Tagami et al. 2008) and are stable enough to impact transcription (see below). Interestingly, productive interactions between Notch and its ligands occur when these are present on neighboring cells (i.e., in trans); when receptor and ligand are present on the same cell (i.e., interactions are in cis), activation is inhibited. cis interactions thus determine whether a cell will signal (the ligand is more abundant than Notch) or receive (Notch is more abundant than the ligand) (Sprinzak et al. 2010). Alternatively, in some cases ligand and receptors can be segregated into different subdomains to allow simultaneous transmission and reception of signals (Luty et al. 2007).
Only one nuclear protein is known to mediate the bulk of Notch signals: CSL (Kopan and Ilagan 2009). CSL is a DNA-binding adaptor that interacts with many proteins to build either repressor complexes, which include histone deacetylases (HDACs) that preserve a closed chromatin conformation, or activating complexes, which contain NICD, along with other proteins including histone acetyltransferases (HATs) that open up chromatin. In canonical, CSL-mediated Notch signaling, NICD translocates to the nucleus, binds to CSL, and helps recruit the adaptor protein Mastermind-like (MAML) (Kopan and Ilagan 2009). MAML recruits the HAT p300 and components of the transcription machinery. Thus, every cleaved Notch molecule generates one signaling unit, and tuning the effectiveness of receptor–ligand interaction directly determines the amount of NICD in the nucleus. During the transcriptional activation process, NICD is phosphorylated on a degron within its PEST domain by kinases such as cyclin-dependent kinase 8 (CDK8) and targeted for proteasome-mediated degradation by E3 ubiquitin ligases such as Sel10 (also known as Fbw7). This limits the half-life of a canonical Notch signal and resets the cell for the next pulse of signaling.
In addition to the canonical signals, mounting evidence indicates that CSL-independent activities of Notch also regulate vertebrate (Rangarajan et al. 2001; Demehri et al. 2008) and invertebrate (Ramain et al. 2001) development, but the biochemical details of this aspect of the pathway are yet to be uncovered. In the absence of ligand, Notch may also be involved in other cellular processes, such as regulating the stability of β-catenin (Sanders et al. 2009), a component of the Wnt signaling pathway (Nusse 2012).
Under most physiological conditions, unbound Notch receptors simply recycle or are targeted for lysosomal degradation. With one exception (Mukherjee et al. 2011), only pathological or experimental conditions are known to lead to receptor activation without ligand. These include mutations in the NRR domain (Weng et al. 2004), overexpression of Notch with ADAM proteases, and exposure to calcium chelators (Bozkulak and Weinmaster 2009; van Tetering et al. 2009), all of which expose Notch to shedding by ADAM17 (also known as TACE). Notch can also become activated when ESCRT components are mutated (Moberg et al. 2005; Thompson et al. 2005; Vaccari and Bilder 2005), which delays entry into the lysosome and permits ligand-independent activation. The frequent activation of Notch by mutations in T-cell acute lymphoblastic leukemia and its frequent inactivation in head and neck squamous cell carcinomas (Agrawal et al. 2011; Stransky et al. 2011) illustrate the importance of the pathway for control of cell fate and proliferation and the severe consequences of its dysregulation.
Acknowledgments
Figures adapted with kind permission of Cell Signaling Technology (http://www.cellsignal.com).
Footnotes
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. 2011. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 333: 1154–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkulak EC, Weinmaster G 2009. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol 29: 5679–5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, Grigsby PW, Miner JH, Farr AG, Kopan R 2008. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol 6: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, L’Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, et al. 2009. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE 4: e6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, Dionne HM, White JQ, Wani K, Boxem M, et al. 2008. OSM-11 facilitates LIN-12 Notch signaling during C. elegans vulval development. PLoS Biol 6: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX 2009. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, Lebail O, Jarriault S, Seidah NG, Israël A 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci 95: 8108–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luty WH, Rodeberg D, Parness J, Vyas YM 2007. Antiparallel segregation of Notch components in the immunological synapse directs reciprocal signaling in allogeneic Th:DC conjugates. J Immunol 179: 819–829 [DOI] [PubMed] [Google Scholar]
- Moberg KH, Schelble S, Burdick SK, Hariharan IK 2005. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell 9: 699–710 [DOI] [PubMed] [Google Scholar]
- Mukherjee T, Kim WS, Mandal L, Banerjee U 2011. Interaction between Notch and Hif-α in development and survival of Drosophila blood cells. Science 332: 1210–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Nusse R 2012. Wnt signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P, Khechumian K, Seugnet L, Arbogast N, Ackermann C, Heitzler P 2001. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr Biol 11: 1729–1738 [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J 20: 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders PG, Munoz-Descalzo S, Balayo T, Wirtz-Peitz F, Hayward P, Arias AM 2009. Ligand-independent traffic of Notch buffers activated armadillo in Drosophila. PLoS Biol 7: e1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB 2010. Cis-interactions between Notch and Delta generate mutually exclusive signaling states. Nature 465: 86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. 2011. The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S, Okochi M, Yanagida K, Ikuta A, Fukumori A, Matsumoto N, Ishizuka-Katsura Y, Nakayama T, Itoh N, Jiang J, et al. 2008. Regulation of Notch signaling by dynamic changes in the precision in S3 cleavage of Notch-1. Mol Cell Biol 28: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM 2005. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell 9: 711–720 [DOI] [PubMed] [Google Scholar]
- Vaccari T, Bilder D 2005. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating Notch trafficking. Dev Cell 9: 687–698 [DOI] [PubMed] [Google Scholar]
- van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M 2009. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem 284: 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP IV, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306: 269–271 [DOI] [PubMed] [Google Scholar]