Abstract
Translation initiation in eukaryotes is a highly regulated and complex stage of gene expression. It requires the action of at least 12 initiation factors, many of which are known to be the targets of regulatory pathways. Here we review our current understanding of the molecular mechanics of eukaryotic translation initiation, focusing on recent breakthroughs from in vitro and in vivo studies. We also identify important unanswered questions that will require new ideas and techniques to solve.
The assembly of an 80S initiation complex on a mRNA, poised to enter the elongation phase of protein synthesis, involves complex interactions and rearrangements and requires at least 12 initiation factors.
This work aims to present the current state of our knowledge of the molecular mechanics of translation initiation in eukaryotes. We focus on advances that have taken place over the last few years and, because of space limitations, assume readers will be able to find references to the foundational literature for the field (published before 2000) in the more recent works that are cited here. As always, we apologize for not having the space to cite many important works. Please view this as merely an introduction to the field rather than a complete summary.
OVERVIEW OF THE INITIATION PATHWAY
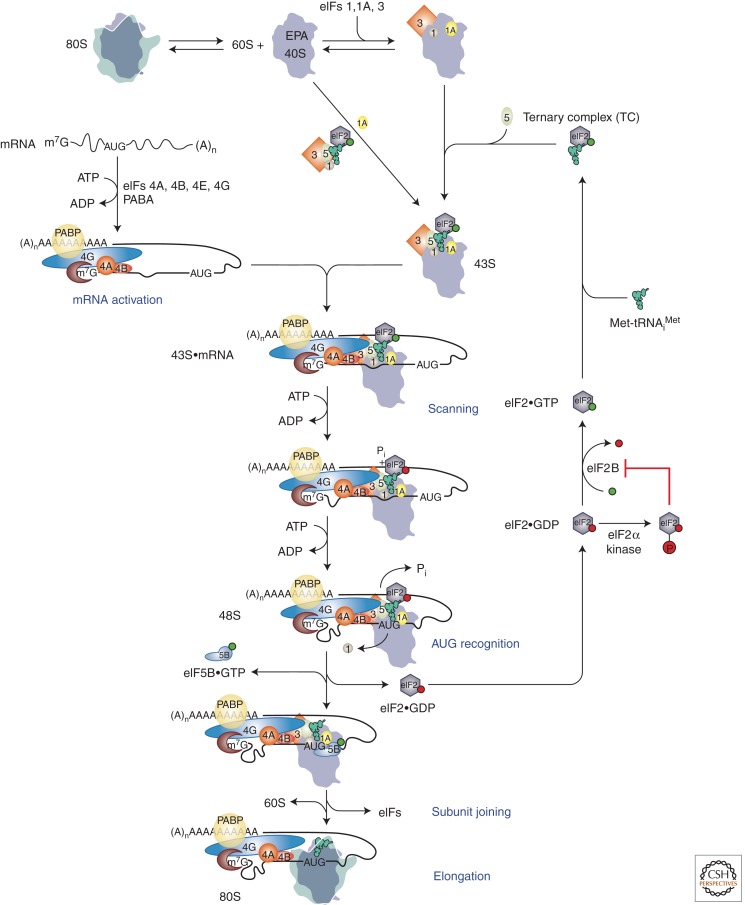
Figure 1 presents the basic outline of the eukaryotic cap-dependent initiation pathway, and the reader is referred to a number of recent reviews that summarize the evidence supporting the current paradigm outlined below (Hinnebusch et al. 2007; Pestova et al. 2007; Lorsch and Dever 2010; Hinnebusch 2011; Parsyan et al. 2011). Identification of the initiation codon by the eukaryotic translational machinery begins with binding of the ternary complex (TC) consisting of initiator methionyl-tRNA (Met-tRNAi) and the GTP-bound form of eukaryotic initiation factor 2 (eIF2) to the small (40S) ribosomal subunit to form the 43S preinitiation complex (PIC). Binding of the TC to the 40S subunit is promoted by eIFs 1, 1A, 5, and the eIF3 complex (Fig. 1). A network of physical interactions links eIFs 1, 3, 5, and TC in a multifactor complex (MFC) in yeast (Asano et al. 2000), plants (Dennis et al. 2009), and mammals (Sokabe et al. 2011). In budding yeast, the MFC enhances the formation or stability of the 43S PIC in vivo (reviewed in Hinnebusch et al. 2007). The 43S PIC binds to the messenger RNA (mRNA) near the 5′-7-methylguanosine cap in a process facilitated by eIF3, the poly(A)-binding protein (PABP), and eIFs 4B, 4H (in mammals), and 4F. The eIF4F complex is comprised of the cap-binding protein eIF4E, eIF4G, and the RNA helicase eIF4A. eIF4G is a scaffold protein that harbors binding domains for PABP, eIF4E, eIF4A, and (in mammals) eIF3. Both yeast and human eIF4G also bind RNA. The binding domains for eIF4E and PABP in eIF4G, along with its RNA-binding activity, enable eIF4G to coordinate independent interactions with mRNA via the cap, poly(A) tail, and sequences in the mRNA body to assemble a stable, circular messenger ribonucleoprotein (mRNP), referred to as the “closed-loop” structure. The eIF4G–eIF3 interaction is expected to establish a protein bridge between this “activated mRNP” and the 43S PIC to stimulate 43S attachment to the mRNA, and the helicase activity of eIF4A is thought to generate a single-stranded landing pad in the mRNA on which the 43S PIC can load (reviewed in Hinnebusch et al. 2007; Pestova et al. 2007; Lorsch and Dever 2010; Hinnebusch 2011).
Figure 1.
Model of canonical eukaryotic translation initiation pathway. The pathway is shown as a series of discrete steps starting with dissociation of 80S ribosomes into subunits. Binding of factors is depicted both as a single step via the multifactor complex and as two separate steps, with eIFs 1, 1A, and 3 binding first followed by binding of ternary complex and eIF5. The resulting 43S preinitiation complex (PIC) is then loaded onto an activated mRNP near the 5′ cap. Subsequent scanning of the mRNA allows recognition of the start codon, which triggers downstream steps in the pathway including eIF1 release from the PIC, Pi release from eIF2, and conversion to the closed, scanning-arrested state of the complex. eIF2·GDP released after subunit joining is recycled back to eIF2·GTP by the exchange factor eIF2B. eIF5B in its GTP-bound form promotes joining of the 60S subunit to the preinitiation complex, which triggers release of eIF5B·GDP and eIF1A to form the final 80S initiation complex, which can begin the elongation phase of protein synthesis. Throughout, GTP is depicted as a green ball and GDP as a red ball. (Modified from Hinnebusch 2011; reproduced, with permission, from the author.)
Once bound near the cap, the 43S PIC scans the mRNA leader for an AUG codon in a suitable sequence context. Base-pairing between the anticodon of Met-tRNAi and the AUG in the peptidyl-tRNA (P) site of the 40S subunit is the initial event in start codon recognition (Lomakin et al. 2006; Kolitz et al. 2009; Hinnebusch 2011). AUG recognition causes arrest of the scanning PIC and triggers conversion of eIF2 in the TC to its GDP-bound state via gated phosphate (Pi) release and the action of the GTPase-activating (GAP) factor eIF5. Following release of eIF2·GDP and several other eIFs present in the PIC, joining of the large (60S) subunit is catalyzed by eIF5B to produce an 80S initiation complex (IC) containing Met-tRNAi base-paired to AUG in the P site and ready to begin the elongation phase of protein synthesis (Fig. 1) (reviewed in Hinnebusch et al. 2007; Pestova et al. 2007; Hinnebusch 2011).
RECRUITMENT OF Met-tRNAi TO THE 40S RIBOSOMAL SUBUNIT
eIF2 Is a G-Protein Switch that Carries Met-tRNAi onto the Ribosome
The Met-tRNAi is delivered to the 40S subunit in the TC with eIF2·GTP. The affinity of Met-tRNAi is greater for eIF2·GTP than for eIF2·GDP, and this affinity switch depends on the methionine moiety on the Met-tRNAi (Kapp and Lorsch 2004). This contribution of methionine, plus the stimulatory role of the unique A1:U72 base pair (bp) in the acceptor stem of tRNAi in binding eIF2 (Kapp and Lorsch 2004; Pestova et al. 2007), presumably act to prevent binding of elongator tRNAs to the factor. This specificity, coupled with the requirement for eIF2 to load tRNA onto the 40S subunit, is thought to eliminate the need for a mechanism to reject elongator tRNAs during PIC assembly, a process in bacteria that relies heavily on IF3 (Hershey and Merrick 2000). (As described below, a structural homolog of IF3 is absent in eukaryotes, but eIF1 acts similarly to ensure selection of AUG as a start codon.) Understanding the structural basis for the stimulatory effects of methionine, the A1:U72 bp, and GTP versus GDP on initiator tRNA binding to eIF2 would be advanced by high-resolution structural analysis of the complete TC. Whereas the crystal structure of the archaeal ortholog (aIF2) has been solved, as well as various aIF2 subcomplexes bound to GDP or GTP analogs (reviewed in Schmitt et al. 2010), no crystal structures or cryo-EM (electron microscopy) models of heterotrimeric eIF2 have been described.
eIF2γ binds directly to both GTP and Met-tRNAi and it appears that the α and β subunits each increase the affinity of the eIF2 complex for Met-tRNAi by ∼100-fold (Naveau et al. 2010), but it is unknown whether this stimulatory effect involves direct contacts between Met-tRNAi and eIF2α or eIF2β. Based on the crystal structure of a heterotrimer of aIF2β, aIF2γ, and a portion of aIF2α (Yatime et al. 2007) it has been proposed that the α and β subunits allosterically induce a conformation in aIF2γ with high affinity for Met-tRNAi (Naveau et al. 2010). Evidence consistent with an allosteric mechanism, at least for eIF2α, comes from directed hydroxyl radical cleavage mapping of Met-tRNAi binding to yeast eIF2 in reconstituted PICs. Met-tRNAi was cleaved by free radicals generated from particular positions in eIF2γ or eIF2β, but not from eIF2α, suggesting the latter does not make direct contact with the tRNA. Interestingly, the patterns of cleavage imply a mode of initiator binding to eIF2γ dramatically different from that seen in crystal structures of the EF-Tu·GDPNP·Phe-tRNAPhe TC, which delivers aminoacylated tRNAs to the A site during elongation. In contrast to the latter complex, domain III of eIF2γ, the subunit homologous to EF-Tu, does not contact the T stem of Met-tRNAi; instead the sole contact is with the methionylated acceptor end of the tRNA in a pocket in eIF2γ formed between the G domain and domain II (Shin et al. 2011). A recent crystal structure of the TC formed by an archaeal aIF2, GDPNP, and E. coli Met-tRNA(fMet) also demonstrated that the tRNA is bound by aIF2 in a manner dramatically distinct from that of elongator tRNA binding to EF-Tu (Schmitt et al. 2012). Consistent with previous models (Schmitt et al. 2010; Shin et al. 2011), the acceptor end of the tRNA binds to aIF2γ according to the EF-Tu paradigm; however, the T-stem minor groove does not contact aIF2 and, instead, the T-loop in the tRNA “elbow” interacts with regions of the aIF2α subunit. As these last contacts were not detected in the hydroxyl radical probing of the eIF2 TC (Shin et al. 2011), it remains to be seen whether they are important in solution and conserved in eukaryotic TC.
Importantly, the patterns of free radical-induced cleavages of 18S rRNA observed in this last study suggested that eIF2γ domain III interacts with h44 of 18S rRNA, but no other contacts between eIF2 and 18S rRNA were detected. Using the cleavage data to dock eIF2γ onto h44 and the 3′ end of Met-tRNAi, making use of high-resolution structures of a bacterial 70S·tRNA·mRNA complex (Selmer et al. 2006), the 40S·eIF1 complex (Rabl et al. 2011), and aIF2αγ and aIF2βγ heterodimers (Yatime et al. 2006, 2007) (among others), a structural model of the 43S PIC was constructed (Shin et al. 2011).
Although this model represents an important step, high-resolution crystal structures and cryo-EM reconstructions of free TC and TC bound to the 43S PIC remain critical goals. In addition, the model does not include known interactions of the eIF2β N-terminal domain (NTD) (lacking in aIF2β) with eIFs 1 and 5 in the MFC (Asano et al. 2000; Singh et al. 2004), and there might be contacts between eIF2α or eIF2β with 40S ribosomal proteins not detected by the hydroxyl radical mapping. Identifying mutations in yeast eIF2 subunits, ribosomal proteins, and 18S rRNA that reduce TC binding to the PIC should help identify the eIF2·40S contacts most critical in vivo. Only one such mutation has been identified in domain III of eIF2γ (R439A) and it produces a synergistic reduction in TC binding to reconstituted 43S·mRNA complexes when combined with an 18S rRNA substitution in helix 28 (A1152U) (Shin et al. 2011) that likely weakens interaction of the anticodon stem loop (ASL) of Met-tRNAi with the 40S P site (Dong et al. 2008). Identifying h44 mutations with these phenotypes would provide valuable support for the Shin et al. model of the 43S PIC.
Binding of TC to the 40S Subunit Is Promoted by Other Factors
TC does not bind to the 40S subunit on its own, and instead requires the assistance of eIFs 1, 1A, 5, and the eIF3 complex (Asano et al. 2001; Algire et al. 2002; Majumdar et al. 2003; Kolupaeva et al. 2005; Pestova et al. 2007). All of these factors, except for eIF1A, are components of the MFC. As all MFC components can bind directly to the 40S subunit (Hinnebusch et al. 2007; Pestova et al. 2007; Sokabe et al. 2011), they would be expected to bind cooperatively in the context of the MFC. Indeed, there is considerable evidence that disrupting particular contacts between MFC components reduces the rate or stability of TC binding to 40S subunits in yeast cells (Valášek et al. 2002, 2004; Nielsen et al. 2004; Singh et al. 2005, 2006; reviewed in Hinnebusch et al. 2007). Recent studies on reconstituted mammalian MFC indicate that the rate of MFC binding to 40S·eIF1A complexes is indistinguishable from TC binding to 40S·eIF1A complexes preloaded with eIFs 1, 3, and 5 (Sokabe et al. 2011), suggesting that the stimulatory effects of other MFC components on TC binding can be exerted outside of the preformed MFC. Thus, it is important to determine in vivo whether TC generally binds to the 40S subunit in the context of the MFC or, rather, the MFC represents only one possible pathway for TC recruitment (as depicted in Fig. 1), or serves another function. It is intriguing that dissociation of Met-tRNAi from eIF2·GDP is enhanced when eIF2 resides in the mammalian MFC, and eIF5 figures prominently in this activity (Sokabe et al. 2011). This might implicate eIF5 in the final step of AUG selection, release of Met-tRNAi from eIF2·GDP, in addition to its function in promoting GTP hydrolysis in the TC. As discussed below, the physical connections among MFC components also function in AUG recognition in yeast cells. Interestingly, these connections might be regulated in plants by phosphorylation of the interacting segments in eIF3c, eIF5, and eIF2β by casein kinase 2 (Dennis et al. 2009).
The unstructured amino-terminal tail (NTT) of yeast eIF1A interacts with eIF2 and eIF3 (Olsen et al. 2003), which could also help stabilize TC binding to the 40S. In addition to physically contacting eIF2, however, eIF1A and eIF1 stimulate TC binding indirectly by stabilizing an “open” conformation of the 40S subunit that is permissive for rapid TC loading (Passmore et al. 2007). As discussed below, it is likely that Met-tRNAi binds differently to the PIC in this open conformation than in the “closed,” scanning-arrested state that prevails after AUG recognition. Presumably, TC binding to the PIC in the model produced by Shin et al. (2011) represents the closed conformation because it was generated using reconstituted 43S·mRNA complexes in which start codon recognition has already taken place (Kolitz et al. 2009). Hence, determining the locations of TC in the free 43S PIC and open, scanning conformation of the 43S·mRNA complex should also be important goals for future research.
Recycling of eIF2·GDP
In the course of initiation, the GTP in TC is hydrolyzed to GDP, and eIF2·GDP must be recycled to eIF2·GTP for renewed TC assembly, a reaction catalyzed by the heteropentameric eIF2B complex. The essential exchange reaction is catalyzed by the carboxy-terminal segment of eIF2Bε, which interacts directly with the G domain of eIF2γ and with lysine-rich regions of eIF2β (Gomez and Pavitt 2000; Gomez et al. 2002; Alone and Dever 2006; Hinnebusch et al. 2007). The other eIF2B subunits, notably the α-β-δ “regulatory” subcomplex also contribute to eIF2·GDP binding through interactions with eIF2α (Dev et al. 2010), and this latter interaction is enhanced by phosphorylation of Ser51 by one of the eIF2α kinases, which are activated in stress conditions to down-regulate general initiation (Krishnamoorthy et al. 2001; Hinnebusch et al. 2007). It was proposed that tighter binding of eIF2α-P to the eIF2B regulatory subcomplex disrupts productive interaction of the catalytic (ε) subunit with eIF2γ, rendering phosphorylated eIF2α·GDP a competitive inhibitor of eIF2B that impedes recycling of unphosphorylated eIF2. Although this model is consistent with a large body of biochemical and genetic data, it should be tested further by structural analysis of eIF2B in complexes with phosphorylated and unphosphorylated eIF2·GDP. There is in vivo evidence in yeast that the recycling of eIF2·GDP by eIF2B is negatively regulated by formation of a competing eIF5·eIF2·GDP complex (Singh et al. 2006). Moreover, eIF5 contains a segment in the linker region connecting its amino- and carboxy-terminal domains that interacts with eIF2γ and inhibits GDP release from eIF2, serving as a GDP dissociation inhibitor (GDI) (Jennings and Pavitt 2010).
eIF2-Independent Met-tRNAi Recruitment
Recent studies indicate that in mammalian reconstituted systems, the protein Ligatin/eIF2D can deliver Met-tRNAi to the 40S subunit independently of eIF2·GTP in the case of certain specialized mRNAs (internal ribosome entry sites [IRES] containing, leaderless, or with A-rich 5′ untranslated regions [UTRs]) in which the AUG can be placed directly in the P site independently of scanning (Dmitriev et al. 2010; Skabkin et al. 2010). This could explain the ability of certain viral mRNAs containing IRESs to maintain translation in the face of eIF2α phosphorylation, a host defense mechanism triggered by many viruses (Dever et al. 2007). It was proposed that Ligatin/eIF2D can increase both on and off rates of tRNA binding in the P site, which could explain its other known activity of dissociating deacylated elongator tRNAs from recycled ribosomes after termination (Skabkin et al. 2010). There is also evidence that the protein eIF2A can bind to the IRES of hepatitis C virus (HCV) and enhance Met-tRNAi loading to the 40S subunit when eIF2 is phosphorylated, and knockdown of eIF2A reduces HCV proliferation in cells (Kim et al. 2011).
mRNA RECRUITMENT TO THE 43S PIC
eIF4F Actively Promotes Loading of mRNA onto the PIC
A critical aspect of the scanning mechanism concerns the reactions involved in directing the 43S PIC to the 5′ end of the mRNA. eIF4F stimulates this step through interaction of eIF4E with the cap structure, recruiting eIF4A to the 5′ UTR (Pestova et al. 2007). eIF4G holds eIF4A in its active conformation (Oberer et al. 2005; Schutz et al. 2008; Hilbert et al. 2011; Nielsen et al. 2011; Ozes et al. 2011), enabling it to unwind the mRNA and produce a single-stranded binding site for the 43S PIC near the 5′ cap. It is believed that eIF4G also helps to recruit the 43S PIC directly, via physical interactions with eIF3 or eIF5 in the PIC (Asano et al. 2001; Pestova et al. 2007). There is genetic and biochemical evidence implicating eIF4A and eIF4F in promoting 43S attachment to mRNAs, in some cases even if they contain relatively short 5′ UTRs without obvious secondary structures. As might be expected, a greater requirement for these factors has been observed for mRNAs with more structured 5′ UTRs (Svitkin et al. 2001; Pestova and Kolupaeva 2002; Mitchell et al. 2010; Hinnebusch 2011). In addition, 43S attachment to model mRNAs expected to lack any structure in the 5′ UTR can occur in reconstituted systems without eIF4F (Pestova and Kolupaeva 2002; Mitchell et al. 2010).
Simultaneous binding of eIF4E to the cap, PABP to the poly(A) tail, and eIF4E and PABP to their separate binding sites in the eIF4G NTD enables circularization of the mRNA (Pestova et al. 2007), and it is frequently assumed that this “closed-loop” conformation is crucial for efficient recruitment of the 43S PIC. However, the importance of the PABP–eIF4G interaction seems to vary with the cell type. Eliminating the PABP–eIF4G interaction by deleting or mutating the PABP-binding domain in eIF4G is not lethal in yeast (Tarun et al. 1997); and even if the eIF4G–eIF4E interaction is impaired, deleting the PABP-binding domain has no effect on yeast cell growth provided that the RNA-binding region in the amino terminus of eIF4G1 (RNA1) is intact (Park et al. 2011a). It appears that RNA1 and the PABP- and eIF4E-binding domains in yeast eIF4G collaborate to promote stable association of eIF4G with mRNA near the cap, and formation of a closed loop may be incidental to the efficiency of 43S attachment. Impairing the PABP–eIF4G interaction had only a modest effect on translation in rabbit reticulocyte lysates (RRLs) (Hinton et al. 2007) but a dramatic effect was seen in Krebs-2 cell extracts, in which it reduced eIF4E binding to the cap, 48S assembly, and 60S subunit joining (Kahvejian et al. 2005). The PABP independence of RRLs likely results from the high ratio of eIF4F to general RNA-binding proteins, as addition of the RNA-binding protein YB-1 to RRLs confers PABP dependence (Svitkin et al. 2009). In addition, tight binding of mammalian eIF4F to the capped 5′ end of mRNA requires the RNA-binding domain in the middle of eIF4G (Yanagiya et al. 2009). Thus, interaction of eIF4G with PABP bound to the poly(A) tail might be critical only when YB-1 or other general RNA-binding proteins effectively compete with eIF4G for direct binding to the mRNA—a situation that apparently does not exist in yeast cells under favorable culture conditions. Interestingly, although the eIF4E–cap interaction adds little to the binding affinity of eIF4F for mRNA in vitro (Kaye et al. 2009), the eIF4E–cap interaction with eIF4G should provide eIF4F with yet another way to circumvent competition with general RNA-binding proteins. In general, it appears that a number of the interactions among the components of the mRNP are redundant and may serve to safeguard the system against failure at a single point and to give the mRNA recruitment machinery an advantage over competing RNA–protein and protein–protein interactions rather than playing central mechanistic roles.
In yeast eIF4G, there are two other RNA-binding domains in the middle and carboxyl terminus (RNA2 and RNA3, respectively) (Berset et al. 2003), which appear to perform critical functions downstream from eIF4F mRNA·PABP assembly (Park et al. 2011a). Interestingly, the RNA3 domain contains a binding site for the DEAD-box RNA helicase Ded1/Ddx3 (Hilliker et al. 2011), an essential protein in yeast implicated in ribosomal scanning (Berthelot et al. 2004; Abaeva et al. 2011; Hinnebusch 2011). Although eliminating the eIF4G-binding domain in the carboxyl terminus of Ded1 impairs translation in vitro, it does not affect cell growth (Hilliker et al. 2011), implying either that the RNA3 domain has another critical function in vivo besides Ded1 recruitment or that Ded1 can be recruited by a redundant pathway. It is clearly important to identify the molecular functions of RNA2 and RNA3 in mRNA recruitment and/or ribosomal scanning. In this regard, a recent study has shown that the three RNA-binding sites in yeast eIF4G work together to impart a strong preference on the eIF4F complex for unwinding RNA duplexes with 5′-single-stranded overhangs over duplexes with 3′-overhangs (Rajagopal et al. 2012). This polarity may be important for establishing the 5′-3′ directionality of scanning by the PIC.
eIF4A is not a processive helicase and is thought to melt short helices in the mRNA by binding in its ATP-bound form to an unpaired RNA strand, with ATP hydrolysis serving either to disrupt the neighboring duplex or to release eIF4A for subsequent rounds of RNA binding and melting (Sengoku et al. 2006; Liu et al. 2008; Bulygin et al. 2010; Parsyan et al. 2011). In the crystal structure of free eIF4A, its amino- and carboxy-terminal RecA-like domains are widely separated and a functional active site does not exist (Caruthers et al. 2000). Interaction with the “HEAT” domains of eIF4G holds the RecA-like domains of eIF4A near each other in a conformation that may be poised to interact with substrates and release products (Oberer et al. 2005; Schutz et al. 2008; Hilbert et al. 2011). It seems clear that eIF4A undergoes a cycle of conformational and ligand-affinity changes driven by ATP hydrolysis and/or nucleotide binding and release and that the conformation of the enzyme is modulated by interactions with other proteins (Oberer et al. 2005; Pestova et al. 2007; Schutz et al. 2008; Marintchev et al. 2009; Hilbert et al. 2011). Exactly how these changes result in RNA unwinding is not yet clear, nor is the stoichiometry of events. It is noteworthy that eIF4A is the most abundant initiation factor; at a concentration of 50 µM in yeast it exists in fivefold excess over ribosomes (von der Haar and McCarthy 2002) and at a concentration similar to that of actin. Thus, it is possible that multiple eIF4A molecules act during recruitment of an individual mRNA to the PIC, both within eIF4F and outside of it. A full understanding of the mechanism of action of eIF4A will require additional structural and biophysical studies, including use of ensemble and single-molecule kinetics approaches.
In addition to recruiting and activating eIF4A, there is evidence that a segment of mammalian eIF4G helps to recruit the 43S PIC to the mRNA 5′ end by its interactions with the e subunit of eIF3 (Korneeva et al. 2000; LeFebvre et al. 2006). This conclusion is based on the inhibitory effects of overexpressing eIF3e (presumably to out-compete eIF3 binding to eIF4G) on translation initiation and on eIF4G and eIF2 association with native PICs. It would be valuable to extend the analysis to include cells depleted of eIF3e, as it is possible that the e subunit does not make the sole (or even most critical) contact between eIF3 and eIF4G. Neither eIF3e nor the eIF3-binding segment of eIF4G is present in yeast (Marintchev et al. 2009), and yeast eIF3 and eIF4G do not directly interact (Asano et al. 2001; Mitchell et al. 2010). However, yeast eIF4G and eIF5 interact directly (Mitchell et al. 2010), and the carboxy-terminal domain (CTD) of eIF5 can bridge interaction between eIF4G2 and the eIF3c-NTD (eIF5’s direct partner in yeast eIF3) and stimulate eIF4G–eIF3 association in yeast cell extracts (Asano et al. 2001). Although a mutation in the eIF5-CTD that disrupts its interaction with eIF4G impaired 43S binding to mRNA in extracts, this defect was not seen in living cells, possibly because the eIF5-CTD mutation also reduces eIF5 GAP function and blocks the downstream conversion of PICs to 80S ICs (Asano et al. 2001). A stimulatory function of eIF5 on mRNA recruitment to the PIC was not observed in a reconstituted yeast system, however (Mitchell et al. 2010). Hence, more work is required to determine whether the eIF4G–eIF5 interaction significantly enhances 43S binding to mRNAs in yeast cells, and if a similar interaction in mammalian cells is redundant with the eIF3–eIF4G interaction.
The Mysterious eIF4B
Mammalian eIF4B binds in vitro to eIF3a through its internal “DRYG” repeats and thus could potentially form a protein bridge between the eIF4F·mRNP and 43S PIC, functioning redundantly with the eIF3–eIF4G interaction. It has been proposed that mammalian eIF4B can also stimulate 43S binding to mRNA more directly by binding simultaneously to mRNA, through its carboxy-terminal, arginine-rich RNA-binding domain, and to 18S rRNA through its amino-terminal RNA recognition motif (RRM). Yeast eIF4B (Tif3) also appears to possess a single-stranded RNA (ssRNA)-binding domain located carboxy-terminal to the conserved amino-terminal RRM, and it was concluded that both halves of Tif3 are required for its ability to stimulate translation in vitro and in vivo; hence, the putative mRNA–rRNA bridging mechanism could apply to yeast eIF4B as well (reviewed in Pestova et al. 2007; Hinnebusch 2011). It is important to test the effects of disrupting the eIF3a- and RNA-binding domains of eIF4B or Tif3 on the efficiency of 43S binding to mRNA both in vitro and in vivo.
Mammalian eIF4B is best known for its function in stimulating the helicase activity of eIF4A—an activity it shares with a homolog, eIF4H (Pestova et al. 2007; Rozovsky et al. 2008; Parsyan et al. 2011). Consistent with this, PIC binding and scanning of structured mRNAs in an in vitro mammalian system was shown to be highly dependent on eIF4B (Dmitriev et al. 2003). A recent study suggests that eIF4B increases the efficiency with which eIF4G-stimulated ATP hydrolysis is coupled to RNA duplex unwinding by eIF4A, and that eIF4H is less efficient than eIF4B in this respect (Ozes et al. 2011). This is consistent with the finding that the carboxy-terminal, RNA-binding region of mammalian eIF4B is required for stimulation of helicase activity (Rozovsky et al. 2008) and the fact that eIF4H is shorter and lacks most of the carboxy-terminal region found in eIF4B.
The mechanism by which eIF4B stimulates eIF4A helicase activity remains unclear. There is evidence that eIF4B stimulates binding of both ATP and RNA by eIF4A (Bi et al. 2000; Rozovsky et al. 2008; Marintchev et al. 2009; Nielsen et al. 2011), possibly by enhancing interdomain closure in the manner described for eIF4G. eIF4B could also load onto single-stranded RNA extensions to stabilize eIF4A binding to the duplex-containing substrate (Rozovsky et al. 2008), it could capture the single-stranded RNA products of the helicase reaction to prevent reannealing, or it could stabilize a conformation of the eIF4A-RNA complex incompatible with duplex formation. Presumably, eIF4H is incapable of one or more of these activities, rendering it less effective than eIF4B in stimulating eIF4A helicase activity (Ozes et al. 2011).
Cross-linking studies have indicated that eIF4B, eIF4H, and eIF4A are bound to mRNA from 12 nucleotides to at least 52 nucleotides from the cap and suggest that multiple molecules of each factor interact with a single mRNA both near the eIF4F-cap complex and further downstream from it (Lindqvist et al. 2008). It is currently unclear, however, whether eIF4A and eIF4B directly interact with each other and whether eIF4B can bind to eIF4G (Marintchev et al. 2009; Nielsen et al. 2011).
As described above, the current model for mRNA recruitment posits that the eIF4 factors and PABP cooperatively assemble on an mRNA and mediate unwinding to produce the activated mRNP. The activated mRNP then binds to the 43S PIC via interactions between the eIF4 factors and factors associated with the PIC, and the 5′ end of the mRNA is loaded into the mRNA-binding channel of the 40S subunit. One alternative to this prevailing model is that the eIF4 factors assemble on the PIC to form a “holoPIC,” which then directly recruits an mRNA. In this model unwinding of the mRNA actually takes place on the holoPIC, allowing the unwound segments of the 5′ UTR to be directly fed into the mRNA-binding channel of the 40S subunit. This model is appealing because it is unclear how the unwound 5′ end of the activated mRNP can be handed off to the PIC without refolding occurring first. Distinguishing between the activated mRNP and holoPIC mechanisms will require the development of new, quantitative assays to directly measure unwinding and mRNA loading in the presence of different combinations of initiation components.
eIF3, a Central Hub in mRNA Recruitment
In addition to the eIF4 group of factors, eIF3 also plays a critical role in mRNA recruitment to the PIC. eIF3 is a large complex of 13 nonidentical subunits (a–m) in mammals, and only 6 subunits (a, b, c, g, h, and j) in budding yeast. There is accumulating evidence that eIF3 interacts primarily with the solvent-exposed, “backside” of the 40S, that it spans the entry and exit pores of the mRNA-binding channel, and that it likely interacts with the mRNA itself at these locations to stabilize 43S attachment or to regulate scanning (Pestova et al. 2007; Hinnebusch 2011). Recent findings from Cate et al. indicate that the bulk of the density visible in cryo-EM models of mammalian eIF3·40S complexes is contributed by the so-called PCI/MPN octamer, which represents only ∼1/2 of the mass of holo-eIF3 and lacks homologs of the essential yeast eIF3 subunits b, g, and i. The PCI/MPN octamer can bind the HCV IRES, 40S subunits, eIF1 and eIF1A, but cannot stimulate 48S PIC assembly, additionally requiring the b-g-i subcomplex for this key activity (Sun et al. 2011). Clearly, a high-resolution model of eIF3 binding to the 40S subunit is an important goal for future research. It has long been known that both yeast and mammalian eIF3 promote 43S binding to mRNA, but because eIF3 also stimulates 43S assembly, it was unclear if it acts directly in 43S attachment to mRNA. Consistent with a direct role are findings that 40S binding of mammalian eIF3 is stimulated by ssRNAs that can likely occupy the mRNA-binding channel of the 40S, although the stabilizing effect of mRNA on eIF3·40S interaction might play a greater role following AUG recognition and release of eIF2·GDP than in 43S attachment to the 5′ UTR (Unbehaun et al. 2004; Kolupaeva et al. 2005). However, recent findings indicate a direct role for yeast eIF3 in 43S binding to capped, native mRNA in vitro, even more critical than that of eIF4F and eIF4B (Mitchell et al. 2010), and conserved residues in the carboxyl terminus of eIF3a have been implicated in this function both in vivo (Chiu et al. 2010) and in vitro (AG Hinnebusch and JR Lorsch, unpubl. observations).
UV-cross-linking data indicate direct interactions of mammalian eIF3a and eIF3d at the mRNA exit channel (Pisarev et al. 2008), which is consistent with the role of yeast eIF3a in reinitiation on GCN4 mRNA (Szamecz et al. 2008). It is thought that these eIF3 subunits comprise an extension of the mRNA exit channel. Consistent with this, yeast eIF3 more strongly enhanced 43S binding to a model mRNA with a long leader upstream of AUG (that would protrude from the exit channel) than one containing a short leader but a long 3′ extension (that would protrude from the entry channel) (Mitchell et al. 2010). Yeast eIF3a substitutions that impair 43S attachment to mRNA also produce phenotypes in vivo indicating defects in scanning and AUG recognition (Chiu et al. 2010). Considering evidence that the yeast eIF3a (Tif32) CTD interacts directly with 40S structural elements (h16 and Rps3) (Valášek et al. 2003; Chiu et al. 2010) that promote the open conformation of the mRNA channel latch (Passmore et al. 2007), it was suggested that the eIF3a CTD facilitates opening of the latch, although it could also help to recruit a helicase that functions at the entry channel to remove secondary structure. Clearly, more detailed structural information about interactions of eIF3 subunits with the ribosome, mRNA, eIF4G, and eIF4B are required to develop a molecular picture of its manifold roles in 43S attachment to mRNA.
Knocking out the Model?
As might be expected from their established biochemical functions, eIF4E, eIF4G, and eIF4A are all essential proteins in yeast. Moreover, mutational analysis indicates that the eIF4E and eIF4A interactions with their respective domains in eIF4G are also essential for yeast cell viability (Hinnebusch et al. 2007). However, depletion of eIF4G1 to undetectable levels in a yeast strain lacking the other isoform (eIF4G2) does not abolish translation initiation, reducing it by only ∼75% and leaving a considerable fraction of polysomes intact (Jivotovskaya et al. 2006), even though cell division is blocked. In contrast, a similar depletion of eIF3 subunits virtually eliminates polysomes and detectable translation. In addition, microarray analysis of polyribosomal mRNAs after eIF4G depletion revealed substantial alterations in translational efficiencies for only a fraction of cellular mRNAs (Park et al. 2011b). These results suggest that eIF4G is rate enhancing, rather than fundamentally crucial, for translation initiation on the large majority of yeast mRNAs in cells, and raise the question of how 43S PICs are directed to the 5′ ends of mRNAs in the absence of cap–eIF4F interaction. It is possible that the RNA-binding sites in yeast eIF4G, which impart a 5′ end dependence on eIF4F, might provide an additional means for directing PICs to the 5′ ends of mRNAs (Rajagopal et al. 2012). Additionally, perhaps the A + U bias and lack of strong secondary structure for the majority of yeast 5′ UTRs (Shabalina et al. 2004; Lawless et al. 2009) renders them intrinsically permissive for 43S attachment, albeit at rates significantly below those possible with eIF4F present. This conclusion is consonant with findings from the yeast reconstituted system, in which omitting eIF4G from reactions containing eIF3, eIF4A, and eIF4B (in addition to eIFs 1, 1A, and TC) reduced the rate of mRNA recruitment by 20-fold, but did not alter the end point of 48S PIC assembly, at least for one native mRNA tested (RPL41A), whereas no mRNA recruitment was observed without eIF3 (Mitchell et al. 2010). Interestingly, the group of mRNAs displaying the largest reductions in translational efficiencies in eIF4G-depleted yeast cells was among the most efficiently translated in wild-type cells and displayed shorter than average 5′-UTR lengths (Park et al. 2011b). Furthermore, none of the yeast mRNAs predicted to contain strong secondary structures in their 5′-UTRs (Lawless et al. 2009) were found to be unusually dependent on eIF4G, suggesting that another factor(s), possibly DEAD-box helicases Ded1 or Dbp1, can substitute for eIF4G to enable 43S attachment or scanning on structured 5′ UTRs in yeast.
The significant G + C bias of mammalian mRNAs (Shabalina et al. 2004) might be expected to impart a much stronger requirement for eIF4F for translation initiation. However, substantial siRNA-mediated depletion of both eIF4GI and eIF4GII simultaneously in mammalian cells had only a moderate effect on translation rates (Ramirez-Valle et al. 2008), and simultaneous depletion of eIF4GI and the eIF4G-like protein DAP5 left >30% of translation intact. The possibility that the residual pool of eIF4G was rendered more active by a compensatory reduction in a negative regulator of eIF4F seems unlikely because assembly of eIF4F was strongly reduced; however, it remains a possibility that depletion of eIF4GI, eIF4GII, and DAP5 simultaneously would more severely impact translation. These studies underscore the importance of examining the in vivo consequences of depleting initiation factors that are deemed to be essential for translation initiation purely on the basis of work performed using in vitro systems. They may also point again to a redundancy of function in the translational machinery, ensuring system failure cannot come from disruption at a single point.
SCANNING AND AUG RECOGNITION
Once the 43S PIC has been loaded onto the 5′ end of an mRNA it scans the 5′ UTR for the start codon, using complementarity with the anticodon of Met-tRNAi to identify the AUG. There are two key aspects of this process that, to some extent, are mechanistically distinct. The first concerns the factors that promote a conformation of the 43S PIC that is competent for threading along the mRNA with base-by-base inspection of the nucleotide sequence for an AUG in suitable context, and which trigger irreversible hydrolysis of GTP in the TC on AUG recognition (refer to Figs. 2 and 3). The second aspect, discussed further below, concerns the requirement to unwind secondary structure in the mRNA 5′ UTR to enable the mRNA to pass through the 40S mRNA entry channel in single-stranded form for base-by-base inspection in the P site. There is also the issue of how the 5′–3′ directionality of the scanning process is established.
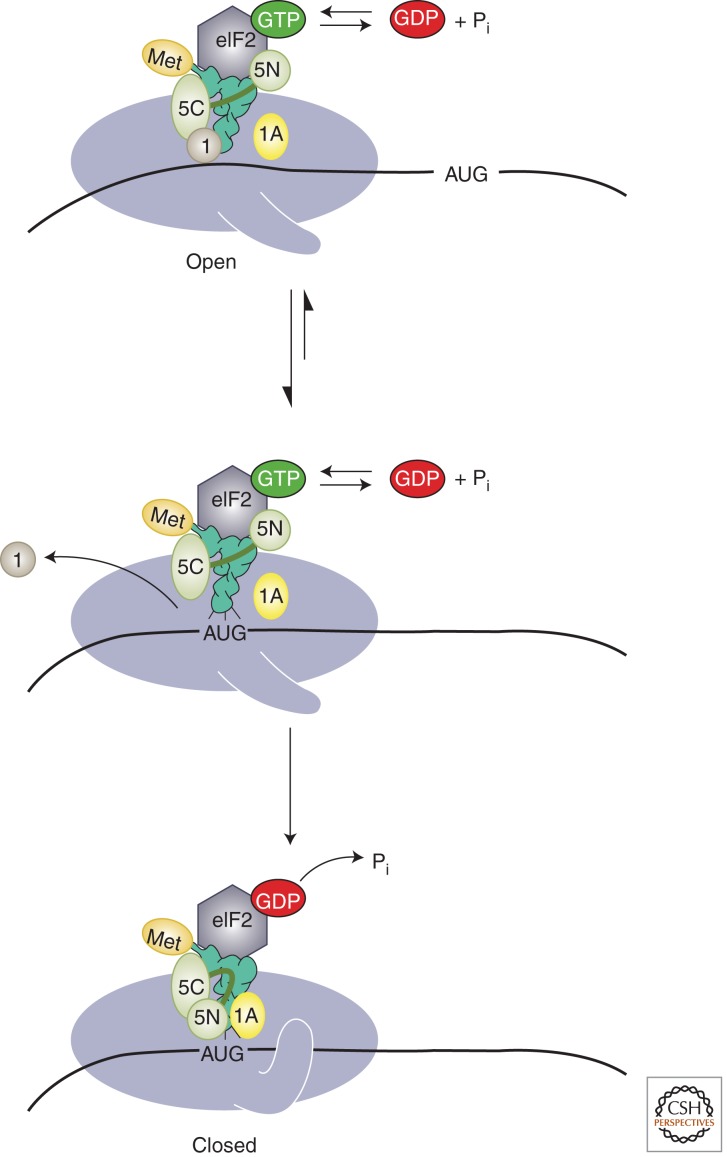
Figure 2.
Model of structural rearrangements in the PIC accompanying start codon recognition. (Top) Before start codon recognition, the PIC exists in an open conformation, promoted by eIF1 and eIF1A, which is capable of scanning the mRNA. (Middle) Base pairing between the anticodon of the initiator tRNA and the start codon promotes movement of the tRNA from the Pout to Pin states and release of eIF1 from the complex. (Bottom) Ejection of eIF1, in turn, triggers release of Pi from eIF2, converting it to its GDP-bound form. Because eIF1 stabilizes the open state of the PIC, its departure also results in conversion of the complex to the closed, scanning-arrested conformation (shown as the closure of a latch on the mRNA entry site). Release of eIF1 is promoted by eIF5, possibly by competition between one of eIF5’s domains (depicted here as the amino-terminal domain; 5N) and eIF1 for the same binding site in the PIC. Start codon recognition also induces an interaction between eIF1A and eIF5, which further stabilizes the closed state of the complex. (Modified from Hinnebusch 2011; reproduced, with permission.)
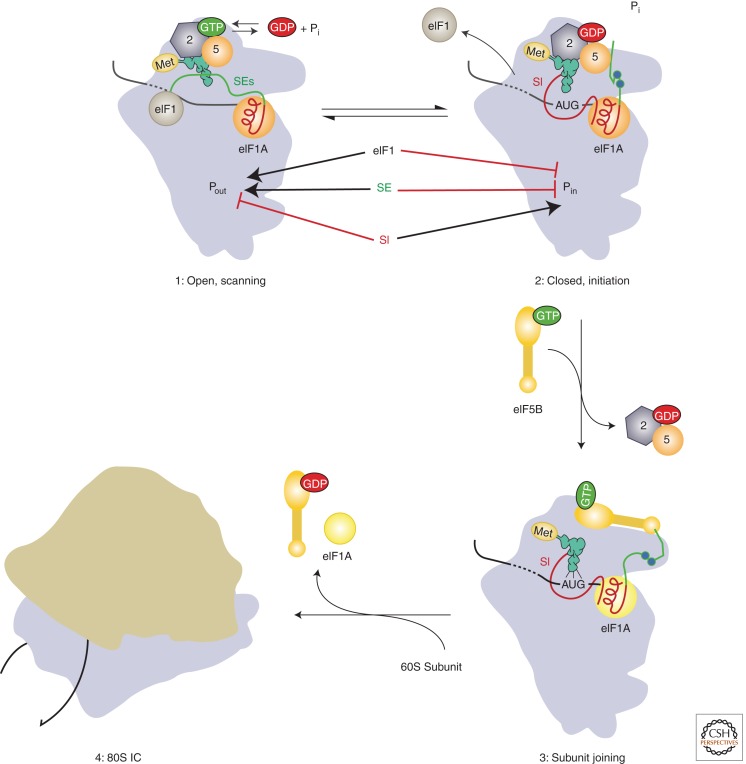
Figure 3.
Model of the roles of eIF1A’s amino- and carboxy-terminal tails in mediating start codon recognition and later steps in eukaryotic translation initiation. Before start codon recognition (complex 1) the amino- and carboxy-terminal tails (CTTs; shown in red and green, respectively) are both in the P site of the 40S subunit. On start codon recognition (complex 2), the initiator tRNA moves from the Pout to Pin state, causing both eIF1 and the CTT of eIF1A to be ejected from the P site. eIF1 stabilizes the open conformation of the PIC and the Pout state of the initiator tRNA. The scanning enhancer (SE) elements in the CTT of eIF1A (shown as blue balls) stabilize the open state of the PIC relative to the closed state. Conversely, the scanning inhibitor (SI) element in eIF1A’s NTT destabilizes the open state, thus promoting closed complex formation. The CTT of eIF1A may interact directly with eIF5 after start codon recognition, and it is hypothesized that this interaction triggers Pi release from eIF2. After Pi release and dissociation of eIF2·GDP and eIF5 from the PIC (complex 3), the CTT of eIF1A is free to interact with eIF5B·GTP, recruiting it to the complex and promoting subunit joining. Release of eIF5B·GDP and eIF1A from the resulting 80S IC produces the final translation-competent ribosome, poised at the start codon to commence decoding of the mRNA. (Modified from Hinnebusch 2011; reproduced, with permission.)
An Open and Shut Case: eIFs 1, 1A, and 5 Mediate Conformational Changes Required for Start Codon Recognition
Toe-printing experiments in the mammalian reconstituted system suggested that eIF1 and eIF1A stabilize an “open” conformation of the 43S PIC conducive to scanning, and that eIF1 impedes formation of a closed state required for progression to downstream steps in the pathway in a manner that is overcome efficiently only when an AUG in preferred sequence context occupies the P site (Pestova and Kolupaeva 2002). Subsequently, it was found that eIF1 is ejected from the PIC on start codon recognition (Maag et al. 2005), consistent with this proposal (Fig. 2). The structure of the Tetrahymena 40S subunit bound to eIF1 appears to explain the mechanism of eIF1 release on start codon recognition, as modeling of tRNA into the P site indicates that it sterically clashes with eIF1 (Rabl et al. 2011). These results support the notion that the anticodon end of the tRNA is not deeply bound in the P site during scanning (Pout state) and only fully engages in the site (Pin state) on codon:anticodon pairing (Yu et al. 2009; Saini et al. 2010), which in turn drives eIF1 out of the site owing to the steric clash.
Ejection of eIF1 triggers release of Pi from eIF2 in the PIC. GTP hydrolysis by eIF2 occurs nearly as fast before start codon recognition as after it, but Pi is only released rapidly once eIF1 has been ejected from the complex (Algire et al. 2005). The connection between eIF1 release and Pi release was shown by the fact that mutations in eIF1 that slow or speed up release of the factor from the complex correspondingly slow or speed up Pi release, which occurs at the same rate as eIF1 release in all cases (Algire et al. 2005; Cheung et al. 2007; Nanda et al. 2009). Supporting the central role of eIF1 as a gatekeeper in start codon recognition, substitutions in the factor that increase initiation at near-cognate UUG codons in vivo (Sui− phenotype) generally weaken eIF1 binding to 40S subunits and accelerate release of eIF1 and Pi from reconstituted PICs, whereas a substitution in eIF1A that suppresses UUG initiation (Ssu− phenotype) retards eIF1 dissociation in vitro (Cheung et al. 2007). In addition, overexpressing wild-type eIF1 suppresses UUG initiation in Sui− mutants (Valasek et al. 2004), consistent with a requirement for its release to trigger downstream events following initial start codon:anticodon pairing in the P site.
eIF1 and eIF1A bind directly and cooperatively to the 40S subunit (Maag and Lorsch 2003), with eIF1 occupying the platform near the P site (Lomakin et al. 2003; Rabl et al. 2011), and the globular (OB-fold) domain of eIF1A most likely occupying the A site (Yu et al. 2011) in the manner observed for its bacterial ortholog (IF1) (Carter et al. 2001). Cryo-EM analysis shows that when eIF1 and eIF1A are bound simultaneously to the 40S, they provoke a structural rearrangement that involves an open conformation of the “latch” of the mRNA entry channel, which is thought to render the PIC competent for scanning. In contrast, the 40S·eIF1A complex, which might be expected to resemble the PIC following eIF1 release, is in a closed state with the latch in a locked conformation that might impede scanning (Passmore et al. 2007). Importantly, eIF1 and eIF1A together stimulate the rate of TC binding to the 40S, but TC binds more tightly to the PIC in the absence of eIF1, leading to the deduction that TC binds to the “open” conformation of the PIC in a metastable state (Pout) conducive for scanning but incompatible with start codon recognition. On AUG recognition, the TC achieves a more stable interaction with the P site (Pin) through an isomerization that requires eIF1 dissociation and rearrangement to the closed 40S conformation (Passmore et al. 2007; Kolitz et al. 2009). The physical basis for the stimulatory effects of eIF1 and eIF1A and the open conformation of the 40S subunit on the rate of TC binding are not understood, although for eIF1A this function has been localized to its unstructured carboxy-terminal tail (CTT) (Saini et al. 2010). As described above, the crystal structure of the Tetrahymena 40S·eIF1 complex suggests that eIF1 may physically block full entry of the anticodon end of the tRNA into the P site (Rabl et al. 2011), preventing conversion to the Pin state before start codon recognition. Additional structural studies will be required to fully elucidate the modes of action of eIF1 and eIF1A in the conformational transitions associated with scanning and start codon recognition.
As mentioned above, the HEAT domain in the eIF5-CTD interacts with the eIF2β-NTD, which could help stabilize TC binding to the 43S PIC in the open (Pout) conformation. It appears that eIF5 also promotes rearrangement to the more stable Pin conformation on AUG recognition by enhancing the dissociation of eIF1 (Nanda et al. 2009). One possibility is that the eIF5-CTD stimulates eIF1 eviction by weakening one of the contacts that anchors eIF1 to the MFC. At least in yeast, however, it appears that the interactions of eIF1 and the eIF5-CTD with the eIF2β-NTD (Singh et al. 2004) and the eIF3c-NTD (Singh et al. 2004) are not mutually exclusive. Another possibility is that the eIF5 NTD facilitates ejection of eIF1 by competing with it for binding to a common site in the PIC—an idea prompted by eIF5-NTD’s structural similarity with eIF1 (Conte et al. 2006). The NTD of eIF5 interacts with eIF2γ via its eIF1-like folded region (Alone and Dever 2006) and also contains a long, unstructured amino-terminal tail (NTT) that includes the key arginine residue required for GTPase activation (Das et al. 2001; Paulin et al. 2001). Nanda et al. (2009) speculated that either the NTD or CTD of eIF5 might move into eIF1’s binding site in the PIC after the latter is released following start codon recognition. This movement could in turn affect the position of the NTT of eIF5 on eIF2γ, allowing Pi release from the TC (Figs. 2 and 3). An attractive feature of this model is that it provides a mechanism for coupling Pi release to eIF1 dissociation, as there is currently no evidence that eIF1 can interact with the eIF2γ G domain and block Pi release directly. Testing the models will require determination of which domain of eIF5 is responsible for promoting eIF1 release and data concerning the positions of eIF5’s domains in the PIC before and after start codon recognition.
Consistent with the role of eIF5 in promoting eIF1 dissociation from the PIC (Nanda et al. 2009), in both mammals and yeast, overexpressing eIF5 reduces the requirement for optimal context and AUG start codon for efficient initiation, whereas overexpressing eIF1 has the opposite effect (Nanda et al. 2009; Ivanov et al. 2010, 2011; Martin-Marcos et al. 2011). These activities are used to negatively autoregulate translation of eIF1 in yeast (Martin-Marcos et al. 2011) and both eIF1 and eIF5 in mammals (Ivanov et al. 2010; Loughran et al. 2012). High-level eIF1 decreases recognition of its own start codon, which has a poor context. High-level eIF5 expression increases initiation at uORFs in its mRNA leader (whose AUGs are in poor context), which then blocks initiation at the main eIF5 ORF. Overexpressing eIF1 has the opposite effect on eIF5 production by promoting leaky scanning of the upstream open reading frames (uORFs) and attendant increased recognition of the main ORF AUG, which has a favorable context. Thus, the opposing effects of eIF5 and eIF1 on accuracy of start codon selection underlie a network of auto- and cross-regulatory interactions that should stabilize the stringency of start codon selection.
At least in yeast, the NTT and CTT of eIF1A display opposing activities that enable the PIC to toggle between the open and closed 40S conformations (Fekete et al. 2007), and also the Pin and Pout modes of Met-tRNAi binding to the 40S. Two 10-amino acid repeats in the CTT, dubbed SE elements, cooperate with eIF1 to promote the open 40S conformation and accelerate TC loading in the Pout state (Saini et al. 2010). Based on directed hydroxyl radical cleavage-mapping, the (mammalian) eIF1A CTT occupies the P site in a manner incompatible with canonical P-site binding of Met-tRNAi (Yu et al. 2011). Hence, the CTT (with its SE elements) must presumably be ejected from the P site for isomerization to the stable Pin state upon AUG recognition. In this view, the SE elements resemble eIF1 in having to be removed from their binding site in the open 40S conformation to achieve the closed state. Consistent with this, mutations inactivating the SE elements show the same Sui− phenotype (elevated UUG:AUG initiation ratio) (Saini et al. 2010) produced by eIF1 mutations that provoke abnormally rapid eIF1 dissociation from the PIC (Cheung et al. 2007). As the eIF1A CTT functionally interacts with the eIF5 GAP domain on AUG recognition (Maag et al. 2006), it is attractive to envision that ejection of the CTT from the P site is stabilized by physical interaction with the eIF5-NTD (Fig. 3). The yeast eIF1A NTT and a helical domain adjacent to the CTT contain SI elements that appear to oppose SE function by destabilizing the open, Pout conformation, thereby promoting the closed, Pin configuration for AUG recognition. Accordingly, SI mutations suppress UUG initiation in Sui− mutants (Fekete et al. 2007; Saini et al. 2010). The eIF1A NTT also seems to occupy the P site, but should not clash with Met-tRNAi in the Pin state (Yu et al. 2011).
Genetics Brings It All Together: Other Factors Participate in Start Codon Recognition
In addition to eIF1 and eIF1A, the subunits of eIF2 and eIF5 have been implicated in stringent AUG recognition by the isolation of Sui− mutations (Donahue 2000). Biochemical analysis suggested that certain Sui− mutations in eIF2γ weaken Met-tRNAi binding to eIF2·GTP, which might allow inappropriate release of initiator tRNA into the P site at near-cognate start codons. More recently, however, other mutations in eIF2γ that weaken binding of Met-tRNAi have been found that increase rather than decrease the stringency of start codon recognition, suggesting that the orientation of tRNA binding to eIF2, rather than simply its affinity, may be crucial for proper start codon recognition (Alone et al. 2008). Sui− mutations in eIF2β, in contrast, appear to primarily elevate eIF5-independent GTP hydrolysis by the TC, and the SUI5 mutation was reported to increase the GAP function of eIF5 (Donahue 2000). Both of the latter effects might accelerate Pi release, and thereby enhance initiation, at near-cognate triplets. As the eIF2β Sui− mutations map to the zinc-binding domain (ZBD), and the ZBD likely interacts directly with the eIF2γ G domain (Yatime et al. 2007), the affected ZBD residues could control access of the GAP domain of eIF5 to the eIF2γ GTP-binding pocket or prevent Pi release at non-AUG codons. As noted above, in addition to promoting start codon selection by its GAP function, there is evidence that eIF5 also functions to stabilize the closed conformation of the PIC through functional interaction with eIF1A (Maag et al. 2006) and by accelerating eIF1 release (Nanda et al. 2009); however, the physical basis of these non-GAP activities is unknown. Their importance is indicated, however, by the fact that the G31R Sui− mutation (SUI5) in eIF5 completely inverts the preference for AUG and UUG codons in an assay measuring the effect of the factor on the open:closed complex equilibrium (Maag et al. 2006).
There is also genetic evidence that the interactions between eIF3c, eIF1, and eIF5 promote the opposing functions of the latter two factors in start codon recognition, as eIF3c-NTD mutations that weaken its binding to eIF1 or eIF5 confer Sui− or Ssu− phenotypes, respectively (Valasek et al. 2004). More work is needed to determine whether eIF1 binds simultaneously to the 40S subunit and the eIF3c-NTD to stabilize the open conformation, as both interactions seem to require helix α1 in eIF1 (Reibarkh et al. 2008; Rabl et al. 2011), or whether eIF1 toggles between these alternative binding sites at different stages of the initiation pathway (i.e., before and after start codon recognition). There is also evidence that a patch of basic residues on the surface of eIF1 that encompasses helix α2 (the “KH” region) interacts with the eIF2β-NTD and eIF5-CTD to promote the open conformation of the PIC (Cheung et al. 2007; Reibarkh et al. 2008); however, the relative importance of these interactions in maintaining 40S association of eIF1 in the scanning complex, or in stimulating eIF1 release at the start codon, remains to be determined. In addition, a mutation in eIF4G that reduces its interaction with eIF1 has been found to produce a weak Sui− phenotype in yeast, suggesting that eIF4G might also act to stabilize binding of eIF1 to the scanning PIC (He et al. 2003). Use of FRET probes on eIF1 and each of its potential binding partners might reveal the dynamics of its myriad interactions during the transition from the open to closed PIC conformations (Maag et al. 2005).
How Does the Sequence Context around the Start Codon Influence Initiation Site Recognition?
There is evidence that eIF2α mediates the stimulatory effect of a permissive sequence context surrounding the AUG codon and interacts with the −3 nucleotide of the mRNA, the key contextual determinant of AUG recognition (Pisarev et al. 2006). Interaction of eIF2α with the 40S E site, in which the −3 nucleotide would reside when AUG is in the P site, would require a substantial reorientation of eIF2α from its location in the 43S model of Shin et al. (Shin et al. 2011); however, crystal structures reveal that domains 1–2 of the aIF2α are highly flexible (Stolboushkina et al. 2008), so perhaps this is possible. eIF2β and eIFs 1 and 1A also function in discriminating against poor AUG context, and the same domains/residues in these factors involved in this activity also discriminate against non-AUG start codons, supporting the model that the AUG and context nucleotides act as a unit to stabilize the closed PIC conformation (Pisarev et al. 2006; Ivanov et al. 2010; Martin-Marcos et al. 2011). Although sequence context regulates AUG recognition in animals and yeast, with the exception of A at −3, the preferred nucleotides differ (Shabalina et al. 2004). In addition to residues immediately upstream of the AUG, it seems that a 5′ UTR of sufficient length to occupy the mRNA exit channel (∼12 nt) and extend some distance from the backside of the 40S is also required to stabilize the closed PIC conformation (Pestova and Kolupaeva 2002). It will be important to determine whether segments of 18S rRNA or ribosomal proteins in the 40S E site or mRNA exit channel (Rabl et al. 2011) interact directly with the context nucleotides and 5′-UTR sequences in a manner that regulates initiation efficiency or accuracy.
What Roles Do rRNA and tRNA Play in Start Codon Recognition and Scanning?
In addition to the various eIFs that regulate scanning and start codon recognition, there is evidence that specific residues in the 18S rRNA and in Met-tRNAi (beyond the anticodon) function in TC binding and AUG recognition. Substitutions that perturb the location or identity of the “bulge” residue in h28 of 18S rRNA decrease the rate and stability of TC binding to mutant 40S subunits in vitro. The bulge G contacts the +1 position of the P-site codon (A of AUG) in crystal structures of bacterial 70S·mRNA·tRNA complexes. Substitutions in other 18S residues predicted to contact the AUG or the ASL of Met-tRNAi also confer in vivo phenotypes indicating unstable TC binding to the scanning PIC or the PIC arrested at AUG (Dong et al. 2008). Thus, the contacts with the codon or ASL of P-site tRNA seen in bacterial elongation complexes are likely also critical for stabilizing Met-tRNAi binding to eukaryotic PICs. Genetic analysis also implicated A1193 in the h31 loop, located directly below the codon–anticodon duplex in bacterial 70S complexes, and G875 in h22, in stabilizing Met-tRNAi binding at the AUG in the closed complex (Pin), either directly (A1193U) or indirectly by reducing the 40S association of eIFs that promote TC binding (G875A) (Nemoto et al. 2010). Further structural studies are required to determine whether bases or proteins in the 40S subunit itself play a direct role in responding to codon–anticodon pairing in the P site, as bacterial bases A1492, A1493, and G530 do in the A site during tRNA selection in the elongation phase of protein synthesis.
tRNAiMet contains highly conserved bases (identity elements) that are not present in elongator tRNAs (Marck and Grosjean 2002). These identity elements appear to play important roles in allowing specific binding of the initiator tRNA by initiation factors (e.g., eIF2) and preventing its binding to elongation factors. They may also be involved in transmitting the signal that the start codon has been located from initial codon:anticodon pairing to the rest of the PIC, promoting downstream events. The ASL of tRNAi contains three consecutive G:C pairs that are conserved from bacteria to eukaryotes. In yeast, changing the first and third of these G:C pairs to their identities in elongator tRNAeMet, eliminates the strong thermodynamic coupling between TC binding to the PIC and an AUG codon in the mRNA (Kapp et al. 2006), suggesting a role for these bases in start codon recognition. Base-pair substitutions at these positions also destabilized mammalian 48S PICs, at least following eIF5-stimulated GTP hydrolysis in the TC (Lomakin et al. 2006).
Strikingly, substitution of two other identity bases in eukaryotic tRNAMetiMet, A54, and A60 in the T loop, with the cognate elongator tRNA residues was found to suppress the loss of coupling between the AUG codon and TC in binding to the PIC caused by the elongator/initiator base swap at the third G:C pair in the ASL (G31:C39; Kapp et al. 2006). It was suggested that the A54,A60 replacements might lower the energy barrier to a structural rearrangement of Met-tRNAi necessary for its full accommodation in the P site (i.e., Pin state), and thereby compensate for the effect of the G31:C39 replacement, which may decrease the ability of the tRNA to fully engage in the P site (Kapp et al. 2006). In this view, structural features of the tRNAiMet contribute with eIF1 and the SE elements in the eIF1A CTT to block the rearrangement from the Pout to Pin state in the absence of a perfect AUG duplex in the P site. Testing the hypothesis that the tRNAiMet body plays an active role in the proper response to start codon recognition will require additional mutagenesis studies to probe the functions of the unique structural elements in the initiator tRNA.
N6-threonylcarbamoyl modification of A37 (t6A37) of tRNAiMet, immediately adjacent to the anticodon triplet, is thought to stabilize the first base pair of the codon:anticodon duplex for ANN or UNN codons (Agris 2008), which includes decoding of AUG by tRNAiMet. Consistent with this, inactivation or elimination of yeast proteins that catalyze t6A37 formation, including Sua5 (El Yacoubi et al. 2009; Lin et al. 2009) and subunits of the EKC/KEOPS complex Kae1 and Pcc1 (Daugeron et al. 2011; El Yacoubi et al. 2011; Srinivasan et al. 2011) evokes phenotypes indicating impaired recognition of AUG codons by the scanning 43S PIC.
Untangling the RNA Helicases Involved in Translation Initiation
As mentioned above, in addition to achieving a ribosomal conformation conducive to scanning, there is also a need to remove secondary structures to allow the mRNA to pass through the 40S mRNA entry or exit channels and permit its base-by-base inspection in the P site. In addition, assuming scanning is a directional process (5′ to 3′) one or more factors must transduce the energy required to impart this directionality, presumably using ATP hydrolysis. By analyzing the effect of increasing the 5′ UTR length on the time required to complete the first round of translation of a reporter mRNA in cell extracts, it was concluded that scanning occurs at a frequency of 6–8 bases/sec in wheat germ and mammalian cell extracts, even in the presence of a strong secondary structure in the 5′ UTR (Vassilenko et al. 2011), which is similar to the figure of ∼10/sec calculated for yeast extracts (Berthelot et al. 2004). The linear relationship between the translation time and 5′-UTR length implies a substantial bias toward 5′–3′ movement at each base; although a certain amount of backward scanning cannot be excluded (Berthelot et al. 2004; Vassilenko et al. 2011). The fact that increasing leader length does not reduce translational efficiency in yeast cells was taken to indicate that multiple PICs can load and scan simultaneously on long 5′ UTRs to compensate for the increased time required for each PIC to reach the AUG codon in extended leaders (Berthelot et al. 2004).
Multiple DEAD-box RNA helicases have been implicated in translation initiation, including eIF4A, Dhx29, and Ded1/Ddx3. Although it is not fully understood which ones participate in scanning, there is increasing evidence that eIF4A stimulates both ribosome attachment at the 5′ end and scanning through structured 5′ UTRs, whereas Dhx29 and Ded1 act primarily in scanning and are particularly important for negotiating long or highly structured 5′ UTRs.
A model of the scanning complex that takes into account much of the accumulated data regarding the role and position of eIF4F has been presented by Marintchev and colleagues. In this model, eIF4G spans the mRNA exit and entry channels on the 40S, positioning eIF4A and eIF4B at the entry channel for unwinding structure ahead of the ribosome. eIF4F can remain engaged with the cap as scanning proceeds, with the scanned nucleotides forming a loop between the cap-eIF4F assembly and the ribosome (Marintchev et al. 2009). The location of the HEAT-1 domain of eIF4G in this model, below the 40S platform, differs from that predicted from a cryo-EM reconstruction (Siridechadilok et al. 2005), but it is consistent with hydroxyl radical cleavage mapping of eIF4G HEAT-1 in reconstituted mammalian 43S PICs (Yu et al. 2011).
The notion that the eIF4 factors are positioned on the downstream side of scanning PICs needs to take into account the lack of evidence for factors stably associated with the mRNA 3′ of native 48S complexes, whereas residues 5′ of the ribosome are protected from RNAse digestion (Spirin 2009). The latter is more compatible with eIF4G working exclusively at the mRNA exit channel to “pull” the mRNA through the 40S subunit (Siridechadilok et al. 2005). Spirin has suggested that, from this location, eIF4G could deliver eIF4B–eIF4A·ATP complexes to single-stranded mRNA as it emerges from the exit channel, and the eIF4B would remain bound to the mRNA and block backward movement by the PIC. When the ribosome moves again, the cycle would repeat itself, constituting a “Brownian ratchet” with eIF4B as the pawl (Spirin 2009). It is also possible that unwinding takes place at the entrance channel and RNA structures reannealing after emerging from the exit channel serve to prevent backward motion of the PIC and confer 5′–3′ directionality. There is not enough experimental evidence currently available to reject or select any one of these models of the scanning PIC.
Genetic evidence from yeast indicates that the essential Ded1/Ddx3 protein cooperates with eIF4F in translation initiation (Hinnebusch 2011), and suggests that Ded1 and Dbp1 are more critical than eIF4A for efficient scanning through a long 5′ UTR in vivo (Berthelot et al. 2004). Using the reconstituted mammalian system, it was found that Dhx29, and to a lesser extent yeast Ded1, enable scanning through highly stable stem loops in a manner that is not possible with eIF4F alone. Dhx29 and Ded1 could not replace eIF4F for initiation on the β-globin 5′ UTR, which was attributed to their inability to stimulate 43S PIC attachment to an mRNA with cap-proximal structure. Thus, Dhx29 and Ded1 might specifically stimulate scanning through secondary structures, whereas eIF4F would enhance both 43S attachment and scanning through secondary structures of moderate stability (Pisareva et al. 2008; Abaeva et al. 2011). Considering the substantial reduction in total protein synthesis evoked by knockdown of Dhx29 in mammalian cells (Parsyan et al. 2009), it seems likely that Dhx29 promotes translation of a large fraction of the cell’s mRNAs and not only those burdened with highly stable 5′-UTR structures.
SUBUNIT JOINING
After the start codon has been recognized, the remaining factors must dissociate from the complex and the 60S subunit must join with the 40S subunit to form the final 80S IC with Met-tRNAi and the start codon in the P site. Subunit joining is facilitated by a second GTPase initiation factor, eIF5B (Pestova et al. 2000), which is the ortholog of the bacterial translation initiation factor IF2. Below we describe the current state of knowledge of these final stages of eukaryotic translation initiation.
As discussed above, start codon recognition results in conversion of eIF2 to its GDP-bound state, which has a low affinity for Met-tRNAi (Kapp and Lorsch 2004). eIF2·GDP must dissociate from the tRNA and 40S subunit before 60S subunit joining. Experiments in the reconstituted mammalian system showed that the presence of eIF5B and 60S subunits enhanced release of eIF2·GDP from PICs following start codon recognition, most likely by driving the equilibrium toward dissociation by capturing the vacated 40S subunits as 80S complexes (Unbehaun et al. 2004). Whether eIF5B or the 60S subunit actively accelerates release of eIF2·GDP is not yet clear. It seems likely that eIF5, which binds tightly to eIF2 in both its GTP- and GDP-bound forms (Algire et al. 2005; Singh et al. 2007), leaves the PIC along with eIF2. Determining whether eIF2·GDP dissociates on its own from the 40S complex after start codon recognition or if its release is actively promoted by an initiation factor or the 60S subunit will require the development of new kinetic assays for this step in the initiation pathway. Mutations in eIF2 that reduce subunit joining, which should produce constitutive derepression of GCN4 translation (Martin-Marcos et al. 2007) without decreasing TC occupancy of 43S PICs, would also be useful to probe factor release from the PIC.
After eIF2·GDP dissociation, eIF5B·GTP binds to the complex and accelerates the rate of 60S subunit joining (Acker et al. 2006, 2009). The ability of eIF5B to facilitate this latter step requires an interaction between the extreme carboxyl terminus of eIF1A (DIDDI) and the CTD of eIF5B (Marintchev et al. 2003; Olsen et al. 2003; Acker et al. 2006; Fringer et al. 2007). The requirement for this interaction underscores the importance of eIF1A and, in particular, its CTT, in the initiation pathway. Before start codon recognition, the CTT is bound in the P site, which presumably sequesters the DIDDI sequence, preventing it from recruiting eIF5B to the complex prematurely. When start codon recognition occurs, the CTT is ejected from the P site. As outlined above, it may then be involved in an interaction with eIF5 that serves to promote Pi release from eIF2. Once eIF2·GDP and eIF5 dissociate from the PIC, the DIDDI end of the CTT is freed to recruit eIF5B·GTP to the complex and promote joining of the 60S subunit. This series of events suggests that the CTT of eIF1A is a key controller of the timing of the steps in the initiation pathway (Fig. 3).
After subunit joining, eIF5B hydrolyzes its bound GTP, which lowers its affinity for the 80S IC and triggers its release (Lee et al. 2002; Shin et al. 2002, 2007). Release of eIF1A occurs only after dissociation of eIF5B from the 80S IC, making eIF1A the only factor to remain on the ribosome during the entire initiation pathway, with the possible exception of eIF3 (Acker et al. 2009). GTP hydrolysis by eIF5B appears to alter the conformation of the 80S IC, as release of eIF1A occurs slowly in the presence of a mutant form of eIF5B that does not hydrolyze GTP but has weakened affinity for the ribosome and thus can still dissociate after subunit joining (eIF5B-T439A,H505Y). A role for eIF5B GTP hydrolysis in altering the final conformation of the IC is consistent with recent single-molecule FRET studies in the bacterial system that indicated a similar role for GTP hydrolysis by the orthologous factor IF2 (Marshall et al. 2009), and with the leaky scanning and Gcn− phenotypes produced in yeast expressing the eIF5B-T439A,H505Y mutant (Shin et al. 2002).
eIF1A has been reported to be the lowest abundance initiation factor in yeast cells (von der Haar and McCarthy 2002). This observation might be explained by the fact that eIF1A still binds with reasonable affinity to the final 80S IC (Acker et al. 2009) and presumably could compete with incoming eEF1A·GTP·aa-tRNA complexes for binding to the A site if present at high concentrations. In addition, the central role played by the CTT of eIF1A in all stages of initiation might also have put selective pressure on cells to keep the factor’s concentration low to prevent the formation of spurious interactions that could prematurely trigger transition from one stage of initiation to another.
Although we now know a considerable amount about these late events in translation initiation, a number of important mysteries remain. We do not know when eIF5B interacts with the 60S subunit. Does it bind to it before it interacts with the PIC and carry it along like a tugboat pulling a ship or does it bind first to the PIC and then wait for an encounter with the 60S to dock the two together? We also know very little about how eIF5B and its interaction with eIF1A accelerate the rate of subunit joining. Understanding the molecular mechanics of this process will require additional structural studies of the 80S IC bound to eIF5B before and after GTP hydrolysis as well as additional mutagenesis and kinetics experiments to elucidate the key regions of each component required to accelerate subunit joining.
FUTURE PROSPECTS
The combination of in vitro and in vivo studies used by the field over the past 10 years has allowed dramatic advances in our understanding of the roles of the individual components of the translation initiation machinery. We have also begun to gain an understanding of what different regions of each of these components do. Obtaining a clearer picture of the molecular mechanics of this critical stage of gene expression will require the continued coupling of increasingly sophisticated in vitro and in vivo approaches, including additional advances in structural biology (Ben-Shem et al. 2011) and kinetic and thermodynamic measurements, forays into single-molecule studies, and genome-wide analyses such as ribosome profiling (Ingolia et al. 2009). The focus will be on understanding how the parts work together to coordinate and facilitate the complex interactions and rearrangements required for the assembly of the 80S IC on an mRNA, poised to enter the elongation phase of protein synthesis.
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
- Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV 2011. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J 30: 115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Dever TE, Lorsch JR 2006. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem 281: 8469–8475 [DOI] [PubMed] [Google Scholar]
- Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR 2009. Kinetic analysis of late steps of eukaryotic translation initiation. J Mol Biol 385: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF 2008. Bringing order to translation: The contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Savio P, Acker MG, Tarun SZ Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, et al. 2002. Development and characterization of a reconstituted yeast translation initiation system. RNA 8: 382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Lorsch JR 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262 [DOI] [PubMed] [Google Scholar]
- Alone PV, Dever TE 2006. Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B ε. J Biol Chem 281: 12636–12644 [DOI] [PubMed] [Google Scholar]
- Alone PV, Cao C, Dever TE 2008. Translation initiation factor 2γ mutant alters start codon selection independent of Met-tRNA binding. Mol Cell Biol 28: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG 2000. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev 14: 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valášek L, Donahue TF, Hinnebusch AG 2001. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J 20: 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Berset C, Zurbriggen A, Djafarzadeh S, Altmann M, Trachsel H 2003. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA 9: 871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JE 2004. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol Microbiol 51: 987–1001 [DOI] [PubMed] [Google Scholar]
- Bi X, Ren J, Goss DJ 2000. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 16: 5758–5765 [DOI] [PubMed] [Google Scholar]
- Bulygin KN, Khairulina YS, Kolosov PM, Ven’yaminova AG, Graifer DM, Vorobjev YN, Frolova LY, Kisselev LL, Karpova GG 2010. Three distinct peptides from the N domain of translation termination factor eRF1 surround stop codon in the ribosome. RNA 16: 1902–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501 [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci 97: 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG 2010. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol Cell Biol 30: 4415–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MR, Kelly G, Babon J, Sanfelice D, Youell J, Smerdon SJ, Proud CG 2006. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry 45: 4550–4558 [DOI] [PubMed] [Google Scholar]
- Das S, Ghosh R, Maitra U 2001. Eukaryotic translation initiation factor 5 functions as a GTPase- activating protein. J Biol Chem 276: 6720–6726 [DOI] [PubMed] [Google Scholar]
- Daugeron MC, Lenstra TL, Frizzarin M, El Yacoubi B, Liu X, Baudin-Baillieu A, Lijnzaad P, Decourty L, Saveanu C, Jacquier A, et al. 2011. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res 39: 6148–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MD, Person MD, Browning KS 2009. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J Biol Chem 284: 20615–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev K, Qiu H, Dong J, Zhang F, Barthlme D, Hinnebusch AG 2010. The β/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol Cell Biol 30: 5218–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Dar AC, Sicheri F 2007. The elF2a kinases. In Translational control in biology and medicine (ed. Mathews M, Sonenberg N, Hershey JWB), pp. 319–344 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN 2003. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol Cell Biol 23: 8925–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T 2000. Genetic approaches to translation initiation in Saccharomyces cerevisiae. In Translational control of gene expression (ed. Sonenberg N, Hershey JWB, Mathews MB), pp. 487–502 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Dong J, Nanda JS, Rahman H, Pruitt MR, Shin BS, Wong CM, Lorsch JR, Hinnebusch AG 2008. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes Dev 22: 2242–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V 2009. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V 2011. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30: 882–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini A, Lorsch JR, Hinnebusch AG 2007. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J 26: 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE 2007. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol Cell Biol 27: 2384–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E, Pavitt GD 2000. Identification of domains and residues within the ε subunit of eukaryotic translation initiation factor 2B (eIF2b) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol Cell Biol 20: 3965–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez E, Mohammad SS, Pavitt GD 2002. Characterization of the minimal catalytic domain within eIF2B: The guanine-nucleotide exchange factor for translation initiation. EMBO J 21: 5292–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K 2003. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol 23: 5431–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JWB, Merrick WC 2000. Pathway and mechanism of initiation of protein synthesis. In Translational control of gene expression (ed. Sonenberg N, Hershey JWB, Mathews MB), pp. 33–88 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Hilbert M, Kebbel F, Gubaev A, Klostermeier D 2011. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res 39: 2260–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A, Gao Z, Jankowsky E, Parker R 2011. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell 43: 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Dever TE, Asano K 2007. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. In Translational control in biology and medicine (ed. Mathews M, Sonenberg N, Hershey JWB), pp. 225–268 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM 2007. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem 282: 1695–1708 [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Pavitt GD 2010. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465: 378–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH 2006. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol Cell Biol 26: 1355–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR 2004. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol 335: 923–936 [DOI] [PubMed] [Google Scholar]
- Kapp LD, Kolitz SE, Lorsch JR 2006. Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation. RNA 12: 751–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye NM, Emmett KJ, Merrick WC, Jankowsky E 2009. Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J Biol Chem 284: 17742–17750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30: 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Takacs JE, Lorsch JR 2009. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15: 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva NL, Lamphear BJ, Hennigan FL, Rhoads RE 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J Biol Chem 275: 41369–41376 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy T, Pavitt GD, Zhang F, Dever TE, Hinnebusch AG 2001. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol Cell Biol 21: 5018–5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Pavitt GD, Hubbard SJ 2009. Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics 10: 7–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Pestova TV, Shin BS, Cao C, Choi SK, Dever TE 2002. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc Natl Acad Sci 99: 16689–16694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE 2006. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J Biol Chem 281: 22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CA, Ellis SR, True HL 2009. The Sua5 protein is essential for normal translational regulation in yeast. Mol Cell Biol 30: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist L, Imataka H, Pelletier J 2008. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA 14: 960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E 2008. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci 105: 20209–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV 2006. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE 2010. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Lorsch JR 2003. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol 330: 917–924 [DOI] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275 [DOI] [PubMed] [Google Scholar]
- Maag D, Algire MA, Lorsch JR 2006. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol 356: 724–737 [DOI] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U 2003. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem 278: 6580–6587 [DOI] [PubMed] [Google Scholar]
- Marck C, Grosjean H 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G 2003. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: A new interaction between old partners. Proc Natl Acad Sci 100: 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Edmonds K, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136: 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Puglisi JD 2009. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell 35: 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Hinnebusch AG, Tamame M 2007. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining and repression of GCN4 translation. Mol Cell Biol 27: 5968–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Cheung YN, Hinnebusch AG 2011. Functional elements in initiation factors 1, 1A and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol 31: 4814–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR 2010. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and block an alternative pathway. Mol Cell 39: 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol 394: 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveau M, Lazennec-Schurdevin C, Panvert M, Mechulam Y, Schmitt E 2010. tRNA binding properties of eukaryotic translation initiation factor 2 from Encephalitozoon cuniculi. Biochemistry 49: 8680–8688 [DOI] [PubMed] [Google Scholar]
- Nemoto N, Singh CR, Udagawa T, Wang S, Thorson E, Winter Z, Ohira T, Ii M, Valasek L, Brown SJ, et al. 2010. Yeast 18 S rRNA is directly involved in the ribosomal response to stringent AUG selection during translation initiation. J Biol Chem 285: 32200–32212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Szamecz B, Valasek L, Jivotovskaya A, Shin BS, Hinnebusch AG 2004. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J 23: 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Behrens MA, He Y, Oliveira CL, Sottrup Jensen L, Hoffmann SV, Pedersen JS, Andersen GR 2011. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res 39: 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberer M, Marintchev A, Wagner G 2005. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev 19: 2212–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DS, EM S, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG 2003. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J 22: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozes AR, Feoktistova K, Avanzino BC, Fraser CS 2011. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J Mol Biol 412: 674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Walker S, Lee J, Rothenburg S, Lorsch J, Hinnebusch A 2011a. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1 PABP mRNPs in vivo. EMBO J 30: 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG 2011b. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics 12: 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, Mathonnet G, Tettweiler G, Hellen CU, Pestova TV, et al. 2009. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc Natl Acad Sci 106: 22217–22222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N 2011. mRNA helicases: The tacticians of translational control. Nat Rev Mol Cell Biol 12: 235–245 [DOI] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Paulin FE, Campbell LE, O’Brien K, Loughlin J, Proud CG 2001. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr Biol 11: 55–59 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CUT 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CUT 2007. The mechanism of translation initiation in eukaryotes. In Translational control in biology and medicine (ed. Mathews M, Sonenberg N, Hershey JWB), pp. 87–128 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV 2006. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV 2008. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J 27: 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736 [DOI] [PubMed] [Google Scholar]
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR 2012. Specific domains in yeast eIF4G strongly bias the RNA unwinding activity of the eIF4F complex towards duplexes with 5′-overhangs. J Biol Chem 10.1074/jbc.M112.347278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ 2008. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 181: 293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K 2008. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem 283: 1094–1103 [DOI] [PubMed] [Google Scholar]
- Rozovsky N, Butterworth AC, Moore MJ 2008. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 14: 2136–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG 2010. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev 24: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Naveau M, Mechulam Y 2010. Eukaryotic and archaeal translation initiation factor 2: A heterotrimeric tRNA carrier. FEBS Lett 584: 405–412 [DOI] [PubMed] [Google Scholar]
- Schmitt E, Panvert M, Lazennec-Schurdevin C, Coureux PD, Perez J, Thompson A, Mechulam Y 2012. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat Struct Mol Biol 19: 450–454 [DOI] [PubMed] [Google Scholar]
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U 2008. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci 105: 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300 [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Rogozin IB, Koonin EV, Lipman DJ 2004. Comparative analysis of orthologous eukaryotic mRNAs: Potential hidden functional signals. Nucleic Acids Res 32: 1774–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Maag D, Roll-Mecak A, Arefin MS, Burley SK, Lorsch JR, Dever TE 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111: 1015–1025 [DOI] [PubMed] [Google Scholar]
- Shin BS, Acker MG, Maag D, Kim JR, Lorsch JR, Dever TE 2007. Intragenic suppressor mutations restore GTPase and translation functions of a eukaryotic initiation factor 5B switch II mutant. Mol Cell Biol 27: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE 2011. Initiation factor eIF2γ promotes eIF2-GTP-Met-tRNA(i)(Met) ternary complex binding to the 40S ribosome. Nat Struct Mol Biol 18: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, He H, Ii M, Yamamoto Y, Asano K 2004. Efficient incorporation of eukaryotic initiation factor 1 into the multifactor complex is critical for formation of functional ribosomal preinitiation complexes in vivo. J Biol Chem 279: 31910–31920 [DOI] [PubMed] [Google Scholar]
- Singh CR, Curtis C, Yamamoto Y, Hall NS, Kruse DS, He H, Hannig EM, Asano K 2005. Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol Cell Biol 25: 5480–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J 25: 4537–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Udagawa T, Lee B, Wassink S, He H, Yamamoto Y, Anderson JT, Pavitt GD, Asano K 2007. Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J Mol Biol 370: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JW 2011. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res 40: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS 2009. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian ratchet model. Biochemistry 48: 10688–1069219835415 [Google Scholar]
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R 2011. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolboushkina E, Nikonov S, Nikulin A, Blasi U, Manstein DJ, Fedorov R, Garber M, Nikonov O 2008. Crystal structure of the intact archaeal translation initiation factor 2 demonstrates very high conformational flexibility in the α- and β-subunits. J Mol Biol 382: 680–691 [DOI] [PubMed] [Google Scholar]
- Sun C, Todorovic A, Querol-Audí J, Bai Y, Villa N, Snyder M, Ashchyan J, Lewis CS, Hartland A, Gradia S, et al. 2011. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc Natl Acad Sci 108: 20473–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin Y, Pause A, Haghighat A, Pyronnet S, Witherell GW, Belsham G, Sonenberg N 2001. The requirement for eukaryotic initiation factor 4A (eIF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Evdokimova VM, Brasey A, Pestova TV, Fantus D, Yanagiya A, Imataka H, Skabkin MA, Ovchinnikov LP, Merrick WC, et al. 2009. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J 28: 58–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valasek L 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22: 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Wells SE, Deardorff JA, Sachs AB 1997. Translation initiation factor eIF4G mediates in vitro poly (A) tail-dependent translation. Proc Natl Acad Sci 94: 9046–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Nielsen KH, Hinnebusch AG 2002. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J 21: 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Mathew A, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG 2003. The Yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol 24: 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilenko KS, Alekhina OM, Dmitriev SE, Shatsky IN, Spirin AS 2011. Unidirectional constant rate motion of the ribosomal scanning particle during eukaryotic translation initiation. Nucleic Acids Res 39: 5555–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, McCarthy JE 2002. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol 46: 531–544 [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N 2009. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol 29: 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatime L, Mechulam Y, Blanquet S, Schmitt E 2006. Structural switch of the γ subunit in an archaeal aIF2αγ heterodimer. Structure 14: 119–128 [DOI] [PubMed] [Google Scholar]
- Yatime L, Mechulam Y, Blanquet S, Schmitt E 2007. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc Natl Acad Sci 104: 18445–18450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV 2009. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res 37: 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Abaeva IS, Marintchev A, Pestova TV, Hellen CU 2011. Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res 39: 4851–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]