Summary
Spinal muscular atrophy (SMA) is a lethal human disease characterized by motor neuron dysfunction and muscle deterioration due to depletion of the ubiquitous Survival Motor Neuron (SMN) protein. Drosophila SMN mutants have reduced muscle size and defective locomotion, motor rhythm and motor neuron neurotransmission. Unexpectedly, restoration of SMN in either muscles or motor neurons did not alter these phenotypes. Instead, SMN must be expressed in proprioceptive neurons and interneurons in the motor circuit to non-autonomously correct defects in motor neurons and muscles. SMN depletion disrupts the motor system subsequent to circuit development and can be mimicked by the inhibition of motor network function. Furthermore, increasing motor circuit excitability by genetic or pharmacological inhibition of K+ channels can correct SMN-dependent phenotypes. These results establish sensory-motor circuit dysfunction as the origin of motor system deficits in this SMA model and suggest that enhancement of motor neural network activity could ameliorate the disease.
Introduction
Animal behaviors such as locomotion depend upon the coordinated activity of neuronal networks. Disruption of individual components of neural circuits by injury or disease can produce a cascade of deleterious secondary effects upon other networked neurons. It has been hypothesized that the chronic dysfunction of neuronal circuits may ultimately lead to degeneration of neurons within the network, both exacerbating the damage and masking the primary cause of the disorder (Palop and Mucke, 2010). Identifying the molecular, cellular and physiological basis of disease is central to understanding the adult motor neuron disorder amyotrophic lateral sclerosis (ALS) (Rothstein, 2009) and the juvenile disease spinal muscular atrophy (SMA) (Burghes and Beattie, 2009). The genetics of ALS are complex with only a minority of cases due to the inheritance of mutations in a diverse range of genes. By contrast, SMA, the most common inherited cause of infant mortality (Pearn, 1978), is both recessive and monogenic. Both ALS and SMA are characterized by motor neuron functional alterations and degeneration, which has focused research on cell autonomous changes in motor neurons themselves. However, recent studies of ALS mouse models have identified contributions of other spinal cord cells such as astrocytes to disease pathology, suggesting that interactions between motor neurons and other partner cells may be an important contributing factor to motor neuron disease (Ilieva et al., 2009).
SMA is caused by recessive mutations in the survival motor neuron 1 (SMN1) gene (Lefebvre et al., 1995) that are not compensated for by SMN2, a human-specific gene (Burghes and Beattie, 2009). While SMN2 is almost identical to SMN1, nucleotide differences alter its splicing pattern resulting in a ~90% reduction of full-length SMN mRNA expression (Lorson et al., 1999; Monani et al., 1999). Therefore SMA is caused by low levels of SMN as opposed to the complete loss of SMN (Burghes and Beattie, 2009). SMN is a multifunctional protein that has been implicated in a variety of cellular processes linked to RNA metabolism (Pellizzoni, 2007). SMN is ubiquitously expressed and has been highly conserved during evolution (Schmid and DiDonato, 2007). In genetic model organisms, complete removal of SMN results in loss of cell viability. In contrast, the reduced level of SMN found in SMA patients does not appear to significantly perturb the majority of organ systems (Crawford and Pardo, 1996). However, SMA patients develop motor problems and muscle weakness, with the proximal limb and trunk muscles stereotypically the most severely affected, progressing eventually to respiratory insufficiency and death (Swoboda et al., 2005). Postmortem studies show that SMA patients have pathologically abnormal motor neurons and evidence of motor neuron loss (Simic, 2008), however it is currently unclear if this the primary origin of motor system dysfunction or a terminal consequence.
Reduction of SMN to low levels, similar to the situation in human SMA, has been studied using several animal models (Schmid and DiDonato, 2007). In the SMA mouse model SMN-Δ7, death occurs two weeks after birth (Le et al., 2005). This is accompanied by a modest loss of motor neurons however these mice have a profound early impairment of motor behavior well before this loss occurs (Park et al., 2010a). Examination of the neuromuscular junctions (NMJ) of SMN-Δ7 mice reveals that most terminals are innervated though some have structural abnormalities (Kariya et al., 2008; Kong et al., 2009; Ling et al., 2011; McGovern et al., 2008). NMJ neurotransmission is aberrant in these mutants with a ~50% reduction in quantal content (Kariya et al., 2008; Kong et al., 2009), however the high safety factor of the neuromuscular junction means these terminals still produce normal muscle twitch tension (Ling et al., 2010). Therefore, given the early severe defects observed in the motor behavior of SMA mice, the disruption of motor neuron function is surprisingly modest.
SMN mutant mice have been used to grossly assess the tissue requirements for SMN by both selective SMN rescue and depletion experiments. Expression of transgenic SMN throughout the nervous system and some muscles (Gavrilina et al., 2008) or through CNS virus delivery (Passini et al., 2010) gives a robust rescue of SMN-Δ7 motor behavior and survival, while muscle specific SMN expression does not (Gavrilina et al., 2008). In contrast, selective genetic reduction of SMN in the motor neurons of mice is not lethal, though NMJ structural and electrophysiological abnormalities are observed (Park et al., 2010b) and genetic restoration of SMN in motor neurons alone does not rescue mutant lethality (Gogliotti et al., 2012; Martinez et al., 2012). In addition to the motor neuron defects, pronounced early deficits of spinal reflexes and reduced numbers of proprioceptive synaptic inputs onto motor neurons have recently been described in SMN-Δ7 mice, although functional contribution of these changes to the SMA phenotype is not yet known (Ling et al., 2010; Mentis et al., 2011). Collectively, these findings suggest the possibility that neurons other than motor neurons could contribute to the motor deficits in SMA.
Here we have exploited Drosophila mutants with low levels of SMN to determine the essential cellular site and requirement for SMN in the motor system of this model. Using previously described loss-of-function smn mutants (Chan et al., 2003; Chang et al., 2008; Rajendra et al., 2007), we confirmed that depletion of SMN in Drosophila results in reduced muscle growth and defective locomotion similar to SMA phenotypes and showed that this was accompanied by aberrant rhythmic motor output and neuromuscular junction neurotransmission. Surprisingly, we found that none of these defects could be rescued by transgenic restoration of SMN in either the muscles or motor neurons of Drosophila smn mutants. Rather, we discovered that SMN must be restored in both proprioceptive neurons and cholinergic interneurons in order to rescue smn mutant phenotypes. Our results reveal that the disruption of motor neurons and muscles is a secondary consequence of a primary dysfunction of sensory-motor network activity in this SMA model and we demonstrate that genetic or pharmacological strategies that increase motor circuit excitability can positively benefit smn mutant phenotypes. Furthermore, in a companion manuscript (Lotti et al., 2012), we demonstrate that an SMN-dependent gene required for normal motor circuit function in Drosophila is also disrupted in the motor circuits of SMN mutant mice. These results suggest that disruption of motor circuit function may be critical to SMA and that strategies designed to manipulate the activity of motor networks might be employed to ameliorate SMA patient symptoms.
Results
Drosophila smn mutants have muscle and motor system defects
To model the low level of SMN found in SMA patients in Drosophila, we utilized the zygotic protein null smn allele, smnX7, which is a small deficiency that removes the entire smn coding region without disrupting nearby loci (Chang et al., 2008). The remaining SMN in these animals is contributed by maternal protein which provided < 6% the level of SMN compared to controls at the third instar larval stage (Supplementary Figure 1A). smnX7 mutants never initiated pupation but instead persisted as third instar larvae, often surviving several days at this stage, consistent with other smn mutant alleles (Chan et al., 2003). To confirm this phenotype was dependent on SMN, we ubiquitously expressed a transgenic UAS flag-tagged SMN construct (Chang et al., 2008) in smnX7 mutants using Actin-Gal4. This restored normal pupation of smnX7 mutants with 100% of larvae initiating pupation and >80% subsequently eclosing to produce adults (data not shown). Thus, smnX7 mutants have low levels of SMN at late larval stages and can be rescued by transgenic SMN. We used this mutant allele for all subsequent experiments except where noted.
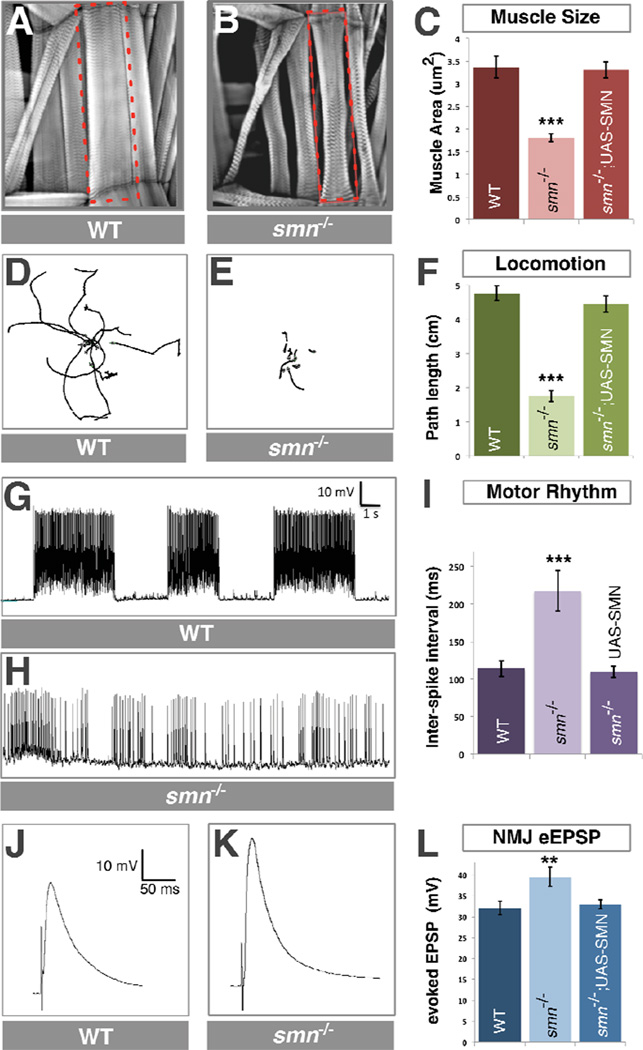
In humans, depletion of SMN affects both muscles and motor function. We noted that Drosophila smn mutant larvae were smaller than control animals and examined if this was associated with a reduction in muscle size. We labeled smn mutant and control larval muscles with phalloidin and found that smn mutants had a 46% (P<0.001) reduction in muscle surface area compared to controls (Figure 1A–C, see also Table S1). This defect was fully rescued by ubiquitous expression of transgenic SMN. smn mutant larvae were also sluggish and moved less frequently than controls. To quantify this defect, we used video capture and tracing software to measure their locomotion. We found that smn mutants showed a 63% (P<0.001) decrease in locomotion velocity compared to control animals which was restored to control levels by ubiquitous expression of transgenic SMN (Figure 1D–F). Thus, similar to SMA patients, Drosophila with low levels of SMN have muscle and locomotion defects.
Figure 1. smn mutants have reduced muscle size, decreased locomotion, defective motor rhythm and aberrant NMJ neurotransmitter release.
A–C. Sample images of muscles from segment A3 of control (A) and smnX7 mutant (B) third instar larvae labeled with TRITC-phallodin show a reduction muscle surface area (C) that is fully rescued by ubiquitous expression of UAS-flag-SMN driven by Da-Gal4 (genotype: Da-Gal4/UAS::flagSMN; smnX7/smnX7). D–F. 10 sample superimposed 60 second larval locomotion path traces from control (D) and smnX7 mutants (E). smn mutant larvae have reduced velocity compared to controls that corrected by ubiquitous expression of transgenic SMN (F). G–I. Recordings from muscle 6 in segment A1 of semi-intact larval preparations where the brain, ventral nerve cord and motor neurons are intact. Control larva produce a regular motor rhythm with periodic bursting activity corresponding to peristaltic muscle contractions (G). smn mutant larvae have an irregular motor pattern with short and uncoordinated bursts as shown by an increase in the average inter-spike interval (I) that is rescued by ubiquitous expression of SMN. J–L. Representative traces recorded from muscle 6 of segment A3 in control (J) and smnX7 mutant (K) larva. smnX7 mutants have increased evoked Excitatory Post-Synaptic Potential (eEPSP) amplitude than controls (K). This is corrected by ubiquitous expression of SMN (L). Data are represented as mean +/− SEM, **=p<0.01, ***=p<0.001. See also Figure S1 and Table S1.
Locomotion of Drosophila larvae has been linked to the rhythmic activity of segmental central pattern-generating networks (CPGs) in the ventral nerve cord (VNC)(Fox et al., 2006) which receive inputs from both the brain hemispheres (Cattaert and Birman, 2001) and proprioceptive sensory neurons (Cheng et al., 2010; Hughes and Thomas, 2007; Song et al., 2007) and output activity to motor neurons. To measure the activity of these pattern-generating neurons, we recorded the spontaneous activity of motor neurons in preparations where we left the brain and VNC in situ (Fox et al., 2006). In control animals, we observed periodic bursting of motor activity at regular intervals consistent with previous studies (Cattaert and Birman, 2001; Fox et al., 2006) (Figure 1G). In contrast, this activity was disrupted in smn mutants which had short irregular bursts that varied in duration (Figure 1H). We quantified this defect by measuring the average inter-spike interval between all spontaneous spike events in smn mutants and controls over a fixed time period. Compared to controls, smn mutants showed a 90% (P<0.001) increase of inter-spike interval (Figure 1I). As with locomotion, normal rhythmic motor activity was fully restored by ubiquitous expression of transgenic SMN. Thus, the output of motor circuits is defective in Drosophila smn mutants.
To investigate the neurotransmitter release properties of individual motor neurons, we removed the brain and stimulated motor neurons directly using a suction electrode (Imlach and McCabe, 2009). Compared to controls, we found a 23% (P<0.005) increase of evoked Excitatory Post-Synaptic Potential (eEPSP) amplitude at the NMJs of smn mutants (Figure 1J–L). The increase of NMJ eEPSP amplitude in smnX7 mutants was restored to control levels by ubiquitous expression of transgenic SMN (Figure 1L). We also observed a 60% (P<0.05) increase in miniature Excitatory Post-Synaptic Potential (mEPSP) frequency. In contrast mEPSP amplitude at smn mutant NMJ terminals was not different to controls (Supplementary Figure 2A,B) leading to a 64% (P<0.001) increase in quantal content (Supplementary Figure 2C). These findings are consistent with a presynaptic change in the neurotransmitter release properties of motor neurons in smn mutants. As our electrophysiology results differed from a previous report (Chan et al., 2003), we also examined trans-heterozygous combinations of smnX7 with other smn mutant alleles and confirmed similar changes in eEPSP amplitudes in these mutants that were not found in heterozygous smnX7 animals (Supplementary Figure 2D). When we examined the morphological features of smnX7 mutant NMJs, we observed no significant difference in the number of synaptic boutons compared to controls (Supplementary Figure 2E). In summary, we find that Drosophila smn mutants have increased NMJ evoked neurotransmitter release that is accompanied by defects in muscle growth, locomotion and motor rhythm.
Restoration of SMN in the nervous system rescues smn mutant phenotypes
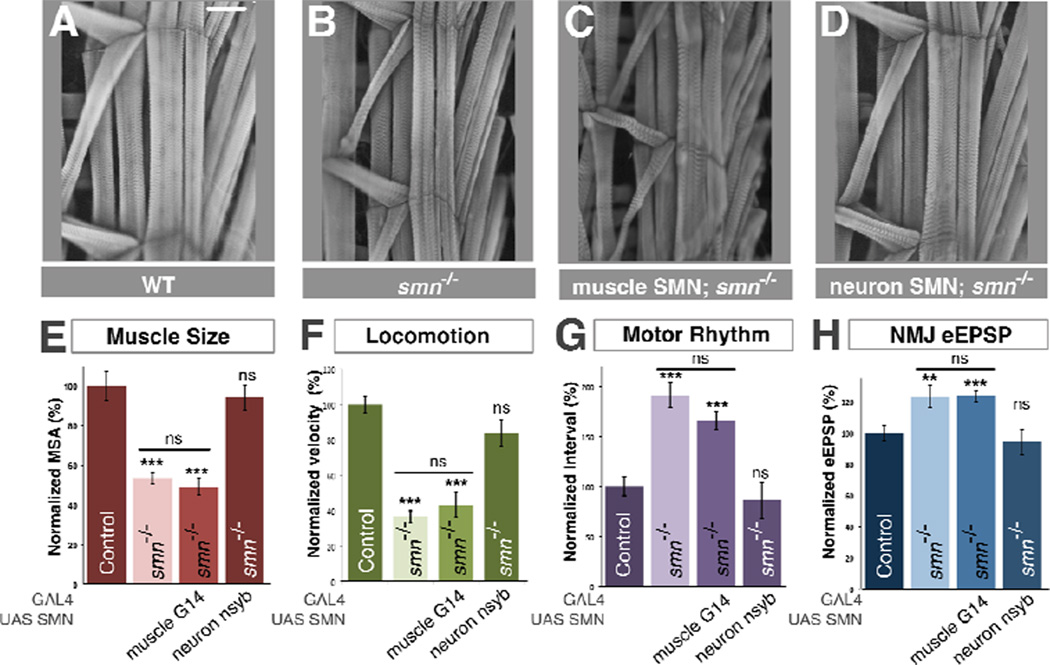
In order to identify the cell autonomous requirement for normal SMN levels, we used iteratively more tissue-restricted Gal4 drivers to assess rescue of smn mutants. We began by expressing transgenic SMN only in the muscles of smnX7 mutants using G14-Gal4, a larval muscle-specific driver. This produced no significant increase in muscle surface area (Figure 2C,E) or effects upon the locomotion, rhythmic motor output and NMJ eEPSP amplitude of smn mutants (Figure 2F–H). We therefore tested SMN restoration only in the nervous system of smn mutants using the neuron-specific nsyb-Gal4 driver. In contrast to muscle restoration of SMN, pan-neuronal restoration of SMN fully rescued the muscle surface area of smn mutants to control levels (Figure 2 B,D,E) and also completely restored their locomotor velocity, rhythmic motor output and NMJ eEPSP amplitudes (Figure 2F–H). Neuron-only rescue of smn mutants was not sufficient to produce viable Drosophila adults (data not shown), presumably due to the SMN level in tissues that are not rescued becoming depleted to the point where cellular viability was compromised. Our results established that the defects of muscle growth in smn mutant larvae are due to a non-autonomous requirement for normal SMN levels in the nervous system rather than in muscle fibers themselves.
Figure 2. SMN expression is required in neurons but not muscles to rescue smn mutants.
A–D. Sample images of muscles from segment A3 of (A) control, (B) smnX7 mutant, (C) smnX7 mutants with transgenic SMN expression only in muscles (G14-Gal4/UAS::flagSMN; smnX7/smnX7) or (D) neurons (nsyb-Gal4/UAS::flagSMN; smnX7/smnX7). Restoration of SMN expression in muscles has no effect on muscle size however restoration in neurons fully rescues muscle surface area. E–H. Quantification of muscle surface area (E), locomotion (F), motor rhythm (G) and NMJ eEPSP amplitude (H) normalized to controls. Expression of transgenic SMN in neurons rescues all of smn mutant phenotypes while expression in muscles does not. Data are represented as mean +/− SEM, **=p<0.01, ***=p<0.001. See also Figure S2.
SMN is required in cholinergic neurons and not in motor neurons
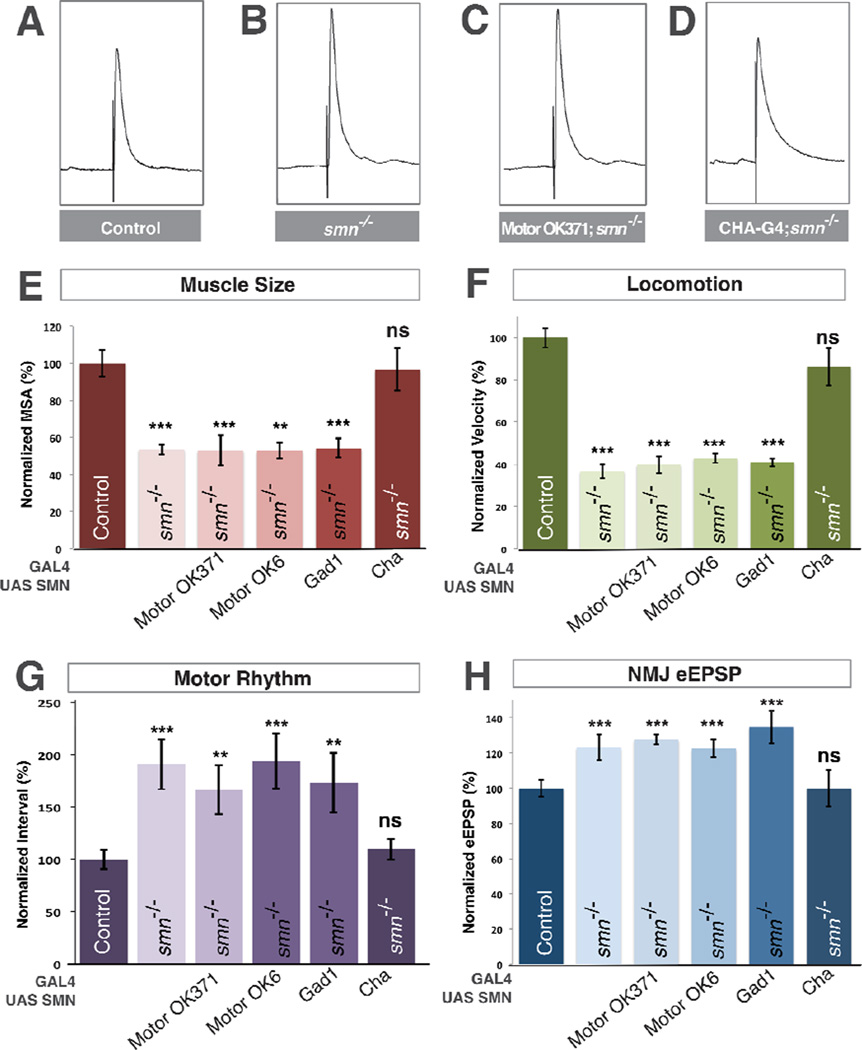
The Drosophila VNC, like the human spinal cord, is populated by neurons with diverse neurotransmitter expression. All Drosophila motor neurons in addition to a subset of central interneurons are glutamatergic (Daniels et al., 2008). Since we observed presynaptic defects in neurotransmitter release at the NMJ in smn mutants, we first tested the ability of transgenic SMN expression in motor neurons to rescue smn mutants. We used OK371-Gal4, an enhancer trap inserted in the vesicular glutamate transporter promoter to express transgenic SMN only in the glutamatergic neurons of smn mutants. This produced no difference in muscle surface area, locomotion velocity or rhythmic motor output compared to smn mutants alone (Figure 3F–G). Surprisingly, we also observed no reduction of the aberrant increase of eEPSP amplitude at the NMJs of these animals (Figure 3B,C,E). We confirmed this unexpected result using a second independent motor neuron-specific driver OK6-Gal4 (Figure 3E–H). Therefore, similar to the requirement for SMN in muscle growth, the aberrant neurotransmitter release at the NMJ of smn mutants is not the result of the cell autonomous loss of SMN in motor neurons. This result prompted us to investigate if SMN was required in other neuron types in the Drosophila motor circuit.
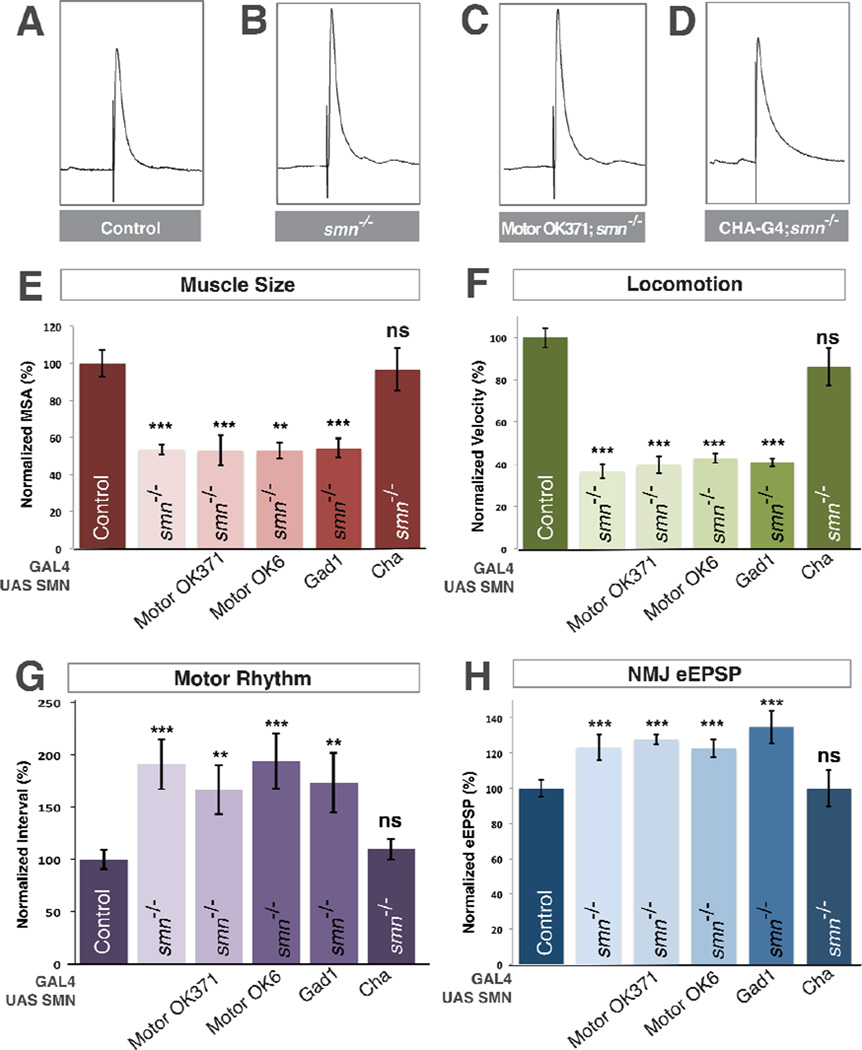
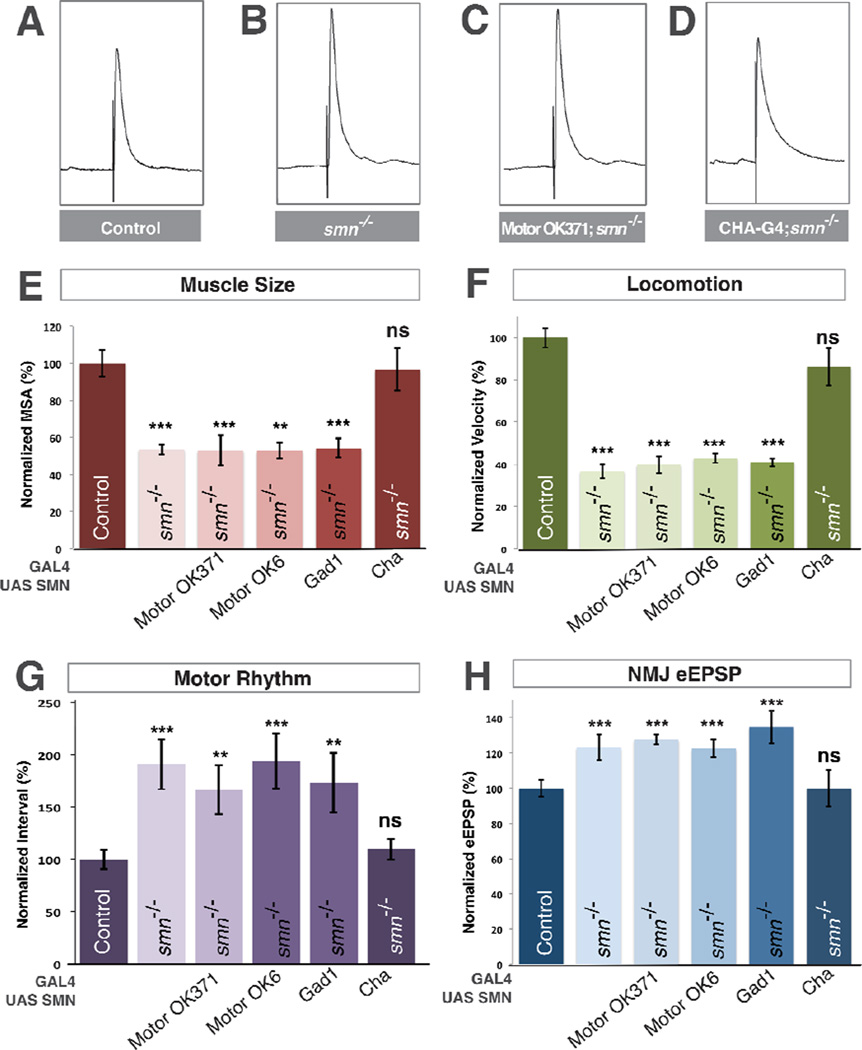
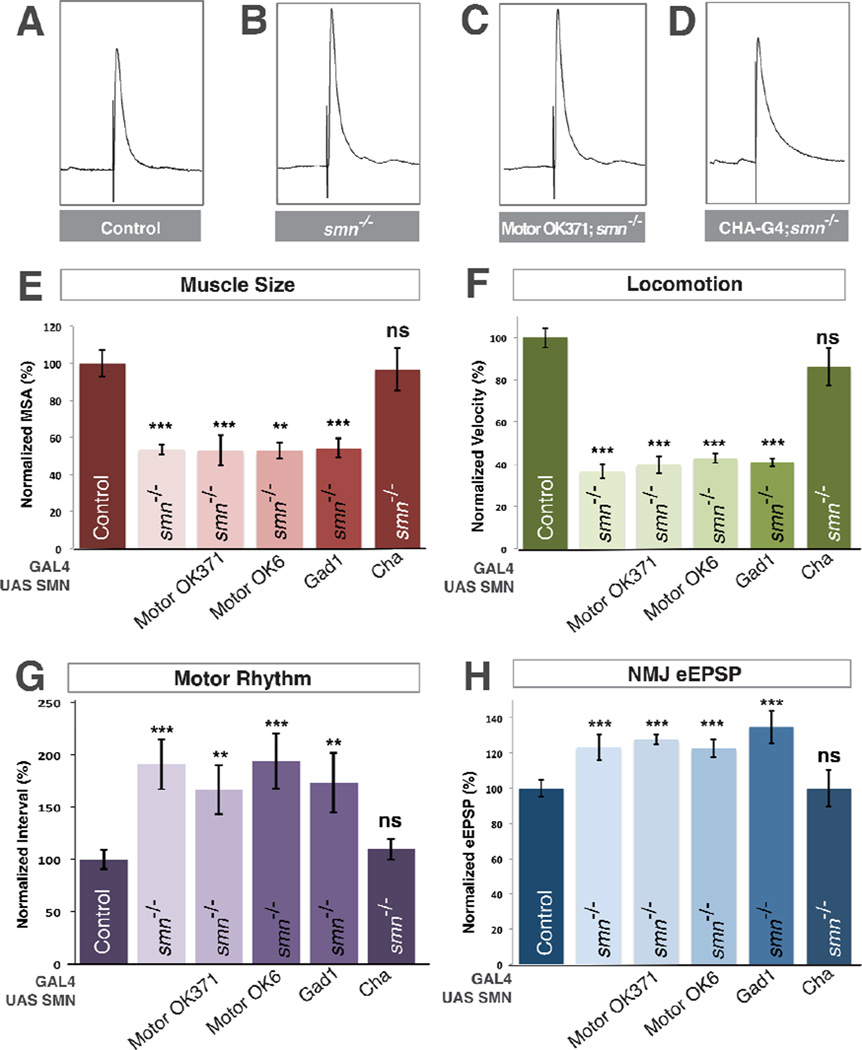
Figure 3. SMN expression is required in cholinergic neurons and not motor neurons.
A–D. Representative traces of control (A), smnX7 mutant (B), transgenic SMN expressed in the motor neurons of smn mutants (OK371-Gal4/UAS::SMN; smnX7/smnX7) (C), transgenic SMN expressed in the cholinergic neurons of smn mutants (Cha-Gal4/UAS::SMN; smnX7/smnX7) (D). Expression of transgenic SMN in motor neurons does not restore NMJ eEPSP amplitudes in smn mutants however expression of SMN on cholinergic neurons does. E–F. Quantification of muscle surface area (E), locomotion (F), motor rhythm (G) and NMJ eEPSP amplitude (H) normalized to controls. Expression of transgenic SMN in the motor neurons of smn mutants with OK371-Gal4 or OK6-Gal4 or in GABAnergic neurons with GAD1-Gal4 does not rescue any phenotype. Expression of transgenic SMN in cholinergic neurons rescues all phenotypes (D). Data are represented as mean +/− SEM, *= p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
Inhibitory inputs are important regulators of motor circuit function (Featherstone et al., 2000) so we next used glutamic acid decarboxylase 1 promoter Gal4 to restore of SMN in gabaergic neurons, however we observed no significant rescue of any smn mutant phenotype (Figure 3E–H). The majority of excitatory neurons in the Drosophila nervous system are cholinergic (Salvaterra and Kitamoto, 2001) and motor neurons receive synaptic input from cholinergic neurons (Baines, 2006). We therefore restored transgenic SMN in smn mutants using choline acetyltransferase (Cha) promoter-driven Gal4. In contrast to glutamatergic and gabaergic drivers, expression of transgenic SMN levels in cholinergic neurons completely rescued the muscle growth, locomotion and rhythmic activity defects of smn mutants (Figure 3E–G). Moreover, expression of SMN in cholinergic neurons also fully rescued eEPSP amplitudes at the NMJ terminals of smn mutants to control levels (Figure 3D,H). Thus, expression of SMN only in cholinergic neurons is sufficient to fully rescue smn mutant phenotypes and can non-autonomously rescue the SMN-dependent defects of both motor neurons and muscles.
SMN is required in both proprioceptive and central cholinergic neurons
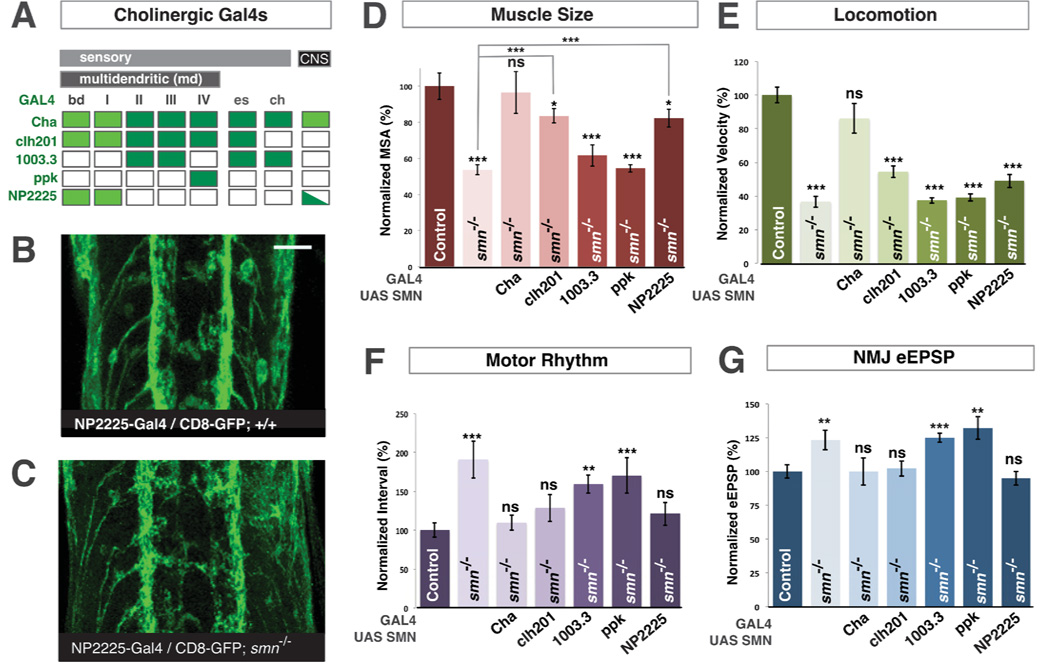
All Drosophila larval sensory neurons in addition to the majority of excitatory central neurons are cholinergic (Salvaterra and Kitamoto, 2001). To dissect the requirement for normal SMN levels between these two populations, we examined the ability of transgenic SMN expression in sensory neurons alone to rescue smn mutant phenotypes. Drosophila sensory neurons are categorized into three major types - multiple dendrite neurons (md) of which there are 5 subclasses (bd, I, II, III and IV), external sense organ neurons (es) and chordotonal neurons (ch). We first used a panel of sensory neuron Gal4 drivers (Figure 4A) to restore SMN only in major types of sensory neurons. We found that when we reinstituted SMN in all md neurons and es sensory neurons but not ch neurons or central neurons both the rhythmic motor output and evoked NMJ eEPSP amplitudes of smn mutants were restored to control levels and muscle surface area was increased to 83.5% of controls (P<0.05) (Figure 4D,F,G). However expression of transgenic SMN with this driver did not significantly change the locomotion of smn mutants (Figure 4E). In contrast, expression of SMN in ch neurons did not rescue any smn mutant phenotype (Figure 4D–G). Using additional Gal4 drivers that are expressed in smaller subsets of md or es sensory neurons (Figure 4A), we found that it was sufficient to restore SMN only in bd and type I md neurons to rescue defects of rhythmic motor output and NMJ neurotransmission and increase the muscle growth of smn mutants (Figure 4D,F,G). Expression of SMN in both the CNS and peripheral cholinergic neurons with Cha-Gal4 fully rescues all phenotypes including locomotion and muscle size (Figure 4D–G). This suggests that, in addition to bd and type I md sensory neurons, SMN must be restored in at least one other additional population of cholinergic neurons that resides within the CNS to completely correct smn mutant locomotion and fully restore muscle size.
Figure 4. SMN is required in both proprioceptive and central cholinergic neurons.
A. Expression of pattern of cholinergic neuron Gal4 lines (dark green). Cha-Gal4 is expressed in both central and sensory cholinergic neurons. Clh201-Gal4 is only expressed in md and es sensory neurons. 1003.3-Gal4, ppk-Gal4 and NP2225-Gal4 are expressed in subsets of md, es or ch sensory neurons. Bright green indicates the ability to rescue of smn mutant phenotypes. B,C. UAS::CD8-GFP labeling the axons of bd and type I md sensory neurons with NP2225-Gal4 in the ventral nerve cord of wild-type (B) or smnX7 mutants (C). Sensory axons project normally into the CNS in smn mutants. D–G. Quantification of muscle surface area (D), locomotion (E), motor rhythm (F) and NMJ eEPSP amplitude (G) normalized to controls (genotype: Gal4/UAS::flagSMN; smnX7/smnX7). Expression of transgenic SMN in both central and sensory cholinergic neurons in smn mutants with Cha-Gal4 fully rescues all phenotypes. Restoration of SMN all sensory neurons or only proprioceptive type I md and bd neurons with NP2225-Gal4 increases muscle size and fully rescues motor rhythm and NMJ eEPSP amplitude but does not rescue locomotion. In contrast, restoration of SMN in with 1003.3-Gal4 or ppk-Gal4 does not rescue any smn mutant phenotype. Scale bar = 10µm. Data are represented as mean +/− SEM, *=p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
Both bd and type I md sensory neurons are required for propioceptive feedback to the motor circuit of Drosophila larvae (Cheng et al., 2010; Hughes and Thomas, 2007). To determine if these neurons were morphologically disrupted by SMN depletion, we examined the sensory or axonal processes of the bd and type I md neurons labeled in smn mutants, however we found no obvious defects in sensory processes (data not shown) and the axons of these neurons projected into the CNS similarly to controls (Figure 4 B,C). Our data therefore suggested that reduced SMN in proprioceptive neurons might disrupt their function rather than their development or connectivity.
smn mutant phenotypes can be rescued after embryogenesis
Drosophila larval neurons develop, connect and become functional during the 24 hours of embryonic development prior to hatching (Baines, 2006). To determine if SMN depletion could have disrupted nervous system assembly during this period we used the ‘geneswitch’ RU486-drug inducible Gal4 system to control the temporal restoration of transgenic SMN. We first asked if smn mutant phenotypes could be rescued by expression of transgenic SMN subsequent to the completion of embryogenesis by exposing smn mutant larvae and controls carrying the neuron-specific elav-geneswitch driver to RU486 containing media immediately after hatching and throughout the subsequent larval period (Figure 5A). When transgenic SMN expression was not induced we found no difference compared to smn mutants alone (Figure 5C–F). In contrast, when SMN expression was induced immediately after embryogenesis, third instar larval muscle size, locomotion, rhythmic motor output and motor neuron eEPSP amplitudes were indistinguishable from control animals (Figure 5B–F). This result established that restoration SMN expression after embryogenesis can rescue smn mutants suggesting they do not have persistent defects in motor circuit assembly.
Figure 5. Restoration of SMN after embryogenesis rescues smn mutants.
A. Schematic of transgenic SMN induction in the nervous system. RU486 is required for the activation of transgene induction by geneswitch Gal4. Elav::geneswitch/UAS::flagSMN; smnX7/smnX7 larva were transferred to either vehicle media or RU486 containing media immediately, 48 hours or 96 hours after hatching. B. Representative traces recorded from smn mutants that were cultured on either vehicle media or RU486 media 0, 48 or 96 hours after hatching. Induction of SMN at every each time-point fully restored normal eEPSP amplitude. C–F. Quantification of muscle surface area (C), locomotion (D), motor rhythm (E) and NMJ eEPSP amplitude (F) normalized to controls. Muscle size, locomotion and motor rhythm is fully rescued if transgenic SMN is induced immediately after hatching, but if SMN induction is delayed rescue is incomplete. Induction of SMN for only 48 hours is however sufficient to completely restore normal NMJ eEPSP amplitude. Data are represented as mean +/− SEM, *=p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
We next delayed SMN expression in the nervous system of smn mutants until progressively later larval stages. When we induced transgenic SMN in smn mutants at 48 or 96 hours hours after embryo hatching, we found intermediate phenotypes where muscle volume, motor rhythm defects and locomotion where only partially restored compared to controls (Figure 5C–D). In contrast, NMJ eEPSP amplitudes were completely restored in smn mutants to control levels by only 48 hours of SMN expression (Figure 5B,F). These results revealed a differential phenotypic sensitivity to the timing of SMN restoration with NMJ neurotransmitter fully corrected by elevating SMN levels at even late stages, while locomotion, motor rhythm and muscle growth required an earlier or longer duration of exposure to increased SMN levels.
Inhibiting cholinergic neuron activity mimics aspects of SMN depletion
During Drosophila embryonic development, complete removal of cholinergic input onto motor neurons results in motor neuron hyperexcitability and increased neurotransmission (Baines et al., 2001). We hypothesized therefore that the non-autonomous changes of motor neuron properties in smn mutants might be explained by defective excitatory input from cholinergic neurons in the motor circuit. Complete loss or inhibition of all cholinergic neuron activity results in embryonic lethality (Kitamoto et al., 2000) so in order to test this hypothesis, we employed transgenes designed to partially inhibit neurotransmission in cholinergic neurons. We used lines that either express moderate levels of the human inward rectifying channel Kir2.1 which inhibits membrane depolarization (Paradis et al., 2001) or express membrane-tethered Plectreurys Toxin II (PLTXII) which inhibits synaptic N-type voltage gated calcium channels (Wu et al., 2008). To determine the effectiveness of this approach, we first expressed these transgenes in motor neurons alone using OK6-Gal4. We found that Kir2.1 reduced eEPSP amplitudes by 32% (P<0.001) while expression of PLTXII reduced eEPSP amplitudes by 96% (P<0.001) indicating the both transgenes were capable of partially inhibiting neurotransmission.
To examine the effects on the motor system of inhibiting cholinergic neuron function, we then expressed each of these Kir2.1 or PLTXII in the cholinergic neurons of wild-type animals using Cha-Gal4. Expression of either transgene in cholinergic neurons had no effect on muscle surface area (Figure 6C), however expression of these transgenes significantly inhibited locomotion by 41% (P<0.001) and 42% (P<0.001) respectively (Figure 6D). They also disrupted spontaneous rhythmic motor activity inducing a 54% (P<0.05) or 59% (P<0.05) increase in average inter-spike intervals (Figure 6B,E). Importantly, inhibition of cholinergic neuron function also resulted in increased eEPSP amplitudes at the NMJs of glutamatergic motor neurons (Figure 6A,F). Expression of Kir2.1 in cholinergic neurons produced a 50% increase (P<0.001) in NMJ eEPSP amplitudes while expression of PLTXII induced a 45% increase (P<0.001) (Figure 6F). Therefore, inhibition of cholinergic neuron activity replicated a number of the features of smn mutants including non-cell autonomous effects on the neurotransmitter release properties of motor neurons, consistent with cholinergic neurons in the motor circuit having reduced function in smn mutants.
Figure 6. Inhibiting cholinergic neuron activity mimics smn mutant phenotypes.
A. Representative traces recorded from the NMJ of control or UAS-human Kir2.1 or UASPLTXII expressed in cholinergic neurons with Cha-Gal4. Inhibiting cholinergic neuron excitability with Kir2.1 or neurotransmitter release with PLTXII increases motor neuron NMJ eEPSP amplitude. B. Expression of Kir2.1 or PLTX in cholinergic neurons disrupts rhythmic motor activity. C–F. Quantification of muscle surface area (C), locomotion (D), motor rhythm (E) and NMJ eEPSP amplitude (F) normalized to controls. Expression of Kir2.1 or PLTXII in cholinergic neurons does not alter muscle size but does reduce locomotor speed, disrupt motor rhythm and increases NMJ eEPSP amplitude. Data are represented as mean +/− SEM, *=p<0.05, ***=p<0.001.
Increasing neuronal excitability rescues smn mutant phenotypes
Building upon the hypothesis that motor circuits have functional deficits in smn mutants, we next asked if increasing the excitability of cholinergic neurons in these animals could increase motor network activity and alter smn mutant phenotypes. Inhibition of the Shaker (Sh) type IA outward K+ current by a dominant negative (SDN) transgene enhances membrane excitability and increases both the amplitude and duration of eEPSPs at synaptic terminals (Mosca et al., 2005). We expressed the SDN transgene in the cholinergic neurons of smn mutants and examined the effect upon mutant phenotypes (Figure 7E–H). Expression of SDN completely restored the muscle surface area, locomotor velocity, rhythmic motor activity and eEPSP amplitudes of smn mutants to levels indistinguishable from control animals (Figure 7 B–H). This striking result established that increasing the activity of cholinergic neurons in the motor circuit of smn mutants can result in robust phenotypic rescue.
Figure 7. Genetic or pharmacological inhibition of K+ channels ameliorates smn mutant phenotypes.
A–C. Locomotion path traces from (A) control, (B) smn mutants and (C) smn mutants expressing a UAS dominant negative Shaker K+ channel (UAS-SDN) in cholinergic neurons with Cha-Gal4. Expressing SDN increases rescues the locomotion of smn mutants. D–G. Quantification of muscle surface area (D), locomotion (E), motor rhythm (F) and NMJ eEPSP amplitude (G) normalized to controls. Expression of SDN in cholinergic neurons with Cha-Gal4 restores muscle size (D), locomotion (E), motor rhythm (F) and NMJ eEPSP (G) of smn mutants to control levels. Addition of 2mM 4-Aminopyridine (4-AP) to culture media throughout larval development does not alter muscle size in control animals but increases the muscle size of smn mutants (D). 4-AP administration inhibits locomotion, motor rhythm and eEPSP size in control animals. Administration of 4-AP to smn mutants corrects locomotion (E) and NMJ eEPSP (G) to levels not significantly different from control 4-AP treated animals and substantially corrects defects in motor rhythm (F). Data are represented as mean +/− SEM, *=p<0.05, **=p<0.01, ***=p<0.001. Significance was calculated versus controls except where otherwise indicated.
As genetic methods to inhibit K+ channel activity benefited smn mutant phenotypes, we next asked if pharmacological antagonists of K+ channels could also be effective. 4-aminopyridine (4-AP) is an FDA approved small molecule inhibitor of voltage activated vertebrate (Hayes, 2007) and Drosophila K+ channels (Wicher et al., 2001). We added 4-AP to larval media and titrated the compound to identify the maximum dosage at which the drug could be tolerated without lethality in wild-type larvae (2 mM). We then examined the effects of exposure of 4-AP throughout the larval period in both control and smn mutants. In control animals, 4-AP had no effect on muscle size but reduced larval locomotion by 35% (P<0.01), decreased rhythmic motor activity by 40% (P<0.01) and reduced NMJ eEPSP amplitudes by 21% (P<0.001) indicating mild systemic toxicity at this dose (Figure 7D–G). Despite this, when we grew smn mutants on 4-AP containing media throughout the larval period, muscle surface area was increased by 66% (P<0.001) compared to untreated smn mutants (Figure 7D). Locomotion was also increased by 55% (P<0.05) and was not significantly different to controls treated with 4-AP (Figure 7E). Defects in rhythmic motor activity in smn mutants were substantially improved with the aberrant increase in inter-spike interval reduced to 31% (p<0.001) above controls treated with 4-AP (Figure 7F). Finally, the increased NMJ EPSP amplitude of smn mutants treated with 4-AP were reduced by 27% (P<0.001) not significantly different to that of 4-AP treated controls (Figure 7G). Thus pharmacological inhibition of K+ channels, similar to genetic inhibition, can benefit smn mutant phenotypes consistent with the defective excitability of motor circuits being a critical consequence of SMN depletion.
Discussion
Across organisms the function of the motor system seems uniquely sensitive to low levels of the ubiquitous protein SMN, the molecular defect responsible for SMA (Burghes and Beattie, 2009; Schmid and DiDonato, 2007). This is also true in Drosophila where smn mutants have reduced muscle size and locomotion which we find is accompanied by defects in rhythmic motor output and motor neuron neurotransmitter release. Surprisingly, restoration of SMN in the motor neurons or muscles of these mutants provided no phenotypic rescue. Instead, SMN must be reinstated in both cholinergic proprioceptive and central neurons to rescue smn mutant phenotypes including non-cell autonomous defects in both motor neurons and muscles. Proprioceptive neurons provide essential inputs to motor circuits (Hughes and Thomas, 2007) and cholinergic interneurons are critical for Drosophila CNS function (Kitamoto et al., 2000), including synaptic output onto motor neurons (Baines et al., 2001). Restoration of SMN after the completion of nervous system development is sufficient to rescue SMN-dependent phenotypes, arguing that is not the connectivity but rather the function of motor circuits that is disrupted by depletion of SMN. Two lines of evidence further support this conclusion. First, inhibiting the activity of cholinergic neurons can mimic a number of smn mutant phenotypes including nonautonomous effects on motor neurons. Second, increasing the excitability of motor circuits through K+ channel inhibition can rescue smn mutant defects. Our results therefore demonstrate that depletion of SMN in Drosophila causes the dysfunction of a select subset of neurons in the motor circuit which consequently disrupts the activity of other networked components of the motor system such as motor neurons and muscles. These findings establish this model of SMA as a paradigm for a neurological disease induced by neuronal circuit dysfunction.
The contribution of cholinergic neurons to Drosophila motor circuits
While our results exclude a cell autonomous requirement for normal SMN levels in Drosophila motor neurons to rescue smn mutants, our data do establish that SMN has to be restored in at least two groups of motor circuit neurons for full rescue of larval phenotypes. One of these groups is bd and type I md sensory neurons which are essential components of a proprioceptive sensory feedback circuit necessary for coordinated contractile locomotion of Drosophila larvae (Hughes and Thomas, 2007). Both bd and type I md subset of sensory neurons express the mechanosensitive NompC TRP channel which is essential for proprioception (Cheng et al., 2010). Sensory feedback does not seem to be necessary for Drosophila larval central pattern generator assembly or basic embryonic and larval movement (Crisp et al., 2008), however without sensory input, both rhythmic motor circuit activity (Fox et al., 2006) and coordinated locomotion behavior is severely disrupted (Hughes and Thomas, 2007; Song et al., 2007). Rescue of SMN in bd and type I md sensory neurons can restore the rhythmic motor output of smn mutants, consistent with an important role for sensory input in regulating this activity (Fox et al., 2006). However, restoration of SMN in proprioceptive neurons alone is not sufficient to correct the locomotion velocity of smn mutants indicating that additional neurons require wildtype levels of SMN in order to restore full mobility.
SMN expression in all cholinergic neurons can completely rescue all smn mutant larval phenotypes including locomotion. Our results therefore implicate an additional cell autonomous requirement for SMN in one or more groups of central cholinergic neurons. Establishing the identity of these central neurons will be a challenge given our limited understanding of central motor circuitry in Drosophila. It is tempting to speculate that these neurons could be descending inputs from the brain (Cattaert and Birman, 2001) or other connections between segmental central pattern generators that promote the coordination necessary for effective locomotion. However, while rescue analysis demonstrates that individual components of the motor circuit can make significant contributions to some smn mutant phenotypes, other phenotypes such as muscle growth additively require SMN in both central and peripheral cholinergic neurons. Therefore, our data suggests that the effect of SMN depletion on the motor network is an amalgam of specific defects in distinct neurons that sum to produce a generalized disruption of the motor system.
Why are cholinergic motor circuit neurons selectively susceptible to SMN depletion? In a companion manuscript (Lotti et al., 2012), we describe a sequence of molecular events that link reduction of SMN to selective motor circuit dysfunction. We show that loss of Drosophila SMN disrupts minor splicing, which is required for the expression of genes with rare U12-type introns (Patel and Steitz, 2003). Through a genome-wide analysis of Drosophila U12 intron-containing genes, we identified a novel transmembrane protein, Stasimon, that has both reduced expression in smn mutants and increased NMJ eEPSP amplitudes when mutated, similar to the smn mutant phenotype. We found that like SMN, Stasimon is required in cholinergic neurons, but not motor neurons to affect NMJ electrophysiology. Furthermore, we demonstrate that expression of a transgenic expression of Stasimon can fully restore normal NMJ eEPSP amplitudes in smn mutants in addition to increasing muscle size. These data establish that reduction of SMN decreases expression of a subset of genes that are particularly sensitive to SMN-dependent splicing disruption. Some of these genes, such as stasimon, are critically required for the normal function of cholinergic motor circuit neurons in Drosophila. These results establish a mechanistic chain linking the role of SMN in RNA splicing to the selective vulnerability of motor circuit function when SMN is depleted.
Parallels between Drosophila and mammalian motor circuits
While the basic elements of motor circuits – proprioceptive neurons, interneurons and motor neurons are conserved between Drosophila and humans, the neuronal constituents and connections that make-up Drosophila central motor circuitry are at present unknown, limiting comparisons with mammalian circuits. However, it is known that the neurotransmitters employed in each system are different (Marder and Rehm, 2005). For example, human and mouse motor neurons are cholinergic while proprioceptive neurons are glutamatergic, the inverse of the neurotransmitters employed in Drosophila motor circuits. Therefore, one possible interpretation of our results is that cholinergic neurons have a particular and conserved sensitivity to the reduced levels of SMN. Neurotransmitter release is defective from the cholinergic motor neurons of SMN-Δ7 mice (Kong et al., 2009; Park et al., 2010a) and this defect does appear to require the cell-autonomous presence of normal SMN levels in these neurons (Park et al., 2010b). Nonetheless, SMN-Δ7 mutants have normal muscle twitch tension (Ling et al., 2010), targeted depletion of SMN in motor neurons does not cause lethality (Park et al., 2010b) and selective restoration of SMN in motor neurons alone produces only a few days of survival benefit to mutant animals (Gogliotti et al., 2012; Martinez et al., 2012). These results imply that if indeed cholinergic neurons are selectively affected by reduction of SMN, additional cholinergic neurons in the mammalian motor circuit must also be involved.
An alternative, though not necessarily exclusive, interpretation is that conserved network elements of motor circuits are vulnerable to low levels of SMN. In support of this, it has recently been shown that SMN-Δ7 mice have early reduced responses to afferent fiber activation (Mentis et al., 2011) which are accompanied by a later decrease in glutamatergic proprioceptive synapses from sensory afferents onto motor neurons (Ling et al., 2010; Mentis et al., 2011). SMA patients have also been reported to have reduced or absent H-reflexes (Renault et al., 1983) which could be consistent with decreased activity of motor reflex circuits. Interestingly, in a companion manuscript (Lotti et al., 2012) we show that the splicing and expression of the SMN-dependent gene Stasimon is preferentially disrupted in the proprioceptive neurons of SMN-Δ7 motor circuits though motor neurons are also affected. The concordant evidence for defective sensory-motor function in both mammalian and Drosophila SMN mutants is striking but also unexpected even with our limited understanding of the central circuitry of both systems. For example, both mouse and human motor neurons receive direct synaptic input on to both somata and dendrites from sensory afferents (Chen et al., 2003) while Drosophila motor neuron dendrites do not appear to contact proprioceptive axon processes (Zlatic et al., 2009). Restoration of SMN in the proprioceptive neurons of Drosophila smn mutants is sufficient to restore normal NMJ neurotransmitter release properties in motor neurons. This suggests that even without direct synaptic contact, increasing SMN in these neurons can influence motor neuron electrophysiological properties, presumably through intermediate interneuron connections. Therefore, it is possible that while the specific details of motor circuit wiring differ between Drosophila and vertebrates, the essential relationships and function of motor networks are conserved and selectively susceptible to depletion of SMN.
Manipulating motor circuit excitability in smn mutants
Drosophila smn mutants have increased NMJ eEPSP amplitude and mEPSP frequency consistent with an increased excitability of motor neurons. Hyperexcitability of motor neurons has also been described in the SMA-Δ7 mouse model (Mentis et al., 2011). In Drosophila, this increase in neurotransmitter release properties is not corrected by restoring SMN in motor neurons themselves but is rescued by expressing SMN in cholinergic neurons. Hyperexcitability of Drosophila motor neurons has previously been reported in embryos where cholinergic neurotransmission is completely inhibited (Baines et al., 2001). Congruent with this, we could replicate the increased evoked neurotransmitter release from smn mutant motor neurons by inhibiting cholinergic neurotransmission in larvae, consistent with a homeostatic compensatory increase in the excitability of motor neurons when synaptic inputs are reduced. A similar phenomenon has recently been described in chicken magnocellular neurons which, when deafferentated by removal of the cochlea, increase in excitability (Kuba et al., 2010). Increasing neuronal excitability by inhibiting K+ channels in smn mutants gave a remarkably robust rescue of muscle size, locomotion, rhythmic motor output and NMJ neurotransmission. The Shaker type IA K+ current plays a critical role in the regulation of membrane excitability in Drosophila neurons and expression of a dominant negative construct inhibiting the Sh current (Mosca et al., 2005) in cholinergic neurons of smn mutants fully rescues all the larval phenotypes we examined. Together these results strongly argue that decreased excitability of motor circuit neurons is a key physiological outcome of reduced levels of SMN.
Treatment with the small molecule K+ channel antagonist 4-AP also showed benefit to Drosophila smn mutant phenotypes. In wild-type animals, 4-AP treatment did not affect muscle size but did reduce locomotion and inhibited NMJ neurotransmitter release as might be anticipated by systemic inhibition of K+ channels, which are present throughout the nervous system and in muscles (Wicher et al., 2001). Nonetheless, administration of 4-AP significantly increased both the muscle area and locomotion of smn mutants and fully corrected defects in rhythmic motor output and NMJ neurotransmission. Treatment with 4-AP has been linked to functional improvement of patients with spinal cord injury, myasthenia gravis and Lambert-Eaton syndrome (Hayes, 2007) and can improve muscle twitch tension in a canine hereditary motor neuron disease (Pinter et al., 1997). A sustained release preparation of 4-AP was recently approved by the FDA for human clinical use in multiple sclerosis (Chwieduk and Keating, 2010). Our data suggest that the efficacy on 4-AP in the Drosophila smn mutant model is likely via its activity upon cholinergic neurotransmission in the sensory-motor circuit. Extrapolating this finding to humans, investigation of compounds like 4-AP that can act within the spinal cord to modify the excitability of motor neural networks could be a new and fruitful therapeutic strategy to ameliorate the symptoms of SMA.
Experimental Procedures
(see also Supplemental Information):
Drosophila stocks
smnX7 (Chang et al., 2008), smn73Ao (Chan et al., 2003), smnE33 (Rajendra et al., 2007). Gal4 and UAS lines are described in Supplemental Information.
Muscle measurement
Muscle area measurements were carried out at muscle 6 segment A3 of phallodin stained muscle fillet preparations (Brent et al., 2009).
Locomotion
Larval locomotion assays were essentially performed as previously described (Suster and Bate, 2002).
Motor rhythm
Spontaneous motor rhythm was recorded as previously described (Fox et al., 2006). To measure the average inter-spike interval, the peak detection feature of MiniAnalysis (Synaptosoft, Inc.) was used detect all spontaneous eEPSPs events that occurred over a 3 minute period.
NMJ electrophysiology
Intracellular recordings from muscle 6, segment A3 were performed as previously described (Imlach and McCabe, 2009).
Drug treatment
Gene-switch GAL4 SMN expression was induced by culturing larvae with RU486 at 10 µg/ml for 148 hours, 96 hours, 72 hours or 48 hours prior to phenotypic measurement in smn mutants (controls were all assayed at wandering L3 stage). SMN induction was confirmed by western blot. For 4-AP treatment, 2mM 4-AP (Sigma) was added to the yeast paste on which larvae were cultured immediately after hatching and throughout the subsequent larval period.
Statistical methods
Significance was tested by ANOVA.
Supplementary Material
Drosophila SMN mutants have multiple motor system defects and reduced muscle growth
Proprioceptive and interneuron SMN rescue corrects motor neuron and muscle defects
Motor network function is disrupted by SMN depletion subsequent to circuit development
SMN mutant phenotypes are corrected by increasing motor circuit excitability
Acknowledgments
We are very grateful to Spyros Artavanis-Tsakonas and Greg Matera for smn stocks and reagents. We also thank Haig Keshishian, Cynthia Hughes, John Thomas, Michael Nitabach, Andrea Pizzo, Jonathan Javitch, Graeme Davis, Julie Simpson and Wesley Grueber for additional stocks and advice. We thank George Mentis and especially Chris Henderson for critical reading of this manuscript. LP was supported by NIH-NINDS R01NS069601 and with BDM by the Columbia University Motor Neuron Center. BDM was supported by DoD-W81XWH-08-1-0009, DoD-W81XWH-11-1-0753 and the Gatsby Initiative in Brain Circuitry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baines RA. Development of motoneuron electrical properties and motor output. Int Rev Neurobiol. 2006;75:91–103. doi: 10.1016/S0074-7742(06)75005-X. [DOI] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent JR, Werner KM, McCabe BD. Drosophila larval NMJ dissection. J Vis Exp. 2009;24:1107. doi: 10.3791/1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaert D, Birman S. Blockade of the central generator of locomotor rhythm by noncompetitive NMDA receptor antagonists in Drosophila larvae. J Neurobiol. 2001;48:58–73. doi: 10.1002/neu.1042. [DOI] [PubMed] [Google Scholar]
- Chan YB, Miguel-Aliaga I, Franks C, Thomas N, Trulzsch B, Sattelle DB, Davies KE, van den Heuvel M. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- Chang HC, Dimlich DN, Yokokura T, Mukherjee A, Kankel MW, Sen A, Sridhar V, Fulga TA, Hart AC, Van Vactor D, et al. Modeling spinal muscular atrophy in Drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Hippenmeyer S, Arber S, Frank E. Development of the monosynaptic stretch reflex circuit. Curr Opin Neurobiol. 2003;13:96–102. doi: 10.1016/s0959-4388(03)00006-0. [DOI] [PubMed] [Google Scholar]
- Cheng LE, Song W, Looger LL, Jan LY, Jan YN. The role of the TRP channel NompC in Drosophila larval and adult locomotion. Neuron. 2010;67:373–380. doi: 10.1016/j.neuron.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwieduk CM, Keating GM. Dalfampridine extended release: in multiple sclerosis. CNS Drugs. 2010;24:883–891. doi: 10.2165/11205910-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Crisp S, Evers JF, Fiala A, Bate M. The development of motor coordination in Drosophila embryos. Development. 2008;135:3707–3717. doi: 10.1242/dev.026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Gelfand MV, Collins CA, DiAntonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508:131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- Featherstone DE, Rushton EM, Hilderbrand-Chae M, Phillips AM, Jackson FR, Broadie K. Presynaptic glutamic acid decarboxylase is required for induction of the postsynaptic receptor field at a glutamatergic synapse. Neuron. 2000;27:71–84. doi: 10.1016/s0896-6273(00)00010-6. [DOI] [PubMed] [Google Scholar]
- Fox LE, Soll DR, Wu CF. Coordination and modulation of locomotion pattern generators in Drosophila larvae: effects of altered biogenic amine levels by the tyramine beta hydroxlyase mutation. J Neurosci. 2006;26:1486–1498. doi: 10.1523/JNEUROSCI.4749-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilina TO, McGovern VL, Workman E, Crawford TO, Gogliotti RG, DiDonato CJ, Monani UR, Morris GE, Burghes AH. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum Mol Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogliotti RG, Quinlan KA, Barlow CB, Heier CR, Heckman CJ, Didonato CJ. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J Neurosci. 2012;32:3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes KC. Fampridine-SR for multiple sclerosis and spinal cord injury. Expert Rev Neurother. 2007;7:453–461. doi: 10.1586/14737175.7.5.453. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Thomas JB. A sensory feedback circuit coordinates muscle activity in Drosophila. Mol Cell Neurosci. 2007;35:383–396. doi: 10.1016/j.mcn.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlach W, McCabe BD. Electrophysiological methods for recording synaptic potentials from the NMJ of Drosophila larvae. J Vis Exp. 2009;24:1109. doi: 10.3791/1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:2552–2569. doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T, Xie X, Wu CF, Salvaterra PM. Isolation and characterization of mutants for the vesicular acetylcholine transporter gene in Drosophila melanogaster. J Neurobiol. 2000;42:161–171. [PubMed] [Google Scholar]
- Kong L, Wang X, Choe DW, Polley M, Burnett BG, Bosch-Marce M, Griffin JW, Rich MM, Sumner CJ. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba H, Oichi Y, Ohmori H. Presynaptic activity regulates Na(+) channel distribution at the axon initial segment. Nature. 2010;465:1075–1078. doi: 10.1038/nature09087. [DOI] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Ling KK, Gibbs RM, Feng Z, Ko CP. Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One. 2010;5:e15457. doi: 10.1371/journal.pone.0015457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F, Imlach WL, Saieva L, Beck ES, Le HT, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, et al. A SMN-Dependent U12 Splicing Event Essential for Motor Circuit Function. Cell. 2012 doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Rehm KJ. Development of central pattern generating circuits. Curr Opin Neurobiol. 2005;15:86–93. doi: 10.1016/j.conb.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Martinez TL, Kong L, Wang X, Osborne MA, Crowder ME, Van Meerbeke JP, Xu X, Davis C, Wooley J, Goldhamer DJ, et al. Survival Motor Neuron Protein in Motor Neurons Determines Synaptic Integrity in Spinal Muscular Atrophy. J Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern VL, Gavrilina TO, Beattie CE, Burghes AH. Embryonic motor axon development in the severe SMA mouse. Hum Mol Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L, Alvarez FJ, Sumner CJ, O'Donovan MJ. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Carrillo RA, White BH, Keshishian H. Dissection of synaptic excitability phenotypes by using a dominant-negative Shaker K+ channel subunit. Proc Natl Acad Sci U S A. 2005;102:3477–3482. doi: 10.1073/pnas.0406164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palop JJ, Mucke L. Amyloid-beta-induced neuronal dysfunction in Alzheimer's disease: from synapses toward neural networks. Nat Neurosci. 2010;13:812–818. doi: 10.1038/nn.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Sweeney ST, Davis GW. Homeostatic control of presynaptic release is triggered by postsynaptic membrane depolarization. Neuron. 2001;30:737–749. doi: 10.1016/s0896-6273(01)00326-9. [DOI] [PubMed] [Google Scholar]
- Park GH, Kariya S, Monani UR. Spinal muscular atrophy: new and emerging insights from model mice. Curr Neurol Neurosci Rep. 2010a;10:108–117. doi: 10.1007/s11910-010-0095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GH, Maeno-Hikichi Y, Awano T, Landmesser LT, Monani UR. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J Neurosci. 2010b;30:12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Bu J, Roskelley EM, Richards AM, Sardi SP, O'Riordan CR, Klinger KW, Shihabuddin LS, Cheng SH. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nature reviews. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter MJ, Waldeck RF, Cope TC, Cork LC. Effects of 4-aminopyridine on muscle and motor unit force in canine motor neuron disease. J Neurosci. 1997;17:4500–4507. doi: 10.1523/JNEUROSCI.17-11-04500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendra TK, Gonsalvez GB, Walker MP, Shpargel KB, Salz HK, Matera AG. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault F, Raimbault J, Praud JP, Laget P. Electromyographic study of 50 cases of Werdnig-Hoffmann disease. Rev Electroencephalogr Neurophysiol Clin. 1983;13:301–305. doi: 10.1016/s0370-4475(83)80042-2. [DOI] [PubMed] [Google Scholar]
- Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65(Suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- Salvaterra PM, Kitamoto T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res Gene Expr Patterns. 2001;1:73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- Schmid A, DiDonato CJ. Animal models of spinal muscular atrophy. J Child Neurol. 2007;22:1004–1012. doi: 10.1177/0883073807305667. [DOI] [PubMed] [Google Scholar]
- Simic G. Pathogenesis of proximal autosomal recessive spinal muscular atrophy. Acta Neuropathol. 2008;116:223–234. doi: 10.1007/s00401-008-0411-1. [DOI] [PubMed] [Google Scholar]
- Song W, Onishi M, Jan LY, Jan YN. Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc Natl Acad Sci U S A. 2007;104:5199–5204. doi: 10.1073/pnas.0700895104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suster ML, Bate M. Embryonic assembly of a central pattern generator without sensory input. Nature. 2002;416:174–178. doi: 10.1038/416174a. [DOI] [PubMed] [Google Scholar]
- Swoboda KJ, Prior TW, Scott CB, McNaught TP, Wride MC, Reyna SP, Bromberg MB. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann Neurol. 2005;57:704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicher D, Walther C, Wicher C. Non-synaptic ion channels in insects--basic properties of currents and their modulation in neurons and skeletal muscles. Prog Neurobiol. 2001;64:431–525. doi: 10.1016/s0301-0082(00)00066-6. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cao G, Pavlicek B, Luo X, Nitabach MN. Phase coupling of a circadian neuropeptide with rest/activity rhythms detected using a membrane-tethered spider toxin. PLoS biology. 2008;6:e273. doi: 10.1371/journal.pbio.0060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic M, Li F, Strigini M, Grueber W, Bate M. Positional cues in the Drosophila nerve cord: semaphorins pattern the dorso-ventral axis. PLoS biology. 2009;7 doi: 10.1371/journal.pbio.1000135. e1000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.