Abstract
Little is known about the nature of the conformational changes that convert G protein-coupled receptors (GPCRs), which bind diffusible ligands, from their resting into their active states. To gain structural insight into this process, various laboratories have used disulfide cross-linking strategies involving cysteine-substituted mutant GPCRs. Several recent disulfide cross-linking studies using the M3 muscarinic acetylcholine receptor as a model system have led to novel insights into the conformational changes associated with the activation of this prototypical class I GPCR. These structural changes are predicted to involve multiple receptor regions, primarily distinct segments of transmembrane helices III, VI, and VII, as well as helix 8. Given the high degree of structural homology found among most GPCRs, it is likely that these findings will be of considerable general relevance. A better understanding of the molecular mechanisms underlying GPCR activation may lead to novel strategies aimed at modulating GPCR function for therapeutic purposes.
Introduction
The superfamily of G protein-coupled receptors (GPCRs) represents the largest group of cell surface receptors found in nature [1]. The transmembrane (TM) core of these receptors, consisting of a bundle of seven TM helices (TM I-VII) shows a very high degree of structural conservation, at least among class I GPCRs [2–6]. Class I GPCRs represent by far the largest GPCR subfamily containing ~670 full-length human receptor proteins [1]. Members of this receptor family, including, for example, different dopamine, serotonin, adrenergic, muscarinic, prostanoid, cannabinoid, opioid, and somatostatin receptor subtypes, are the target of an extraordinarily large number of clinically important drugs [1]. Such drugs are widely used in the treatment of psychiatric and cardiovascular disorders and many other pathophysiological conditions [1].
The conformational changes involved in the activation process of class I GPCRs are currently being studied by many laboratories. The most common biochemical and biophysical approaches that have been used to monitor such structural changes are listed in Box 1 (for a recent review, see ref. 7). The structural insights gained from these studies may lead to the design of novel classes of drugs that can modulate specific GPCR signaling pathways.
BOX 1.
Common biophysical and biochemical approaches used to monitor conformational changes in GPCRs
| Techniquea | Advantagesb | Disadvantagesc |
|---|---|---|
| Site-directed spin labeling (SDSL) studies |
High temporal and, potentially, structural resolution |
Purified receptor proteins in the solution state are usually required Modification with reporter molecule is needed |
| Fluorescence measurements involving the use of fluorescent reporter groups |
High temporal resolution Multiple conformations can be distinguished by fluorescence lifetime studies |
Purified receptor proteins in the solution state are usually required Modification with reporter molecule is needed |
| Disulfide cross-linking scanning analysis |
Studies can be carried out with membrane preparations or intact cells Modification with reporter group is not required |
Trapping of rare receptor conformations may occur Limited temporal resolution |
| Substituted cysteine accessibility method (SCAM) |
Studies can be carried out with membrane preparations or intact cells |
Restricted to residues lining the ligand binding pocket Modification with sulfhydryl-specific reagents is required |
| Fluorescence resonance energy transfer (FRET) studies |
High-resolution kinetic studies can be carried out with live cells |
Limited structural resolution Modification with reporter proteins is needed |
Biochemical and biophysical analysis of bovine rhodopsin, a class I GPCR that is unique in that its endogenous ligand (11-cis-retinal) is covalently bound to the receptor protein, has elucidated the light-induced changes in receptor conformation in considerable detail [8–13]. The vast majority of these studies were carried out with mutant versions of rhodopsin in the solution state.
A recent study [14] reported the crystal structure of a photoactivated intermediate of bovine rhodopsin at relatively low resolution (4.15 Å). The authors concluded that the resting and the activated states of bovine rhodopsin showed only minor structural differences [14]. In contrast, a great amount of biophysical and biochemical evidence suggests that rhodopsin activation is associated with more significant structural changes [8–13]. Possible explanations for the surprising observation that Salom et al. did not detect any large-scale structural changes may be crystal packing constraints [14] or the existence of multiple substates of metarhodopsin II endowed with different degrees of activity [15].
In contrast to the photoreceptor rhodopsin, much less is known about the agonist-induced conformational changes that occur in GPCRs activated by diffusible ligands. Using fluorescence-based biophysical studies carried out with purified, mutationally modified versions of the β2-adrenergic receptor, Kobilka and his colleagues have detected several activity-dependent changes at the intracellular receptor surface [16, 17].
More recently, we employed a disulfide cross-linking strategy that allows the identification of activity-dependent conformational changes using receptors present in their native membrane environment (in situ) [18–24]. For these studies, we used the rat M3 muscarinic acetylcholine (ACh) receptor (M3 mAChR), a prototypic class I biogenic amine GPCR that preferentially activates Gq-type G proteins, as a model system. Similar approaches have recently been developed to study agonist-induced conformational changes in the thyrotropin-releasing hormone receptor type I [25] and parathyroid hormone receptors [26]. The aim of this review is to provide an overview about the outcome of several recent disulfide cross-linking studies carried out with the M3 mAChR and to provide a model of the structural changes associated with M3 mAChR activation. In addition, the conclusions drawn from these studies, including potential caveats associated with disulfide cross-linking approaches, will be discussed in the context of the activation mechanism proposed for bovine rhodopsin, the prototypic class I GPCR.
General strategy used to monitor disulfide cross-link formation in mutant M3 mAChRs
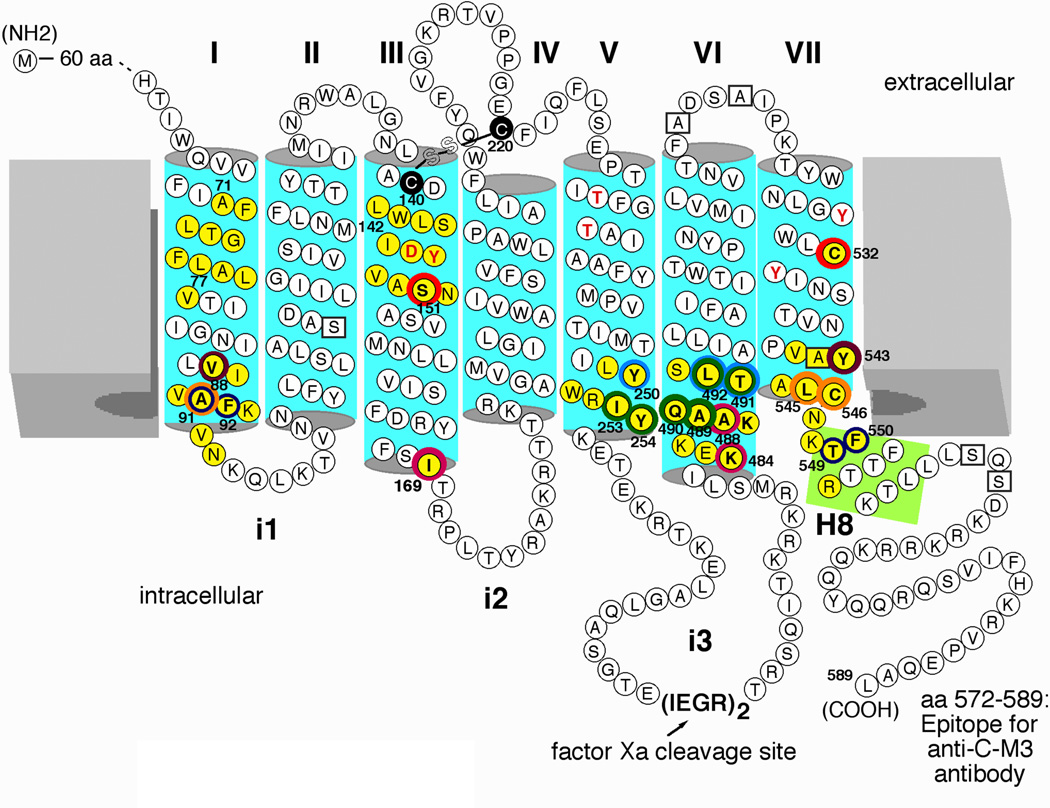
To render the M3 mAChR suitable for disulfide cross-linking studies, the receptor protein was modified as shown in Figure 1. Importantly, the modified M3 receptor lacked most native cysteine (Cys) residues, except for C140, C220, and C532, which proved to be essential for proper receptor expression and function [27]. In addition, the central portion of the third intracellular loop (i3 loop) of the receptor was replaced with two adjacent factor Xa cleavage sites. This mutationally modified M3 mAChR (referred to as M3’(3C)-Xa receptor in the following) shows ligand binding affinities and G protein coupling properties similar to those of the wild-type M3 mAChR [27].
Figure 1.
Agonist-modulated disulfide cross-linking in double Cys mutant M3 mAChRs. All Cys substitutions were introduced into the M3’(3C)-Xa receptor background, a modified version of the rat M3 mAChR [18]. In this construct, most endogenous Cys residues were replaced with either serine or alanine (boxed residues). The M3'(3C)-Xa construct contains only three remaining native Cys residues, C140, C220, and C5327.42 (note that C140 and 220 are engaged in a disulfide bond). In addition, the central portion of the i3 loop (A274-K469) was replaced with two adjacent factor Xa cleavage sites. The five potential N-glycosylation sites present in the N-terminal portion of the receptor protein (N6, N15, N41, N48, and N52) were replaced with Gln residues (not shown). A rabbit polyclonal antibody (referred to as anti-C-M3) was raised against the indicated C-terminal receptor sequence [18]. Muscarinic agonists promoted the formation of disulfide cross-links between Cys residues present at positions 1513.36 and 5327.42 (red rings; [21]), 881.53 and 5437.53 (brown rings; [20]), 911.56 and 5457.55/5467.56 (orange rings; [20]), 1693.54 and 4846.29/4886.33 (purple rings; [22]), 250C5.58 and 491C6.36/4926.37 (light blue rings; [24]), 2535.61 and 4896.34-4926.37 (green rings; [24]), and 2545.62 and 4896.34-4926.37 (green rings; [18]). In contrast, muscarinic agonists inhibited the formation of disulfide cross-links between Cys residues present at positions 911.56/921.57 and 5497.59/5507.60 (dark blue rings) [23]. The residues highlighted by red letters are known to play key roles in the binding of ACh and other conventional muscarinic agonists [33, 34]. Numbers refer to amino acid positions in the rat M3 mAChR sequence [33]. I-VII, TM helices I-VII; i1-i3, the three intracellular loops of the M3 mAChR; H8, helix 8.
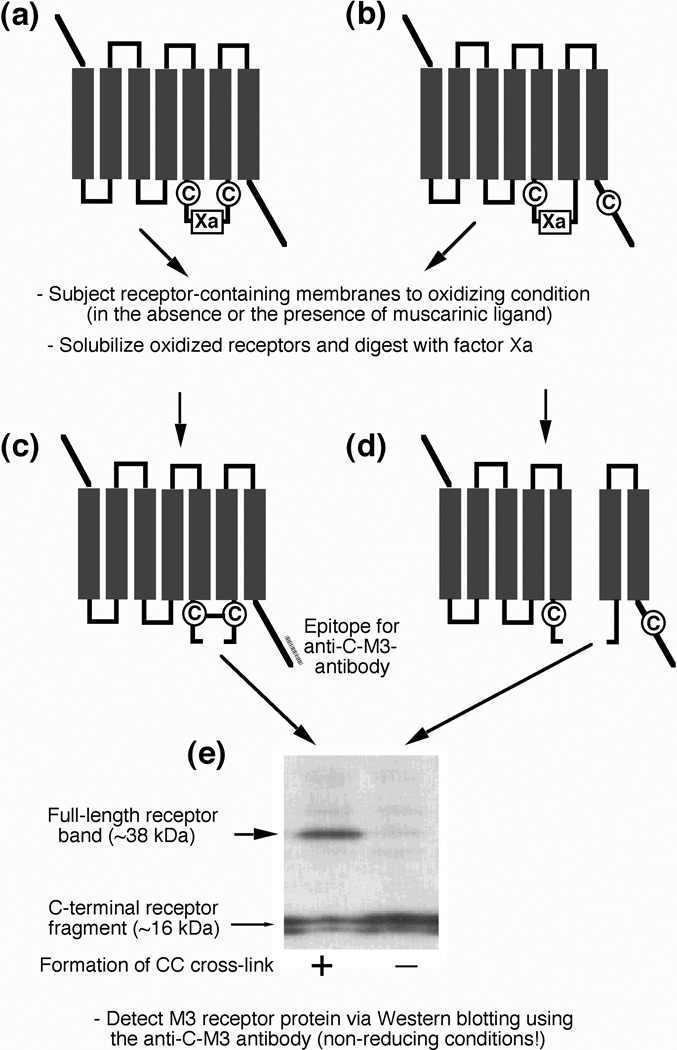
To obtain information about the conformational changes involved in M3 mAChR activation, we reintroduced pairs of Cys residues into the M3’(3C)-Xa background receptor, one Cys N-terminal and the other one C-terminal of the factor Xa cleavage site (Figure 2). When two Cys residues face each other in the three-dimensional (3D) structure of the receptor, they have the potential to form a disulfide bridge (Figure 2), either spontaneously, or in the presence of oxidizing agents such as Cu(II)-(1,10-phenanthroline)3 (referred to as 'Cu-Phen' in the following) or molecular iodine. Upon cleavage with factor Xa, the disulfide bridge will keep the two cleavage products covalently linked. As a result, a full-length receptor band, ~38 kDa in size, will appear on Western blots run under non-reducing conditions (Figure 2).
Figure 2.
General strategy used to detect the formation of disulfide bonds between vicinal Cys residues in mutant M3 mAChRs. Pairs of Cys residues were introduced into the M3’(3C)-Xa background receptor (see Figure 1), one Cys N-terminal and the other one C-terminal of the factor Xa cleavage site. (a, c) When two Cys residues lie adjacent to each other in the 3D structure of the receptor, they have the potential to form a disulfide bridge, either spontaneously or in the presence of oxidizing agents. Following cleavage with factor Xa, the two resulting receptor fragments will remain covalently linked by the disulfide bridge. Consequently, a full-length receptor band (~38 kDa in size) can be detected on Western blots run under non-reducing conditions (e, left lane). (b, d) When the two introduced Cys residues are not close to each other in the 3D structure of the receptor, they are unable to form a disulfide bridge. In this case, factor Xa digestion will yield two separate cleavage products and a full-length receptor band will not appear on Western blots run under non-reducing conditions (e, right lane).
In most cases, the distance between the α–carbon atoms of Cys residues engaged in a disulfide bridge ranges from 4.4 to 6.8 Å [28, 29]. The general assumption therefore is that two Cys residues have the potential to form a disulfide bond (under the appropriate experimental conditions) when the distance between their two α–carbon atoms is less than ~7 Å.
Activity-dependent conformational changes in the M3 mAChR as deduced from disulfide cross-linking studies
During the past few years, we have used the approach summarized in Figure 2 to analyze more than 100 different double Cys mutant M3 mAChRs [18–24], using membrane preparations from transiently transfected COS-7 cells. The positions that were targeted by Cys substitution mutagenesis are highlighted in Figure 1. To ensure that the double Cys mutant receptors were properly folded, all receptors were characterized in radioligand binding and second messenger studies for their ability to bind muscarinic ligands and to mediate agonist-dependent G protein activation, respectively [18–24].
Agonist-induced conformational changes occurring in the immediate vicinity of the ligand binding pocket
Agonist binding to GPCRs involves residues located on the extracellular surface of the receptor proteins [30–32]. Several of the key residues involved in ACh binding to the M3 mAChR (as well as all other mAChR subtypes; [33, 34]) are highlighted in Figure 1 (bold red letters). ACh binding to the M3 mAChR is predicted to trigger conformational changes within the TM receptor core, which are then propagated to the cytoplasmic surface of the receptor which is involved in G protein recognition and activation. The general view is that agonist binding leads to the disruption of existing interhelical (intramolecular) interactions, thereby promoting a set of interactions that leads to a new, energetically favorable conformational state of the receptor [17, 34].
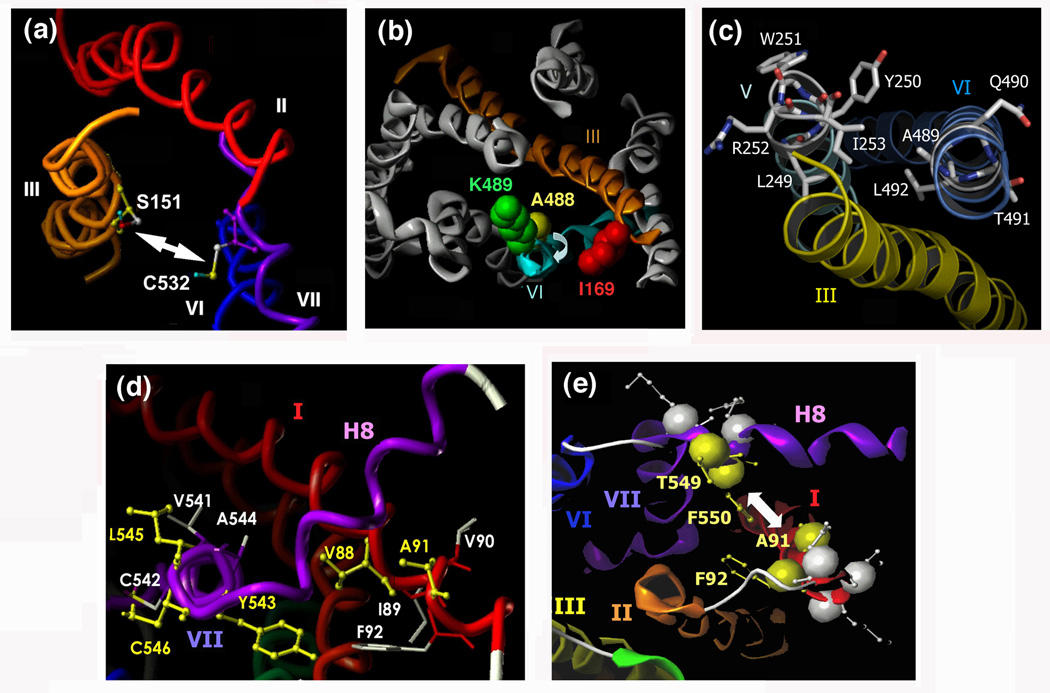
Interestingly, a recent study [21] demonstrated that muscarinic agonists promoted disulfide cross-linking between a Cys residue introduced into TM III at position S151 (position 3.36 [S151C3.36] according to the Ballesteros-Weinstein nomenclature of GPCRs; see Ref. [35]) and a TM VII Cys residue (C5327.42) of the M3 mAChR (Figure 1). A 3D model of the inactive form of the M3 receptor indicates that S151C3.36 and C5327.42 face each other in the interior of the TM receptor bundle (Figure 3a). The observed cross-linking pattern is therefore consistent with a model in which agonist binding to the M3 receptor increases the proximity of the exofacial segments of TM III and VII, thus allowing the formation of a disulfide bridge between S151C3.36 and C5327.42.
Figure 3.
Views of the M3 mAChR depicting regions/amino acids that undergo activity-dependent conformational changes. A 3D model of the inactive state of the rat M3 mAChR was built via homology modeling using the high-resolution X-ray structure of bovine rhodopsin as a template [2, 20]. (a) Extracellular view of the M3 mAChR parallel to the path of the exofacial segment of TM III. As discussed in the text, muscarinic agonists promote the formation of a disulfide bond between C5327.42 and a Cys residue introduced at position 1513.36 [21]. (b) Predicted location of I1693.54, K4846.29, and A4886.33 in the 3D structure of the inactive state of the M3 mAChR (cytoplasmic view). K4846.29 and A4886.33, which are located at the cytoplasmic end of TM VI, are projecting away from I1693.54 present at the bottom of TM III. Disulfide cross-linking data support the view that muscarinic agonists trigger a conformational change that includes a clockwise rotational movement of the cytoplasmic segment of TM VI [22]. (c) Predicted location of L2495.57-I2535.61 (TM V) and A4896.34-L4926.37 (TM VI) in the inactive state of the M3 mAChR (cytoplasmic view). Among the five highlighted TM V residues, only Y2505.58 and I2535.61 are predicted to face TM VI. Disulfide scanning mutagenesis studies strongly suggest that the cytoplasmic end of TM VI, but not the corresponding region of TM V, undergoes a major activity-dependent conformational change (see text for more details; [24]). (d) Cytoplasmic view of the inactive state of the M3 mAChR showing the locations of residues V881.53-F921.57 (TM I) and V5417.51-C5467.56 (TM VII). Cys substitution of the residues highlighted in yellow resulted in double Cys mutant receptors that showed agonist-dependent disulfide cross-linking (881.53/5437.53, 911.56/5457.55, and 911.56/5467.56) [20]. Whereas V881.53 and Y5437.53 directly face each other in the inactive state of the M3 muscarinic receptor, L5457.55 and C5467.56 point towards the lipid face and TM VI, respectively, away from A911.56, indicating that the cytoplasmic end of TM VII undergoes a major activity-dependent structural change during receptor activation (see text for more details; [20]). (e) Cytoplasmic view of a selected region of the intracellular surface of the M3 mAChR. Disulfide cross-linking data suggest that muscarinic agonists increase the distance between Cys residues introduced at a) positions 911.56 and 5497.59 and b) positions 921.57 and 5507.60 (these four residues are highlighted in yellow) [23]. In contrast, inverse muscarinic agonists are predicted to reduce the distance between these residues [23]. The individual figure panels were taken from Refs [21]-[24]. Numbers refer to amino acid positions in the rat M3 mAChR sequence [33].
Figure 1 indicates that S1513.36 and C5327.42 are located just below the ACh ligand binding domain. The positively charged ammonium head group of ACh is known to form a salt bridge with the negatively charged side chain of D1473.32 [33, 34]. This ion pair is predicted to be surrounded by a cage of aromatic amino acids located in TM III, VI, and VII, including Y1483.33, Y5066.51, Y5297.39, and Y5337.43 (Figure 1), which are thought to promote high affinity ACh binding to the receptor via cation-π interactions [33, 34].
Replacement of the three N-methyl groups of ACh with ethyl groups resulted in a muscarinic antagonist which, like the classical muscarinic antagonist atropine, was unable to promote disulfide bond formation between S151C3.36 and C5327.42 [21]. These data support the view that muscarinic antagonists are unable to mimic the proposed ACh-induced 'tightening' of the aromatic cage around the ACh quaternary ammonium group, most likely due to steric hindrance caused by the increased size of their ammonium head groups [21, 34].
The concept that agonist binding to GPCRs increases the proximity of the extracellular ends of different TM domains is consistent with studies in which metal ion (Cu2+ or Zn2+) binding sites (i.e. His or Cys residues) were introduced into the exofacial segments of TM III, VI and/or VII of the β2-adrenergic and the tachykinin NK1 receptors (for a recent review, see Ref. [36]). For example, a recent study [37] demonstrated that metal ions, either alone or in combination with aromatic chelators, can activate a modified version of the β2-adrenergic receptor in which positions Asp1133.32, Phe2896.51 and Asn3127.39 were substituted with His or Cys residues. Distance measurements derived from molecular modeling studies suggested that metal ion-mediated receptor activation involves a movement of the extracellular segments of TM VII and VI inward towards TM III [37], in agreement with the disulfide cross-linking data reported in Ref. [21].
It is tempting to speculate that the proposed inward movement of the extracellular ends of TM VI and VII is a common structural feature of members of the GPCR superfamily. At present, little is known about the series of conformational events that must occur within the TM receptor core in order to propagate these structural changes to the intracellular receptor surface where G protein coupling is known to occur. As has been discussed in detail elsewhere (for a recent review, see Ref. [36]), one possibility is that a rotation and straightening of conserved proline bends found in TM V-VII, particularly the one in TM VI, plays a key role in mediating these conformational changes.
Agonist-induced conformational changes at the cytoplasmic surface of the receptor protein
Changes in TM VI
To study the ability of muscarinic ligands to trigger conformational changes on the cytoplasmic side of the M3 receptor protein, we carried out disulfide cross-linking studies using a large number of mutant receptors containing pairs of Cys residues on the intracellular receptor surface [18, 21–24]. Consistent with the outcome of biochemical and biophysical studies carried out with bovine rhodopsin [8, 12, 13] and the β2-adrenergic receptor [16, 32], these cross-linking studies showed that the C-terminal segment of TM VI undergoes a major conformational change during receptor activation [18, 22, 24]. For example, a recent study [22] demonstrated that the muscarinic agonist, carbachol, promoted disulfide cross-linking in two mutant receptors that contained one Cys residue at the bottom of TM III (I169C3.54) and a second Cys residue at positions K484C6.29 or A488C6.33 within the cytoplasmic segment of TM VI (Figure 3b).
A 3D model of the rat M3 mAChR, which was established via homology modeling using the dark state of bovine rhodopsin as a template [2, 20], predicts that K4846.29 and A4886.33 do not project towards I1693.54 in the inactive state of the receptor (Figure 3b). The observed cross-linking pattern therefore supports a model in which M3 receptor activation is associated with a clockwise rotation of the cytoplasmic end of TM VI (cytoplasmic view), which would allow the formation of disulfide cross-links between I169C3.54 and K484C6.29 or A488C6.33. This conclusion is in agreement with the results of a site-directed spin labeling (SDSL) study carried out with Cys-substituted mutant versions of bovine rhodopsin [8]. The outcome of that study [8] suggested that light-induced rhodopsin activation triggers an 'outward' movement of the cytoplasmic portion of TM VI, accompanied by a clockwise rotation of ~30°, as viewed from the cytoplasm. A similar change in receptor conformation has also been proposed based on biochemical and biophysical studies carried out with mutant versions of the β2-adrenergic receptor [10, 16, 17, 32]. Biochemical studies with several GPCRs have also demonstrated that cross-linking of the cytoplasmic ends of TM III and VI, either via disulfide bonds [8] or via binding of metal ions [10] inhibits receptor/G protein coupling, indicating that the activity-dependent reorientation of the cytoplasmic end of TM VI plays a central role in G protein recognition and activation.
A related study [18] examined the ability of a Cys residue introduced at the TM V/i3 loop boundary (Y254C5.62) to form activity-dependent disulfide bonds with Cys residues introduced into the cytoplasmic portion of TM VI (K4846.29-S4936.38). This analysis demonstrated that agonist treatment promoted the formation of disulfide cross-links in four of the ten analyzed mutant receptors (Y254C5.62/A489C6.34, Y254C5.62/Q490C6.35, Y254C5.62/T491C6.36, and Y254C5.62/L492C6.37). Since the A4896.34-L4926.37 sequence is predicted to form an α-helical turn, this cross-linking pattern suggested that M3 receptor activation involves a more dramatic conformational change of the cytoplasmic end of TM VI, as compared to the corresponding domain of bovine rhodopsin (see next paragraph). The cross-linking data reported in ref. [18] also indicated that agonist binding to the M3 receptor increases the proximity between the cytoplasmic ends of TM V and VI (Figure 4), consistent with the results of fluorescence spectroscopic studies carried out with the β2-adrenergic receptor [38].
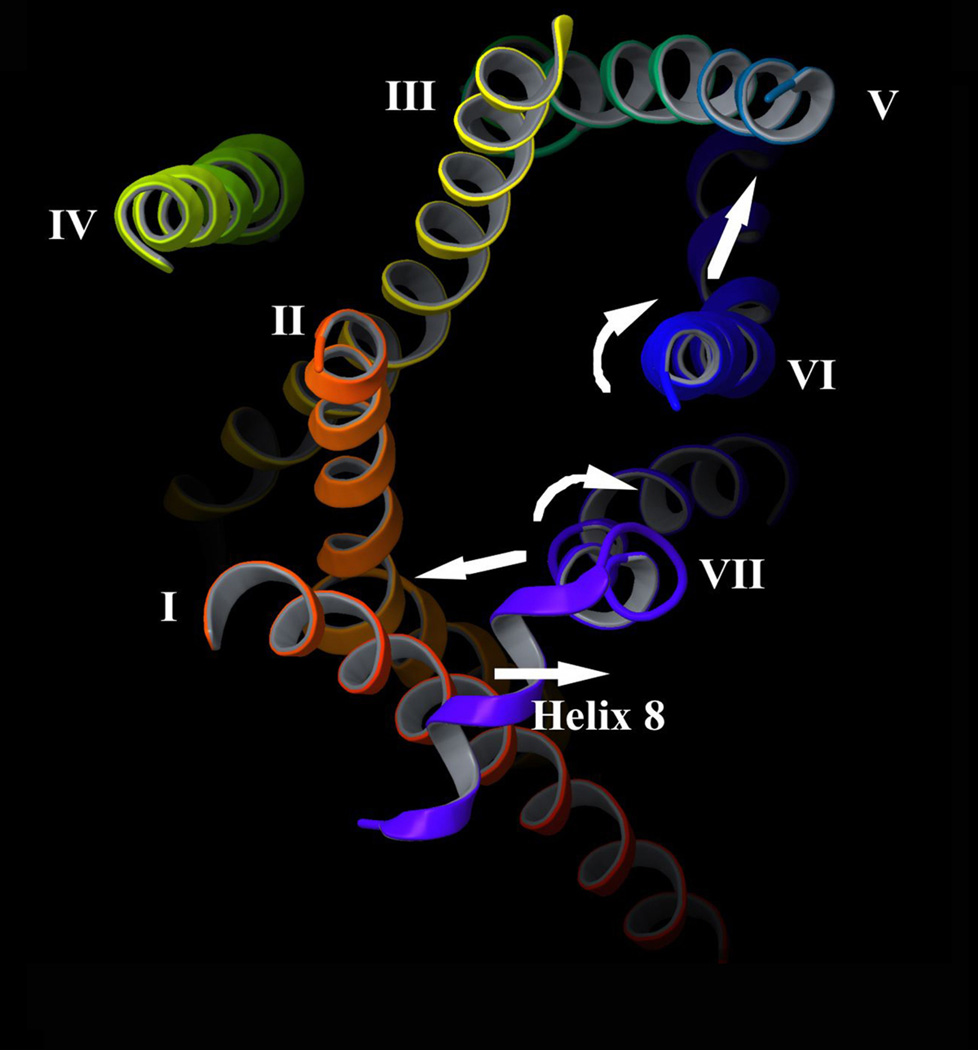
Figure 4.
Summary of activity-dependent structural changes predicted to occur at the cytoplasmic surface of the M3 mAChR. A 3D model of the inactive state of the rat M3 mAChR was built via homology modeling using the high-resolution X-ray structure of the β2-adrenergic receptor [4, 5] as a template (S. Costanzi and J. D. Karpiak; unpublished results). The intracellular surface of the M3 mAChR is viewed from the cytoplasm. As discussed in detail in the text, disulfide cross-linking data suggest that agonist binding to the M3 mAChR causes the following structural changes: 1) The cytoplasmic end of TM VI is predicted to undergo a rotational movement (and perhaps a partial unfolding) and to move closer to the corresponding segment of TM V [18, 22, 24]. 2) The C-terminal portion of TM VII is thought to move closer to the corresponding region of TM I, accompanied by a rotational movement (and perhaps a partial unfolding) of this TM VII segment [20]. 3) The N-terminal portion of helix 8 is predicted to move away from the cytoplasmic end of TM I [23]. The agonist-induced structural changes are thought to expose previously inaccessible receptor residues or surfaces (e.g. on the cytoplasmic ends of TM III, VI, and VII), ultimately resulting in productive receptor/G protein coupling.
To obtain additional structural information regarding activity-dependent changes in the relative orientation of TM V and VI, a recent disulfide cross-linking study [24] examined a series of double Cys mutant M3 receptors harboring one Cys substitution within the cytoplasmic end of TM V (L2495.57-I2535.61) and a second one within the cytoplasmic end of TM VI (A4896.34-L4926.37). This analysis showed that muscarinic agonists promoted the formation of disulfide bonds in six of the twenty analyzed double Cys mutant M3 receptors (Y250C5.58/T491C6.36, Y250C5.58/L492C6.37, I253C5.61/A489C6.34, I253C5.61/Q490C6.35, I253C5.61/T491C6.36, and I253C5.61/L492C6.37), confirming the view that M3 receptor activation increases the proximity between the cytoplasmic ends of TM V and VI. Whereas A4896.34 and L4926.37 are oriented towards Y2505.58 and I2535.61 in the inactive state of the receptor, Q4906.35 and T4916.36 project away from TM V facing the lipid bilayer and TM VII, respectively (Figure 3c). Thus, the ability of muscarinic agonists to promote the formation of disulfide bonds in constructs containing the Q490C6.35 or T491C6.36 mutations further corroborates the concept that M3 receptor activation involves a major structural change at the cytoplasmic end of TM VI [18]). Possible explanations for the observed cross-linking patterns include a pronounced rotational movement and/or a partial unfolding of this helical domain.
Interestingly, in the same study [24], muscarinic agonists failed to induce the formation of disulfide bonds in all double Cys mutant M3 receptors containing the L249C5.57, W251C5.59, or R252C5.60 substitutions in TM V. In contrast to Y2505.58 and I2535.61 (see above), L2495.57, W2515.59, and R2525.60 do not face TM VI but project toward the bottom of TM III (L2495.57) or the lipid bilayer (W2515.59 and R2525.60) in the inactive state of the receptor (Figure 3c). It is therefore likely that the cytoplasmic end of TM V does not undergo major activity-dependent conformational changes in the M3 mAChR.
Changes in TM VII
Much evidence suggests that residues contained within the C-terminal portion of TM VII (including the highly conserved NPXXY motif) play a key role in class I GPCR activation [12, 13, 39–42]. To test the hypothesis that this receptor region undergoes conformational changes during M3 receptor activation, a recent cross-linking study [20] analyzed a series of double Cys mutant M3 receptors containing one Cys substitution within the C-terminal portion of TM VII (V5417.51-S5467.56) and another Cys substitution within the cytoplasmic end of TM I (V881.53-F921.57). These two receptor regions are predicted to lie adjacent to each other in the 3D structure of class I GPCRs [2–6]. In the inactive state of the M3 receptor (in the absence of ligands), only the V88C1.53-Y543C7.53 receptor showed a robust disulfide cross-linking signal, consistent with the observation that V881.53 and Y5437.53 directly face each other at the TM I/TM VII interface (Figure 3d). Interestingly, agonist (carbachol) treatment further enhanced the intensity of the disulfide cross-linking signal displayed by this mutant receptor, suggesting that V881.53 and Y5437.53 move closer to each other following M3 receptor activation. Agonist binding also led to the formation of disulfide bonds in two additional double Cys mutant receptors, A91C1.56/L545C7.55 and A91C1.56/S546C7.56 [20]. In the inactive state of the M3 receptor, L5457.55 and S5467.56 project away from A911.56, facing the lipid bilayer and TM VI, respectively (Figure 3d). The observed cross-linking pattern therefore suggests that M3 receptor activation involves a major conformational change at the cytoplasmic end of TM VII. One possibility is that this region undergoes a significant rotational movement (and/or perhaps a partial unfolding) which would bring residues L5457.55 and S5467.56 within cross-linking distance of position A911.56. Moreover, the cross-linking data suggest that M3 receptor activation increases the proximity of the cytoplasmic portions of TM I and VII (Figure 4). Consistent with these findings, biochemical and biophysical studies with bovine rhodopsin indicate that the C-terminal segment of TM VII undergoes a significant light-induced conformational change [39–41].
TM VII, but not TM I, contains several key amino acids (e.g. Y5297.39 and Y5337.43 in the M3 mAChR) that are critically involved in the binding of muscarinic agonists [33, 34]. Moreover, TM VII contains a highly conserved proline residue (part of the NPXXY motif; corresponding to N5397.49-Y5437.53 in the rat M3 receptor; Figure 1). As discussed elsewhere [21, 36], it is possible that agonist binding triggers a reorientation of the conserved TM VII proline bend, which in turn leads to a conformational rearrangement of the C-terminal segment of TM VII. For these reasons, it is likely that the cytoplasmic end of TM VII moves towards the corresponding region of TM I (Figure 4).
Changes in Helix 8
Helix 8 represents a cytoplasmic α-helical extension of TM VII to which it is connected via a short linker sequence (Figure 1). Considerable evidence suggests that helix 8 plays an important role in productive receptor/G protein coupling [42–46]. A recent study [23] therefore tested the hypothesis that M3 receptor activation involves a reorientation of helix 8. Given the observation that several helix 8 residues are located close to the cytoplasmic end of TM I in bovine rhodopsin [2, 12], this disulfide cross-linking study [23] examined a series of double Cys mutant M3 receptors, all of which contained one Cys substitution within the cytoplasmic end of TM I (A911.56-N951.60) and a second one within the N-terminal segment of helix 8 (K5487.58-R5517.61), assuming that the TM I Cys residues might serve as useful reporters to detect potential ligand-induced movements of helix 8. This analysis showed that muscarinic agonists inhibited constitutive disulfide cross-linking displayed by the A91C1.56/T549C7.59 and F92C1.57/F550C7.60 double Cys mutant receptors [23]. Figure 3e indicates that residues 911.56 and 921.57 lie adjacent to residues 5497.59 and 5507.60 in the inactive state of the M3 receptor, strongly suggesting that M3 receptor activation is accompanied by a separation of the N-terminal segment of helix 8 from the cytoplasmic end of TM I (Figure 4). One likely scenario is that the reorientation of helix 8 is triggered by the agonist-induced conformational rearrangement of the cytoplasmic end of TM VII (see discussion above).
Interestingly, the same study [23] also reported that atropine and Nmethylscopolamine, two classical muscarinic antagonists that have recently been reclassified as inverse agonists [47, 48], enhanced disulfide bond formation in the A91C1.56/T549C7.59 and F92C1.57/F550C7.60 mutant receptors. This finding suggests that inverse muscarinic agonists, in contrast to muscarinic agonists, decrease the distance between the cytoplasmic end of TM I and the N-terminal portion of helix 8. A related study examining the formation of disulfide cross-links between Cys residues introduced at the cytoplasmic ends of TM V and VI also demonstrated that full and inverse muscarinic agonists had distinct effects on the efficiency of disulfide bond formation in specific double Cys mutant M3 receptors [24]. These studies [23, 24] therefore provide a structural basis for the opposing biological effects of muscarinic agonists and inverse agonists, highlighting the usefulness of disulfide cross-linking approaches to study changes in GPCR structure caused by different classes of agonists (also see Refs. [7, 16, 17, 49, 50]).
Summary of conformational changes
Overall, agonist binding to the M3 mAChR is predicted to cause the following structural changes at the intracellular receptor surface (Figure 4; for a summary of disulfide crosslinking data, also see Table 1). 1) The cytoplasmic end of TM VI is thought to undergo a rotational movement (and perhaps a partial unfolding) and to move closer to the corresponding segment of TM V [18, 22, 24]. 2) The cytoplasmic portion of TM VII is predicted to move closer to the corresponding region of TM I, also accompanied by a rotational movement and/or a partial unfolding [20]. 3) The N-terminal portion of helix 8 is likely to move away from the cytoplasmic end of TM I [23]. As discussed in more detail below ('Caveats inherent in the use of disulfide cross-linking strategies'), the possibility exists that at least some of the observed changes in agonist-dependent disulfide cross-linking patterns are also affected by the overall increase in dynamic flexibility of the active receptor confirmation.
Table 1.
Summary of the effects of muscarinic agonists on the formation of disulfide cross-links in double Cys mutant M3 mAChRs
| Residuea | Helix | Residue | Helix | Agonist effect on cross-linkingb |
Ref. |
|---|---|---|---|---|---|
| 881.53 | I | 5437.53 | VII | Stimulation | [20] |
| 911.56 | I | 5457.55 | VII | Stimulation | [20] |
| 911.56 | I | 5467.56 | VII | Stimulation | [20] |
| 911.56 | I | 5497.59 | H8 | Inhibition | [23] |
| 921.57 | I | 5507.60 | H8 | Inhibition | [23] |
| 1513.36 | III | 5327.42 | VII | Stimulation | [21] |
| 1693.54 | III | 4846.29 | VI | Stimulation | [22] |
| 1693.54 | III | 4886.33 | VI | Stimulation | [22] |
| 2505.58 | V | 4916.36 | VI | Stimulation | [24] |
| 2505.58 | V | 4926.37 | VI | Stimulation | [24] |
| 2535.61 | V | 4896.34 | VI | Stimulation | [24] |
| 2535.61 | V | 4906.35 | VI | Stimulation | [24] |
| 2535.61 | V | 4916.36 | VI | Stimulation | [24] |
| 2535.61 | V | 4926.37 | VI | Stimulation | [24] |
| 2545.62 | V | 4896.34 | VI | Stimulation | [18] |
| 2545.62 | V | 4906.35 | VI | Stimulation | [18] |
| 2545.62 | V | 4916.36 | VI | Stimulation | [18] |
| 2545.62 | V | 4926.37 | VI | Stimulation | [18] |
Numbers refer to amino acid positions in the rat M3 mAChR sequence [33]. Cys residues were introduced at the indicated positions, as described in the text.
All cross-linking studies were carried out under oxidizing conditions, either in the absence or presence of agonist, using membranes prepared from COS-7 cells transiently expressing double Cys mutant M3 mAChRs.
It remains unclear at present whether these predicted conformational changes occur in a concerted fashion or whether a primary structural change (e.g. the reorientation of TM VI) triggers secondary changes in the structure and orientation of other TM helices. In any case, the disulfide cross-linking data are consistent with the concept that agonist activation of the M3 mAChR opens a cleft on the intracellular receptor surface that increases the accessibility of various residues located at the cytoplasmic ends of different TM helices including TM III, VI, and VII (Figure 4). These activity-dependent changes are thought to promote receptor binding to heterotrimeric G proteins, ultimately triggering productive receptor/G protein coupling.
Comparison of activity-dependent changes predicted for the M3 mAChR versus bovine rhodopsin
As mentioned in the introduction, biochemical and biophysical studies with bovine rhodopsin, including the use of sophisticated SDSL techniques, have led to a rather detailed view of the activity-dependent structural changes occurring at the cytoplasmic surface of this photoreceptor protein [8–13]. Several of the dynamic changes observed in these studies, including conformational changes at the cytoplasmic ends of TM VI, VII, and helix 8 (summarized in Ref. [12]), are consistent with the outcome of the M3 mAChR disulfide cross-linking studies reviewed above. However, while the cytoplasmic end of TM VII is thought to move away from the TM helical bundle during rhodopsin activation [12], studies with the M3 mAChR suggest that this region moves closer to the intracellular end of TM I following agonist stimulation. Moreover, rhodopsin activation is predicted to be accompanied by small but significant outward movements of the cytoplasmic ends of TM I-III [12], movements that were not observed or not systematically studied in the M3 mAChR. It is unclear at present to which extent these observations reflect true differences between the pattern of structural changes that accompany the activation of these two receptor proteins. However, it is likely that at least some of these phenomena are caused by differences in experimental techniques and conditions used. Whereas the vast majority of the rhodopsin studies were carried out with mutant versions of rhodopsin in the solution state, all disulfide cross-linking studies were carried out with Cys-substituted mutant M3 receptors present in cellular membranes. Rhodopsin has been shown to have increased flexibility in the solution state as compared to native membranes [12, 51], providing a possible explanation for at least some of the differences observed between the two receptor systems. Moreover, while biophysical approaches, including SDSL and fluorescence-based techniques, usually provide information only about average receptor conformations, disulfide cross-linking can occur by trapping rather transient receptor conformations (for example, conformations that are intermediate between the ground state and the fully active state of the receptor) in which the two Cys residues are temporarily close to each other in the 3D structure of the receptor. The two different techniques therefore yield results that are complementary in nature.
Interestingly, a high-resolution crystal structure of ligand-free native opsin from bovine retinal rod cells has been published very recently [52]. When compared to rhodopsin, opsin displays several striking structural changes, resembling in many aspects the structure of the active state of rhodopsin predicted by biophysical and biochemical studies [12]. Many, but not all, of the activity-dependent conformational changes proposed for the M3 mAChR on the basis of disulfide cross-linking studies were also observed in the opsin structure. For example, when compared rhodopsin, the cytoplasmic end of TM VI displays an outward, rotational movement in opsin, the cytoplasmic ends of TM V and VI move closer towards each other, and the C-terminal portion of TM VII undergoes a rotational movement (clockwise as viewed from the cytoplasm; ref. [52])
Caveats inherent in the use of disulfide cross-linking strategies
One caveat associated with the use of disulfide cross-linking approaches is that negative results are difficult to interpret. Previous studies have shown that the efficiency of disulfide bond formation is determined not only by the distance between the two Cys residues under investigation but also by their relative orientation and the environment surrounding the Cys residues which can strongly affect the pKa values of the SH groups [28, 53]. It is therefore possible that two Cys residues, despite being close to each other in the 3D structure of the receptor (distance between α-carbons <7 Å), do not readily form a disulfide bond, even under strong oxidizing conditions.
It is also possible that two Cys residues whose α–carbon atoms are predicted to be relatively far apart in the 3D structure of the receptor (e.g. >7 Å) can be linked by a disulfide bridge [9, 10]. Such cross-links can form when the Cys residues are contained in receptor regions endowed with a high degree of conformational flexibility. For example, disulfide cross-linking studies carried out with Cys-substituted versions of a bacterial chemoreceptor of known structure demonstrated that some disulfide bonds can readily form when the two Cα carbons are up to ~12 Å apart [54]. In fact, structural and biophysical evidence suggests that several cytoplasmic GPCR domains, including the C-terminal tail beyond helix 8 and the central portion of the i3 loop, are characterized by a high degree of structural flexibility [2–5, 16, 17, 55].
In view of the caveats raised above, it is difficult (or impossible) to draw meaningful conclusions from disulfide cross-linking data obtained with only a small number of double Cys mutant receptors. It is therefore recommended to perform systematic scans in which consecutive residues in two receptor regions of interest are replaced, one by one, with Cys. Disulfide cross-linking data obtained with such a larger collection of double Cys mutant receptors are more likely to yield useful structural information.
Cu-Phen is a commonly used oxidizing agent to induce the formation of disulfide bonds in receptor-containing membrane preparations. The mechanism by which Cu-Phen agent promotes disulfide cross-linking is not well understood but may involve the transitory reduction of Cu2+ to Cu+ [56]. One caveat associated with the use of Cu-Phen is that this agent may induce the formation of disulfide cross-links of proteins in non-native conformations when used at high concentrations [9; J. Wess, unpublished observations). To reduce the likelihood of such events, we recommend to use Cu-Phen at relatively low concentrations (2.5 to 100 µM), if possible.
Previous work suggests that Cu-Phen-catalyzed disulfide bond formation does not require the generation of hydrogen peroxide as an oxidizing intermediate [56], suggesting that the Cu-Phen complex must be present in the direct vicinity of the SH groups to be cross-linked. This concept is supported by the results of a previous disulfide cross-linking study suggesting that muscarinic ligands compete with the relatively bulky Cu-Phen moiety for access to the hydrophilic binding crevice [19]. In such cases, the use of oxidizing agents that are smaller in size, such as molecular iodine, may yield more meaningful cross-linking data [19]. Mercury II salts (HgCl2) which can bridge vicinal pairs of Cys residues have also been used successfully to achieve cross-linking between Cys residues buried in the TM receptor core [57, 58].
Concluding remarks and future directions
Conclusions
In conclusion, disulfide cross-linking studies with the M3 mAChR have shed new light on the conformational changes associated with M3 mAChR activation. These structural changes are predicted to involve multiple receptor regions, primarily distinct segments of TM III, VI, and VII, as well as helix 8. Overall, the activity-dependent conformational changes postulated for the M3 mAChR are similar (but not identical) to those observed with the photoreceptor rhodopsin, highlighting the usefulness of disulfide cross-linking strategies to examine dynamic changes in GPCR structure. Future cross-linking studies are likely to provide even more detailed insights into the molecular mechanisms involved in GPCR activation. Since GPCRs represent ideal drug targets, the novel structural information obtained from these studies might facilitate the development of novel classes of therapeutic agents.
Future directions
All disulfide cross-linking studies carried out with the M3 mAChR have so far relied on the use of a receptor construct that contained a protease cleavage site within the i3 loop (Figure 1). As a result, this construct can be used only to detect the formation of disulfide bonds in double Cys mutant receptors where one Cys residue is located N-terminal and the other one C-terminal of the protease cleavage site. To be able to study relative conformational changes within the two receptor regions containing TM I-V and TM VI/VII, it will therefore be necessary to introduce protease cleavage sites into other intra- or extra-cellular loops of the receptor.
Studies with several GPCRs suggest that structurally diverse classes of agonists stabilize different receptor conformations which may differ in their ability to interact with G proteins and other receptor-associated proteins such as arrestins [7, 16, 17]. How these various conformations differ at the molecular level remains unclear at present. The disulfide cross-linking strategy described in this review may therefore prove useful to define the structural differences between these various active receptor states.
Little is known about the sequence of conformational events that link the activity-dependent structural changes in the immediate vicinity of the agonist binding site to the conformational rearrangement of the cytoplasmic receptor surface. It should therefore be highly informative to systematically apply the disulfide cross-linking approach described in this review to the entire TM receptor core.
Various lines of evidence indicate that GPCRs [59], including the M3 mAChR [60, 61], are arranged in dimeric or oligomeric arrays. Moreover, recent data suggest that GPCR activation may lead to changes in receptor quaternary structure. For example, disulfide cross-linking studies with the D2 dopamine [62] and the 5HT2c serotonin receptors [63] have shown that receptor activation is associated with dynamic changes at the dimer interface. These studies indicate that a full understanding of the molecular mechanisms that underlie GPCR activation also requires more detailed information about changes in GPCR quaternary structure.
Acknowledgements
The authors' own research covered in this review was supported by the Intramural Research Program of the NIH, NIDDK. We thank Dr. Stefano Costanzi and Mr. Joel D. Karpiak (NIH, NIDDK) for preparing Figure 4.
Glossary
- Ballesteros-Weinstein nomenclature of GPCRs
In the Ballesteros-Weinstein nomenclature of GPCRs [35], the most highly conserved residue in each TM helix is assigned the number 50 (N1.50, D2.50, R3.50, W4.50, P5.50, P6.50, and P7.50 in TM I-VII, respectively).
- Antagonist
A compound that binds to a receptor and blocks agonist-promoted effects without eliciting a cellular response on its own
- Inverse agonist
A compound that binds to a receptor and inhibits its constitutive activity
- Site-directed spin labeling (SDSL)
SDSL represents a powerful biophysical approach to explore protein structure and dynamics. In most cases, nitroxide spin label probes are introduced at specific positions in the protein. Analysis of the electron paramagnetic resonance (EPR) spectrum of the spin label provides information about the local environment in the protein. Analysis of a sufficiently large set of labeled proteins can provide rather detailed structural information. Importantly, changes in protein structure during function can be followed in real time.
References
- 1.Lagerström MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 2008;7:339–357. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 2.Palczewski K, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen SG, et al. Crystal structure of the human β2 adrenergic G-proteincoupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 4.Cherezov V, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 6.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann C, et al. Conformational changes in G-protein-coupled receptors - the quest for functionally selective conformations is open. BrJPharmacol. 2008;153(Suppl. 1):S358–S366. doi: 10.1038/sj.bjp.0707615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrens DL, et al. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 9.Struthers M, Oprian DD. Mapping tertiary contacts between amino acid residues within rhodopsin. Meth. Enzymol. 2000;315:130–143. doi: 10.1016/s0076-6879(00)15840-9. [DOI] [PubMed] [Google Scholar]
- 10.Meng EC, Bourne HR. Receptor activation: what does the rhodopsin structure tell us? Trends. Pharmacol. Sci. 2001;22:587–593. doi: 10.1016/s0165-6147(00)01825-3. [DOI] [PubMed] [Google Scholar]
- 11.Sakmar TP, et al. Rhodopsin: insights from recent structural studies. Annu. Rev. Biophys. Biomol. Struc. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 12.Hubbell WL, et al. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv. Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 13.Altenbach C, et al. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl. Acad. SciUSA. 2008;105:7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. SciUSA. 2006;103:16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knierim B, et al. Sequence of late molecular events in the activation of rhodopsin. Proc. Natl. Acad. SciUSA. 2007;104:20290–20295. doi: 10.1073/pnas.0710393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobilka BK. G protein coupled receptor structure and activation. Biochim. Biophys. Acta. 2006;768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Ward SD, et al. Use of an in situ disulfide cross-linking strategy to map proximities between amino acid residues in transmembrane domains I and VII of the M3 muscarinic acetylcholine receptor. J. Biol. Chem. 2002;277:2247–2257. doi: 10.1074/jbc.M107647200. [DOI] [PubMed] [Google Scholar]
- 19.Hamdan FF, et al. Use of an in situ disulfide cross-linking strategy to map proximities between amino acid residues in transmembrane domains I and VII of the M3 muscarinic acetylcholine receptor. Biochemistry. 2002;41:7647–7658. doi: 10.1021/bi016029c. [DOI] [PubMed] [Google Scholar]
- 20.Han SJ, et al. Pronounced conformational changes following agonist activation of the M3 muscarinic acetylcholine receptor. J. Biol. Chem. 2005;280:24870–24879. doi: 10.1074/jbc.M500379200. [DOI] [PubMed] [Google Scholar]
- 21.Han SJ, et al. Identification of an agonist-induced conformational change occurring adjacent to the ligand-binding pocket of the M3 muscarinic acetylcholine receptor. J. Biol. Chem. 2005;280:34849–34858. doi: 10.1074/jbc.M506711200. [DOI] [PubMed] [Google Scholar]
- 22.Ward SD, et al. Use of an in situ disulfide cross-linking strategy to study the dynamic properties of the cytoplasmic end of transmembrane domain VI of the M3 muscarinic acetylcholine receptor. Biochemistry. 2006;45:676–685. doi: 10.1021/bi051503q. [DOI] [PubMed] [Google Scholar]
- 23.Li JH, et al. Distinct structural changes in a G protein-coupled receptor caused by different classes of agonist ligands. J. Biol. Chem. 2007;282:26284–26293. doi: 10.1074/jbc.M704875200. [DOI] [PubMed] [Google Scholar]
- 24.Li JH, et al. Ligand-specific changes in M3 muscarinic acetylcholine receptor structure detected by a disulfide scanning strategy. Biochemistry. 2008;47:2776–2788. doi: 10.1021/bi7019113. [DOI] [PubMed] [Google Scholar]
- 25.Huang W, et al. Agonist-induced conformational changes in thyrotropinreleasing hormone receptor type I: disulfide cross-linking and molecular modeling approaches. Biochemistry. 2005;44:2419–2431. doi: 10.1021/bi048808+. [DOI] [PubMed] [Google Scholar]
- 26.Thomas BE, et al. Conformational changes in the parathyroid hormone receptor associated with activation by agonist. Mol. Endocrinol. 2008;22:1154–1162. doi: 10.1210/me.2007-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng FY, et al. Use of a disulfide cross-linking strategy to study muscarinic receptor structure and mechanisms of activation. J. Biol. Chem. 1999;274:16629–16640. doi: 10.1074/jbc.274.23.16629. [DOI] [PubMed] [Google Scholar]
- 28.Kosen PA. Disulfide Bonds in Proteins. In: Ahern TJ, Manning MC, editors. Stability of Protein Pharmaceuticals, Part A: Chemical and Physical Pathways of Protein Degradation. Plenum Press; 1992. pp. 31–67. [Google Scholar]
- 29.Klein-Seetharaman J, et al. Probing the dark state tertiary structure in the cytoplasmic domain of rhodopsin: proximities between amino acids deduced from spontaneous disulfide bond formation between Cys316 and engineered cysteines in cytoplasmic loop 1. Biochemistry. 2001;40:12472–12478. doi: 10.1021/bi010746p. [DOI] [PubMed] [Google Scholar]
- 30.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce KL, et al. Seven-transmembrane receptors. Nat. Rev. Mol. Cell. Biol. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 32.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 33.Wess J. Molecular biology of muscarinic acetylcholine receptors. Crit. Rev. Neurobiol. 1996;10:69–99. doi: 10.1615/critrevneurobiol.v10.i1.40. [DOI] [PubMed] [Google Scholar]
- 34.Lu ZL, et al. Seven-transmembrane receptors: crystals clarify. Trends Pharmacol. Sci. 2002;23:140–146. doi: 10.1016/S0165-6147(00)01973-8. [DOI] [PubMed] [Google Scholar]
- 35.Ballesteros JA, Weinstein H. Integrated methods for modeling Gprotein coupled receptors. Meth. Neurosci. 1995;25:366–428. [Google Scholar]
- 36.Schwartz TW, et al. Molecular mechanism of 7TM receptor activation - a global toggle switch model. Annu. Rev. Pharmacol. Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 37.Elling CE, et al. Metal ion site engineering indicates a global toggle switch model for seven-transmembrane receptor activation. J. Biol. Chem. 2006;281:17337–17346. doi: 10.1074/jbc.M512510200. [DOI] [PubMed] [Google Scholar]
- 38.Ghanouni P, et al. Agonist-induced conformational changes in the G-proteincoupling domain of the β2 adrenergic receptor. Proc. Natl. Acad. SciU.SA. 2001;98:5997–6002. doi: 10.1073/pnas.101126198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdulaev NG, Ridge KD. Light-induced exposure of the cytoplasmic end of transmembrane helix seven in rhodopsin. Proc. Natl. Acad. SciU.SA. 1998;95:12854–12859. doi: 10.1073/pnas.95.22.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altenbach C, et al. Structural features and light-dependent changes in the sequence 306–322 extending from helix VII to the palmitoylation sites in rhodopsin: a site-directed spin-labeling study. Biochemistry. 1999;38:7931–7937. doi: 10.1021/bi9900121. [DOI] [PubMed] [Google Scholar]
- 41.Fritze O, et al. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc. Natl. Acad. SciU.SA. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wess J. Molecular basis of receptor/G-protein-coupling selectivity. Pharmacol. Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- 43.Cai K, et al. Single-cysteine substitution mutants at amino acid positions 306–321 in rhodopsin, the sequence between the cytoplasmic end of helix VII and the palmitoylation sites: sulfhydryl reactivity and transducin activation reveal a tertiary structure. Biochemistry. 1999;38:7925–7930. doi: 10.1021/bi9900119. [DOI] [PubMed] [Google Scholar]
- 44.Ernst OP, et al. Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin α and γ subunits. J. Biol. Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 45.Swift S, et al. Role of the PAR1 receptor 8th helix in signaling: the 7-8-1 receptor activation mechanism. J. Biol. Chem. 2006;281:4109–4116. doi: 10.1074/jbc.M509525200. [DOI] [PubMed] [Google Scholar]
- 46.Delos Santos NM, et al. Characterization of the residues in helix 8 of the human β1-adrenergic receptor that are involved in coupling the receptor to G proteins. J. Biol. Chem. 2006;281:12896–12907. doi: 10.1074/jbc.M508500200. [DOI] [PubMed] [Google Scholar]
- 47.Spalding TA, et al. Pharmacology of a constitutively active muscarinic receptor generated by random mutagenesis. J. Pharmacol. Exp. Ther. 1995;275:1274–1279. [PubMed] [Google Scholar]
- 48.Nelson CP, et al. Constitutive activity and inverse agonism at the M2 muscarinic acetylcholine receptor. J. Pharmacol. Exp. Ther. 2006;316:279–288. doi: 10.1124/jpet.105.094383. [DOI] [PubMed] [Google Scholar]
- 49.Gether U, et al. Fluorescent labeling of purified β2 adrenergic receptor. Evidence for ligand-specific conformational changes. J. Biol. Chem. 1995;270:28268–28275. doi: 10.1074/jbc.270.47.28268. [DOI] [PubMed] [Google Scholar]
- 50.Vilardaga JP, et al. Molecular basis of inverse agonism in a G proteincoupled receptor. Nat. Chem. Biol. 2005;1:25–28. doi: 10.1038/nchembio705. [DOI] [PubMed] [Google Scholar]
- 51.Kusnetzow AK, et al. Conformational states and dynamics of rhodopsin in micelles and bilayers. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JH, et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 53.Shaked Z, et al. Rates of thiol-disulfide interchange reactions involving proteins and kinetic measurements of thiol pKa values. Biochemistry. 1980;19:4156–4166. doi: 10.1021/bi00559a004. [DOI] [PubMed] [Google Scholar]
- 54.Careaga CL, Falke JJ. Thermal motions of surface α-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. J. Mol. Biol. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park PS, et al. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu. Rev. Pharmacol. Toxicol. 2008;48:107–141. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobashi K. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim. Biophys. Acta. 1968;158:239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- 57.Soskine M, et al. Crosslinking of membrane-embedded cysteines reveals contact points in the EmrE oligomer. Proc. Natl. Acad. SciUSA. 2002;99:12043–12048. doi: 10.1073/pnas.192392899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo W, et al. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. SciUSA. 2005;102:17495–17500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milligan G, Bouvier M. Methods to monitor the quaternary structure of G protein-coupled receptors. FEBS J. 2005;272:2914–2925. doi: 10.1111/j.1742-4658.2005.04731.x. [DOI] [PubMed] [Google Scholar]
- 60.Zeng FY, Wess J. Identification and molecular characterization of m3 muscarinic receptor dimers. J. Biol. Chem. 1999;274:19487–19497. doi: 10.1074/jbc.274.27.19487. [DOI] [PubMed] [Google Scholar]
- 61.Goin JC, Nathanson NM. Quantitative analysis of muscarinic acetylcholine receptor homo- and heterodimerization in live cells: regulation of receptor down-regulation by heterodimerization. J. Biol. Chem. 2006;281:5416–5425. doi: 10.1074/jbc.M507476200. [DOI] [PubMed] [Google Scholar]
- 62.Guo W, et al. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. SciUSA. 2005;102:17495–17500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mancia F, et al. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–369. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]