Abstract
Benzo[a]pyrene (BP) is a potent pro-carcinogen and ubiquitous environmental pollutant. Here, we examined the induction and modulation of CYP1A1 and CYP1B1 and 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPdG) adduct formation in DNA from 20 primary normal human mammary epithelial cell (NHMEC) strains exposed to BP (4 μM) in the absence or presence of chlorophyllin (5 μM). Real-time polymerase chain reaction (RT-PCR) analysis revealed strong induction of both CYP1A1 and CYP1B1 by BP, with high levels of inter-individual variability. Variable BPdG formation was found in all strains by r7, t8-dihydroxy-t-9, 10 epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE)-DNA chemiluminescence assay (CIA). Chlorophyllin mitigated BP-induced CYP1A1 and CYP1B1 gene expression in all 20 strains when administered with BP. Chlorophyllin, administered prior to BP-exposure, mitigated CYP1A1 expression in 18/20 NHMEC strains (p < 0.005) and CYP1B1 expression in 17/20 NHMEC strains (p < 0.005). Maximum percent reductions of CYP1A1 and CYP1B1 gene expression and BPdG adduct formation were observed when cells were pre-dosed with chlorophyllin followed by administration of the carcinogen with chlorophyllin (p < 0.005 for CYP1A1 and CYP1B1 expression and p < 0.0005 for BPdG adducts). Therefore, chlorophyllin is likely to be a good chemoprotective agent for a large proportion of the human population.
Keywords: Real-time-PCR, BPDE-DNA chemiluminescence, immunoassay, Chemoprevention, Chlorophyllin, Polycyclic aromatic hydrocarbons, DNA adducts
1. Introduction
Humans are constantly exposed to an array of potentially genotoxic xenobiotics, either through the environment, lifestyle or diet. Benzo[a]pyrene (BP), a polycyclic aromatic hydrocarbon (PAH), is a ubiquitous environmental pollutant formed from the incomplete combustion of fossil fuels, food (notably char-broiled meat and fish), tobacco, and other vegetable material [1]. Exposure can occur through ingestion, inhalation or dermal absorption.
BP, a procarcinogen, is bioactivated by BP inducible cytochrome P450s (CYP1A1 and CYP1B1) to reactive, DNA binding electrophiles [2]. Among the various metabolites formed, r7, t8-dihydroxy-t-9, 10 epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) is highly reactive and capable of binding to deoxyguanosine in DNA to form 10-(deoxyguanosin-N2-yl)-7,8,9-trihydroxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPdG) adducts [3].
Given the ubiquitous nature of PAHs, complete avoidance of exposure is impossible, thus creating a need for intervention strategies to reduce genotoxicity. Chemoprevention, involving the selective modulation of metabolic enzyme pathways, may help to reduce carcinogenic risk. Chlorophyllin, a water soluble copper–sodium metalloporphyrin, derived by treatment of a methanolic solution of the natural pigment chlorophyll with sodium hydroxide or potassium hydroxide [4], is currently used as a food colorant and deodorizer. It has been found to be a safe and effective chemopreventive agent when used in human populations in Africa and China which ingest aflatoxins and are therefore at risk of hepatocarcinoma induction [5,6]. Given its multiple modes of action, which include its role as a cell cycle modulator, Phase II enzyme inducer, antioxidant, carcinogen transport modulator, interceptor molecule and a modulator of Phase I inducers [7–9], we hypothesized that chlorophyllin might be a useful chemopreventive agent for a broad spectrum of the human population.
Normal human mammary epithelial cells (NHMECs) have previously been used as a model for human inter-individual variability in studies examining the metabolism and activation of BP [10]. Normal human breast tissue taken at reduction mammoplasty was used to establish the panel of 20 cell strains used here. In these experiments we examined the induction and inter-individual variability of CYP1A1 and CYP1B1 expression, and BPdG adduct formation upon exposure of NHMECs to BP, in the absence or presence of the chemopreventive agent chlorophyllin.
2. Materials and methods
2.1. Cell strains
Primary NHMEC strains were developed from breast tissues discarded at reduction mammoplasty and obtained through the Cooperative Human Tissue Network (CHTN sponsored by the National Cancer Institute (NCI) and National Disease Research Interchange). The isolation of mammary epithelial cells from breast tissue involved a combination of mechanical disruption and enzymatic digestion and has been described in detail elsewhere [11]. The National Institute for Occupational Safety and Health (NIOSH) Human Studies Review Board (HSRB) was informed about this work, but since no personally identifiable data were collected it was not judged to be in the perview of the HSRB. Information on age and ethnicity of donors, as well as the corresponding deidentified pathology report, were provided by the CHTN.
2.2. Chemicals, media and NHMEC exposures
BP was obtained from the NCI Chemical Carcinogen Reference Standard Repository (Kansas City, MO). Chlorophyllin was purchased from Sigma–Aldrich (St. Louis, MO).
Serum free mammary epithelial basal medium (MEBM medium) was purchased from Clonetics™ (Walkersville, MD). Cells, maintained at 37 °C, 5% CO2 and 95% relative humidity, were grown in MEBM supplemented with Singlequots™ (gentamycin sulfate-amphotericin B, recombinant human epidermal growth factor, bovine insulin, bovine pituitary extract and hydrocortisone) according to the manufacturer’s instructions. All treatments were carried out using 70% confluent cells at passage 6.
The exposure regimens used were as follows: T1-BP (4 μM) for 24 h; T2-BP (4 μM) plus chlorophyllin (5 μM) co-treatment for 24 h; T3–24 h pre-treatment with chlorophyllin (5 μM) followed by BP (4 μM) for 24 h; and T4–24 h pre-treatment with chlorophyllin (5 μM) followed co-treatment for 24 h with chlorophyllin plus BP (4 μM). In preliminary studies chlorophyllin alone (5 μM) was used in addition to the above treatments. All exposures were carried out in duplicate.
2.3. Expression analysis of CYP1A1 and CYP1B1
Total RNA isolated from all treatment groups (RNeasy kit, Qiagen, Valencia, CA) and 1 μg was reverse transcribed to single stranded cDNA (Advantage RT for PCR kit, BD Biosciences, Palo Alto, CA). Transcribed RNA was used for the quantitation of CYP1A1 and CYP1B1 gene expression using an ABI7700 Sequence Detection System, and Taqman® Gene Expression Assays, with Taqman® Universal PCR Master Mix (Applied Biosystems, Foster City, CA). The expression levels of CYP1A1 and CYP1B1 were normalized to that of GAPDH and the fold-change expressed using the 2−ΔΔCT method [12].
2.4. DNA isolation
Following exposure, adherent cells at ~90% confluency were washed once with 1× PBS and incubated in lysis buffer (100 mM Tris pH 8.5, 5 mM EDTA, 0.2% SDS, 200 mM NaCl and 100 μg/ml Proteinase K) at 37 °C for 3 h. DNA from the lysed cells was precipitated directly, using an equal volume of isopropanol, washed twice with 70% ethanol, and dissolved in an appropriate volume of nuclease free water [13].
2.5. BPdG adduct analysis by chemiluminescence immunoassay (CIA)
Antiserum elicited against DNA modified with BPDE [14] was used to measure BPdG adducts in isolated DNA by competitive BPDE-DNA CIA. Briefly, 96-well high-binding LIA plates (Greiner Labortechnik, FRG) coated for 48 h with 1 fmol of sonicated BPDE–DNA (modified to about 1.0%), or calf thymus DNA, were blocked of non-specific binding by incubation with 0.25% casein in phosphate buffered saline containing 0.05% Tween-20 (PBST) for 60 min. Subsequently, serially diluted, heat denatured (95 °C, 4 min) BPDE–DNA standard (1.0 BPdG adduct/106 nucleotides), or sample DNA from treatment groups, were mixed with an equal volume of rabbit anti-BPDE–DNA antibody (diluted to 1:3,000,000 in 0.25% casein), incubated for 15 min at 37 °C, and added to the LIA plate wells. The final mixture was incubated at 37 °C for 90 min. At the end of the incubation, the plates were washed with PBST, incubated with biotinylated anti-rabbit IgG (1:3000 dilution in 0.25% casein in PBST) for 90 min at room temperature and washed again with PBST. The plates were then incubated for 60 min with streptavidin alkaline phosphatase (diluted 1:3000 using 0.25% casein in PBST), washed again with PBST and Tris buffer, and finally incubated with CDP-Star containing Emerald II enhancer (4 °C, overnight). The luminescence was read at 542 nm both at the end of the incubation and on the following day. The limit of detection was 0.3 adducts/109 nucleotides.
2.6. Statistical analysis
Pearson correlation coefficients were used to compare the effects of BP and chlorophyllin treatments in the Excel program [15]. Paired, 2- tailed Student’s t test was used to compare the adducts formed in the T1 group to the adducts formed in the T2, T3 and T4 groups.
3. Results
3.1. CYP1A1 and CYP1B1 gene expression in NHMECs exposed to BP
The NHMEC strains used in this study, derived from 20 donors between 18 and 51 years of age whose pathology reports all indicated normal breast tissues, were exposed (as described in Section 2.2) and examined by RT-PCR for expression of CYP1A1 and CYP1B1. Preliminary studies with exposure to chlorophyllin alone showed no change in the expression of CYP1A1 and CYP1B1 (data not shown). In cells exposed to BP alone (T1 group), induction of CYP1A1 was 3.6- to 95.8-fold (Table 1), and induction of CYP1B1 was 3.5- to 43.4-fold (Table 2). CYP1A1 induction was moderately, but significantly, correlated with CYP1B1 induction (r = 0.530, p < 0.01), replicating data observed previously [12].
Table 1.
CYP1A1 induction (fold-change measured by RT-PCR) in NHMEC strains by BP in the absence (T1a) and presence (T2, T3, T4) of chlorophyllin.
| Cell Strain | T1 | T2 | T3 | T4 |
|---|---|---|---|---|
| M98014 | 4.6 ± 1.5b | 2.8 ± 0.3 | 8.3 ± 1.9 | 5.9 ± 0.9 |
| M98015 | 50.0 ± 6.6 | 44.8 ± 12.1 | 42.6 ± 10.5 | 47.4 ± 9.4 |
| M98011 | 5.4 ± 2.0 | 5.1 ± 1.0 | 5.5 ± 0.6 | 5.0 ± 2.6 |
| M99003 | 14.3 ± 5.0 | 10.2 ± 2.3 | 11.5 ± 1.6 | 13.0 ± 4.0 |
| M99015 | 35.9 ± 4.7 | 20.8 ± 3.0 | 31.8 ± 2.7 | 29.9 ± 9.7 |
| M98019 | 3.6 ± 1.0 | 1.9 ± 0.5 | 3.0 ± 0.6 | 2.4 ± 0.8 |
| M98025 | 11.2 ± 3.2 | 6.0 ± 2.6 | 9.8 ± 1.6 | 6.9 ± 0.7 |
| M99021 | 5.9 ± 1.5 | 3.7 ± 1.6 | 3.8 ± 0.4 | 3.6 ± 0.5 |
| M00012 | 17.1 ± 2.9 | 11.0 ± 1.7 | 12.2 ± 2.8 | 9.8 ± 2.7 |
| M98026 | 52.7 ± 12.9 | 39.0 ± 10.2 | 31.3 ± 11.5 | 30.3 ± 6.0 |
| M99004 | 20.3 ± 2.1 | 10.8 ± 4.0 | 10.5 ± 3.4 | 11.6 ± 1.6 |
| M99016 | 95.8 ± 9.9 | 62.4 ± 20.8 | 62.4 ± 4.8 | 53.5 ± 5.4 |
| M99006 | 6.5 ± 1.5 | 4.2 ± 1.0 | 5.3 ± 0.6 | 3.5 ± 0.8 |
| M00004 | 7.1 ± 2.5 | 4.0 ± 0.5 | 4.4 ± 0.8 | 3.8 ± 1.4 |
| M98035 | 13.8 ± 2.9 | 12.8 ± 2.0 | 9.7 ± 1.9 | 5.3 ± 1.6 |
| M98030 | 44.3 ± 12.4 | 18.5 ± 1.3 | 25.5 ± 2.1 | 16.0 ± 4.1 |
| M99005 | 13.7 ± 1.4 | 7.9 ± 1.7 | 8.1 ± 1.1 | 4.1 ± 1.4 |
| M99025 | 6.6 ± 2.9 | 4.1 ± 1.6 | 2.6 ± 0.5 | 1.7 ± 0.5 |
| M98016 | 32.5 ± 3.4 | 10.0 ± 2.3 | 13.2 ± 1.4 | 6.1 ± 2.0 |
| M98013 | 66.7 ± 12.1 | 16.4 ± 20.8 | 14.7 ± 6.9 | 8.0 ± 5.0 |
T = Treatment groups as defined in Section 2.
Numbers represent fold-change ±SD (n = 2).
Table 2.
CYP1B1 induction (fold-change measured by RT-PCR) in NHMEC strains by BP in the absence (T1a) and presence (T2, T3, T4) of chlorophyllin.
| Cell Strain | T1a | T2 | T3 | T4 |
|---|---|---|---|---|
| M98014 | 3.5 ± 0.6b | 2.9 ± 0.9 | 7.9 ± 1.0 | 6.8 ± 1.8 |
| M98015 | 43.4 ± 4.8 | 35.9 ± 3.3 | 39.7 ± 9.3 | 39.3 ± 6.9 |
| M98011 | 4.7 ± 0.9 | 3.7 ± 1.5 | 5.7 ± 1.5 | 6.3 ± 1.5 |
| M99003 | 19.7 ± 1.9 | 13.0 ± 1.2 | 15.3 ± 1.0 | 16.5 ± 5.2 |
| M99015 | 33.9 ± 1.7 | 13.5 ± 2.3 | 22.6 ± 11.2 | 19.9 ± 3.7 |
| M98019 | 6.1 ± 0.4 | 4.4 ± 0.4 | 7.9 ± 1.0 | 6.8 ± 1.8 |
| M98025 | 8.4 ± 2.2 | 4.4 ± 1.8 | 4.8 ± 1.5 | 3.9 ± 0.5 |
| M99021 | 8.8 ± 1.4 | 5.8 ± 1.0 | 6.3 ± 1.2 | 5.5 ± 0.9 |
| M00012 | 17.5 ± 3.1 | 10.3 ± 1.7 | 14.6 ± 4.4 | 11.9 ± 3.3 |
| M98026 | 31.7 ± 3.3 | 21.1 ± 1.7 | 24.2 ± 8.4 | 23.6 ± 6.4 |
| M99004 | 12.1 ± 1.3 | 7.7 ± 1.3 | 7.0 ± 0.7 | 6.9 ± 0.8 |
| M99016 | 8.9 ± 1.4 | 8.6 ± 1.6 | 8.3 ± 1.1 | 6.9 ± 1.3 |
| M99006 | 8.9 ± 1.6 | 5.5 ± 0.4 | 9.9 ± 3.6 | 4.8 ± 0.8 |
| M00004 | 7.9 ± 2.6 | 5.1 ± 0.5 | 7.3 ± 2.9 | 4.5 ± 2.0 |
| M98035 | 9.5 ± 1.9 | 8.0 ± 1.8 | 7.3 ± 1.8 | 3.3 ± 0.4 |
| M98030 | 16.0 ± 4.1 | 7.0 ± 1.4 | 6.5 ± 1.0 | 4.9 ± 1.8 |
| M99005 | 9.2 ± 1.8 | 7.5 ± 0.4 | 5.8 ± 1.7 | 3.5 ± 1.3 |
| M99025 | 22.9 ± 3.9 | 11.4 ± 3.1 | 9.2 ± 1.2 | 6.5 ± 1.3 |
| M98016 | 14.7 ± 3.0 | 6.2 ± 0.6 | 10.0 ± 5.9 | 3.9 ± 1.1 |
| M98013 | 42.0 ± 6.9 | 14.0 ± 1.5 | 11.5 ± 1.9 | 7.0 ± 3.2 |
T = Treatment groups as defined in Section 2.
Numbers represent fold-change ±SD (n = 2).
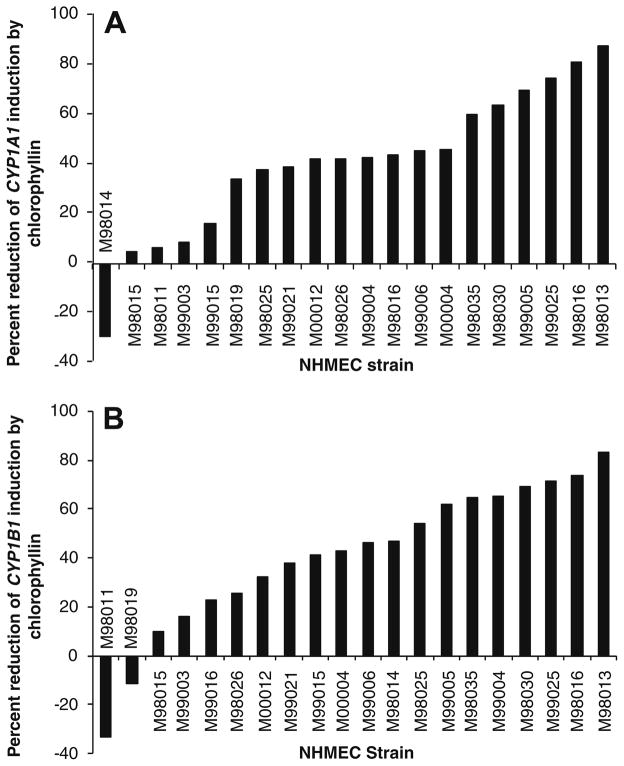
In all cells treated with chlorophyllin (exposure groups T2, T3, T4) in addition to BP, there were generalized reductions in induction of both CYP1A1 and CYP1B1 (Tables 1 and 2, respectively), compared to the T1 group. Of the three different chlorophyllin treatment regimens, T4 was most effective in mitigating induction of both cytochrome P450s. Induction of CYP1A1 was reduced between 5.1% and 87.9% by chlorophyllin, with the exception of strain M98014, in which induction was increased by 29.1% in the presence of chlorophyllin (Fig. 1A). Similarly, CYP1B1 induction was generally reduced (9.6–83.3%), although it was increased by 11.2% and 33.3% in cell strains M98019 and M98011, respectively (Fig. 1B). Fig. 2 shows a high degree of correlation in the chlorophyllin-mediated reduction of CYP1A1 and CYP1B1 induction by BP (r = 0.657, p = 0.0013).
Fig. 1.
(A and B) Bar graphs showing percent reduction of expression (by RT-PCR) of CYP1A1 (A) and CYP1B1 (B) in each primary NHMEC strain when treatment group T1 (BP alone) was compared with treatment group T4 (24 h chlorophyllin followed by 24 h of BP plus chlorophyllin). The magnitude of the bar represents the extent to which chlorophyllin reduced the BP-induced gene expression.
Fig. 2.
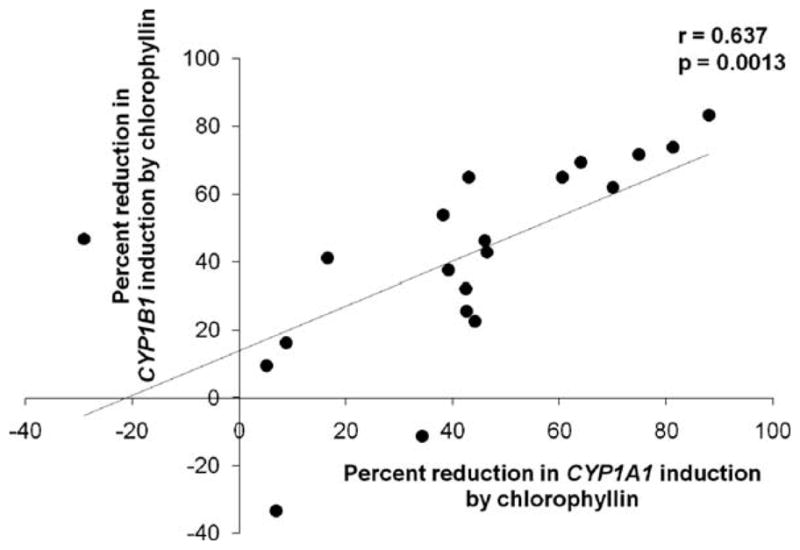
Strain-wise correlation for the percent reduction in expression of CYP1A1 and CYP1B1 in T4 cells compared to T1 cells (r = 0.637, p = 0.0013); the graph shows data for all 20 NHMEC strains.
3.2. DNA adduct levels induced by BP
In NHMEC strains exposed to BP alone (T1 group), BPdG adducts were measured by BPDE-DNA CIA and ranged from 2.5 to 57.5 adducts/108 nucleotides (Table 3). The addition of chlorophyllin resulted in reduced BPdG adduct levels in all three groups (T2, T3, and T4) for all 20 of the NHMEC strains studied, with the greatest decrease in the T4 group (see Table 3 and Fig. 3). Mitigation of BPdG adduct levels was by 25–87% to 1.3–35.4 adducts/108 nucleotides in the T4 group (see last column in Table 3). There was a wide inter-individual variation in response to chlorophyllin, and the 20 cell strains could be divided into three groups: 10 cell strains showing adduct level reductions of 25–50%, 6 cell strains showing adduct level reductions of 50–75%; and, 4 cell strains showing adduct reductions of >75% (Fig. 3).
Table 3.
BPdG adducts formed in NMECs exposed to BP in the absence (T1a) or presence (T2, T3, T4) of chlorophyllin.
| Cell Strain | T1 | T2 | T3 | T4 | % Reduction T1 vs. T4c |
|---|---|---|---|---|---|
| M98014 | 4.7 ± 0.6b | 2.2 ± 0.4 | 2.9 ± 0.2 | 2.1 ± 0.7 | 55.1c |
| M98015 | 8.3 ± 5.0 | 5.5 ± 3.3 | 5.5 ± 4.0 | 4.3 ± 4.1 | 48.2 |
| M98011 | 20.8 ± 13.0 | 15.3 ± 13.1 | 17.8 ± 9.2 | 11.7 ± 9.3 | 43.7 |
| M99003 | 7.3 ± 1.5 | 3.3 ± 0.3 | 3.5 ± 1.2 | 1.3 ± 0.4 | 82.1 |
| M99015 | 57.5 ± 16.2 | 35.0 ± 15.9 | 51.4 ± 12.1 | 35.4 ± 10.8 | 38.4 |
| M98019 | 5.1 ± 0.9 | 3.1 ± 1.3 | 3.2 ± 0.8 | 3.6 ± 1.1 | 29.5 |
| M98025 | 15.1 ± 2.4 | 5.1 ± 0.6 | 5.0 ± 1.8 | 3.7 ± 3.4 | 75.5 |
| M99021 | 13.7 ± 3.2 | 14.3 ± 1.5 | 15.3 ± 3.8 | 10.2 ± 5.4 | 25.2 |
| M00012 | 21.0 ± 4.5 | 14.7 ± 1.8 | 16.4 ± 3.5 | 10.9 ± 3.3 | 48.3 |
| M98026 | 54.5 ± 29.7 | 47.3 ± 26.0 | 42.5 ± 21.4 | 32.5 ± 18.2 | 40.3 |
| M99004 | 4.8 ± 0.5 | 2.8 ± 0.6 | 2.3 ± 1.0 | 1.9 ± 0.9 | 60.0 |
| M99016 | 34.0 ± 3.4 | 23.9 ± 3.6 | 25.5 ± 2.6 | 12.5 ± 1.4 | 63.2 |
| M99006 | 17.3 ± 3.7 | 13.6 ± 3.3 | 15.5 ± 5.8 | 12.6 ± 4.1 | 27.5 |
| M00004 | 33.4 ± 6.1 | 27.1 ± 13.7 | 28.3 ± 9.6 | 19.8 ± 12.3 | 40.7 |
| M98035 | 5.9 ± 3.9 | 2.7 ± 1.1 | 5.7 ± 1.6 | 2.7 ± 1.3 | 54.7 |
| M98030 | 9.5 ± 1.5 | 8.1 ± 3.3 | 5.5 ± 1.8 | 4.0 ± 1.6 | 57.7 |
| M99005 | 2.5 ± 1.3 | 1.3 ± 1.2 | 2.2 ± 2.3 | 1.5 ± 1.7 | 38.8 |
| M99025 | 3.2 ± 0.6 | 3.2 ± 0.2 | 2.5 ± 1.3 | 1.4 ± 0.1 | 57.1 |
| M98016 | 53.4 ± 2.5 | 26.1 ± 2.3 | 25.6 ± 7.0 | 7.0 ± 1.4 | 86.8 |
| M98013 | 23.1 ± 2.0 | 14.5 ± 2.8 | 12.2 ± 1.2 | 3.0 ± 0.9 | 87.0 |
T = Treatment groups as described in Section 2.
Number of adducts per 108 nucleotides.
Percentage (%) reduction in number of adducts in the T4 group compared to the T1 group.
Fig. 3.
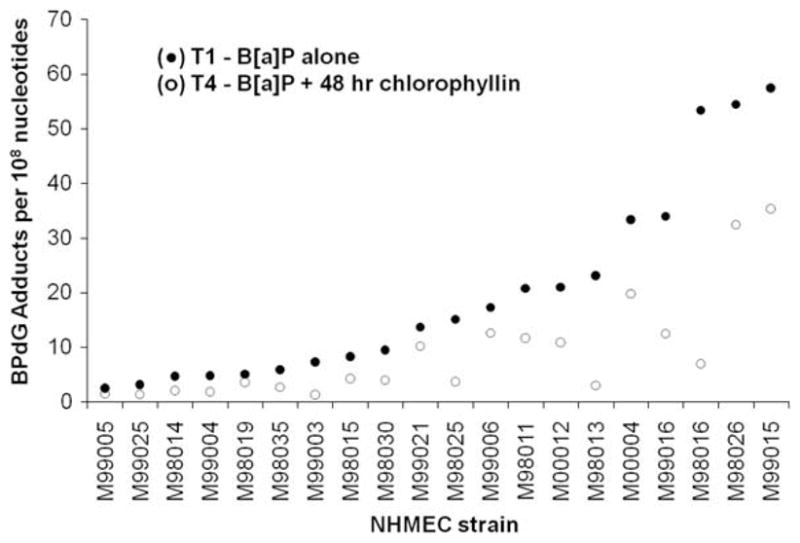
Pairwise BPdG adduct values (per 108 nucleotides) for 20 NHMEC strains exposed to either BP alone (T1 group, ●) and or BP plus chlorophyllin (T4 group, ○). Adduct values for each strain are ranked in order of extent of BPdG formation in the T1 group.
3.3. Correlations between CYP450 expression and BPdG adducts
In this study, in cells exposed to BP alone (T1 group), neither CYP1A1 nor CYP1B1 expression correlated with BPdG adduct formation (r = 0.437 for CYP1A1 and r = 0.306 for CYP1B1, p > 0.10 for both). This result has been observed previously [12]. In addition, in cells exposed to BP with chlorophyllin (T4 group) there were poor correlations for the percentage reduction of BP-induced cytochrome P450 induction with the corresponding percentage reduction in BPdG adduct levels (r = 0.249, p = 0.096 for CYP1A1; and r = 0.409, p = 0.070 for CYP1B1). Similar trends were observed for the other chlorophyllin treatments as well (data not shown). There were good correlations between the numbers of adducts formed by comparison of cells treated with BP alone (T1 group) with cells treated in the presence of chlorophyllin [r = 0.947, p = 0.0008 for T1 vs. T2; r = 0.945, p = 0.0017 for T1 vs. T3 and r = 0.826, p = 0.0004 for T1 vs. T4].
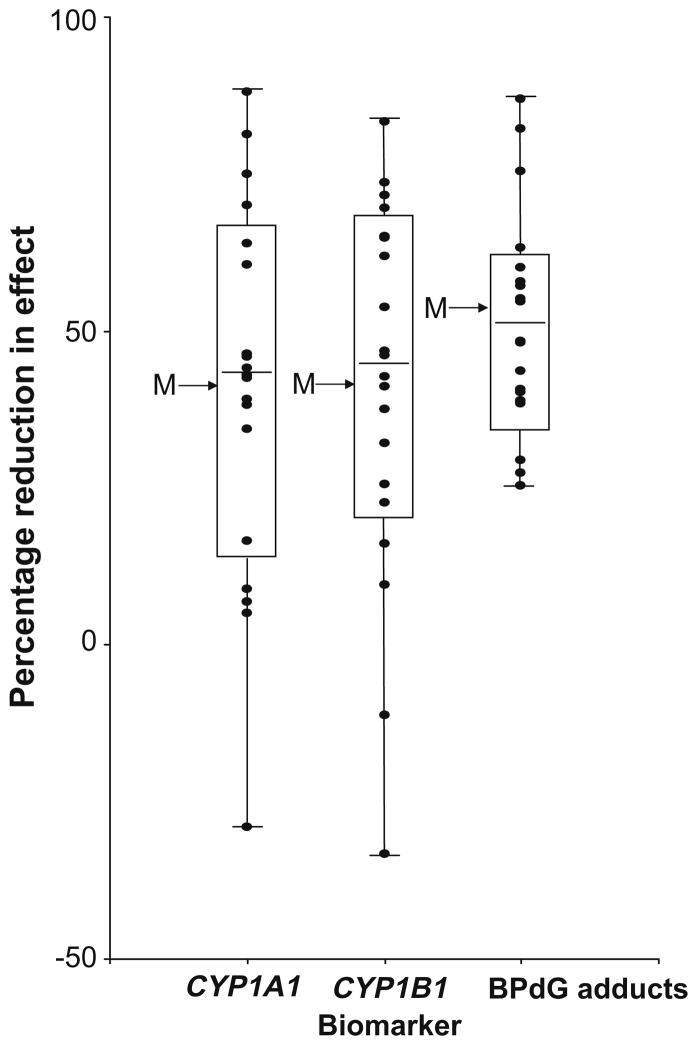
Overall, the cell strained showed wide inter-individual variability in CYP1A1 and CYP1B1 induction, BPdG adduct formation, reduction in CYP1A1 and CYP1B1 induction by chlorophyllin treatment and mitigation of BP-DNA adduct levels by chlorophyllin treatment. Fig. 4 depicts the consistent pattern of reduction in percent CYP1A1 and CYP1B1 gene expression and BPdG adduct formation across most of the 20 NHMEC strains in response to chlorophyllin treatment in group T4 (p < 0.005, p < 0.005 and p < 0.0005 for CYP1A1, CYP1B1 and BPdG adducts, respectively).
Fig. 4.
Percentage reduction in CYP1A1 and CYP1B1 expression levels and benzo[a]pyrene-diol-epoxide-DNA adduct (BPdG) levels in the presence of chlorophyllin for normal human mammary epithelial cells from 20 individual donors that were exposed to benzo[a]pyrene in vitro. This box and whisker plot indicates the median percentage and 25% and 75% percentage changes, and outliers are indicated by the whiskers. The mean values are indicated by arrows marked M, these values are 41.1 (±6.5 SE) for CYP1A1, 41.0 (±6.7 SE) for CYP1B1, and 53% (±4.2 SE) for BPdG. These changes were statistically significant p < 0.005 for CYP1A1, p < 0.005 for CYP1B1 and p < 0.0005 for BPdG adducts.
4. Discussion
In this study we found that exposure of 20 NHMEC strains, from 20 individual donors, to BP resulted in up- regulation of CYP1A1 and CYP1B1 in almost all of the strains, and produced measurable BPdG adducts in all of the strains. The addition of chlorophyllin resulted in lowering almost all the BP-induced increases in CYP1A1 and CYP1B1 expression, and was effective in reducing BPdG levels in all 20 NHMEC strains. The lack of any exceptions to the chlorophyllin-modulated reduction in DNA damage suggests that chlorophyllin may be a useful chemopreventive agent for the human population because most individuals should respond to chlorophyllin treatment with decreased PAH activation although the magnitude may vary.
The gene expression experiments presented here expand our previous work using DNA microarrays in a single NHMEC strain, M98013 [16]. M98013 was used in this study and values for BP-induced CYP1A1 and CYP1B1 expression were similar to those found previously [16], however, the addition of 19 new NHMEC strains revealed considerable variation in levels of BP-induced CYP1A1 and CYP1B1 expression. This wide inter-individual variation might be explained, at least in part by polymorphisms in the genes coding for cytochrome P450s and other Phase I enzymes, as well as genes coding for Phase II or detoxication enzymes and DNA repair functions [11]. Other factors may also be relevant. For example, we found a poor correlation between CYP1 gene expression and donor age (data not shown), in agreement with a previous study showing age-independent variation in CYP1A1 expression in normal breast tissue samples (n = 58) obtained from breast cancer patients as well as individuals with no diagnosis of cancer [17]. In our study, the magnitude of CYP1B1 induction was much less than that found for CYP1A1. CYP1B1 is a dioxin inducible cytochrome P450 isoform thats expressed constitutively in various extrahepatic tissues, including the breast., It exhibits weak induction by aryl-hydrocarbon receptor (AhR) ligands. This may explain the relatively lower induction of CYP1B1 compared to CYP1A1 in our study, and may contribute to the inter-individual variability in inducible expression levels observed [18].
In this study, chlorophyllin suppressed the BP-mediated induction of both CYP1A1 and CYP1B1 expression in almost all 20 of the NHMEC strains. Inhibition was observed in the group of cells co-treated with BP and chlorophyllin (group T2), as well as when cells were pre-treated with chlorophyllin alone, washed and subsequently treated with BP (group T3). Other in vivo studies also found highly effective inhibition when chlorophyllin was co-administered with the carcinogen [19]. These data suggest that chlorophyllin might be acting by at least two different mechanisms. In previous studies, chlorophyllin has been found to form tight molecular complexes with planar aromatic hydrocarbons, possessing at least partial ring structure, when a large molar excess of chlorophyllin was added simultaneously with the carcinogen [20]. In addition, complex formation between BPDE and chlorophyllin facilitates BP tetrol foramtion limiting the amount of BPDE [21], contributing to the reduction of BPdG adducts as well as BP-induced CYP1A1 and CYP1B1 expression when chlorophyllin was given simultaneously with BP. A reduction in expression in most NHMEC strains upon treatment with chlorophyllin prior to carcinogen exposure (treatment group T3) suggests mitigation of cytochrome P450 expression by mechanisms other than just carcinogen sequestration. Chlorophyllin-induced modulation of CYP1 expression via mechanisms independent of “carcinogen sequestration” might occur through inhibition or reduction of the basal levels of NADPH-cytochrome c reductase [21], or modulation of the levels of Phase I enzymes directly or indirectly by altering expression of genes involved in the AHR pathway. These mechanisms could occur as a result of chlorophyllin exposure and continue to be active even after chlorophyllin is removed from the medium and BP is added (as in group T3). Therefore, the reduction of BP-induced CYP1 expression in NHMEC treatment group T4 is consistent with the role of chlorophyllin as an interceptor molecule as well as a possible indirect modulator of CYP1 enzymes.
In conclusion, the lack of correlation between the suppression of CYP1A1 and CYP1B1 induction and reduction in DNA adduct levels suggests that the modulation of human DNA adduct levels may be partially mediated independently of cytochrome P450 induction. Additionally, the extent of adduct formation and sensitivity of detection of adducts may be different from that of CYP1 gene expression thereby at least partially contributing to the discrepancy. In this study, in every single cell strain, chlorophyllin protected against BPdG adduct formation to some degree (25–87%). Therefore, chlorophyllin is likely to be an effective chemoprotective agent for a large proportion of the human population.
Acknowledgments
Thanks to Kathy Boyce and Melanie Moore for clerical assistance. We also gratefully acknowledge the Cooperative Human Tissue Network (sponsored by the National Cancer Institute and the National Disease Research Interchange) for providing normal human mammary tissues with which we developed the NHMEC strains. This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute, NIH (Bethesda, MD), West Virginia University, and the intramural program of the National Institute for Occupational Safety and Health, CDC (Morgantown, WV).
Footnotes
Conflict of interest
None of the authors have any financial or other conflict of interest.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Cancer Institute or the National Institute for Occupational Safety and Health.
References
- 1.Brandt HC, Watson WP. Monitoring human occupational and environmental exposures to polycyclic aromatic compounds. Ann Occup Hyg. 2003;47:349–378. doi: 10.1093/annhyg/meg052. [DOI] [PubMed] [Google Scholar]
- 2.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sims P, Grover PL, Swaisland A, Pal K, Hewer A. Metabolic activation of benzo(a)pyrene proceeds by a diol-epoxide. Nature. 1974;252:326–328. doi: 10.1038/252326a0. [DOI] [PubMed] [Google Scholar]
- 4.Sarkar D, Sharma A, Talukder G. Chlorophyll and chlorophyllin as modifiers of genotoxic effects. Mutat Res. 1994;318:239–247. doi: 10.1016/0165-1110(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 5.Egner PA, Wang JB, Zhu YR, Zhang BC, Wu Y, Zhang QN, Qian GS, Kuang SY, Gange SJ, Jacobson LP, Helzlsouer KJ, Bailey GS, Groopman JD, Kensler TW. Chlorophyllin intervention reduces aflatoxin-DNA adducts in individuals at high risk for liver cancer. Proc Natl Acad Sci USA. 2001;98:14601–14606. doi: 10.1073/pnas.251536898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler TW, Egner PA, Wang JB, Zhu YR, Zhang BC, Lu PX, Chen JG, Qian GS, Kuang SY, Jackson PE, Gange SJ, Jacobson LP, Munoz A, Groopman JD. Chemoprevention of hepatocellular carcinoma in aflatoxin endemic areas. Gastroenterology. 2004;127:S310–s318. doi: 10.1053/j.gastro.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 7.Fahey JW, Stephenson KK, Dinkova-Kostova AT, Egner PA, Kensler TW, Talalay P. Chlorophyll, chlorophyllin and related tetrapyrroles are significant inducers of mammalian phase 2 cytoprotective genes. Carcinogenesis. 2005;26:1247–1255. doi: 10.1093/carcin/bgi068. [DOI] [PubMed] [Google Scholar]
- 8.Kumar SS, Shankar B, Sainis KB. Effect of chlorophyllin against oxidative stress in splenic lymphocytes in vitro and in vivo. Biochim Biophys Acta. 2004;1672:100–111. doi: 10.1016/j.bbagen.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Mata JE, Yu Z, Gray JE, Williams DE, Rodriguez-Proteau R. Effects of chlorophyllin on transport of dibenzo(a, l)pyrene, 2-amino-1-methyl-6-phenylimidazo-[4,5-b]pyridine, and aflatoxin B(1) across Caco-2 cell monolayers. Toxicology. 2004;196:117–125. doi: 10.1016/j.tox.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Grover PL, MacNicoll AD, Sims P, Easty GC, Neville AM. Polycyclic hydrocarbon activation and metabolism in epithelial cell aggregates prepared from human mammary tissue. Int J Cancer. 1980;26:467–475. doi: 10.1002/ijc.2910260412. [DOI] [PubMed] [Google Scholar]
- 11.Keshava C, Whipkey D, Weston A. Transcriptional signatures of environmentally relevant exposures in normal human mammary epithelial cells: benzo[a]pyrene. Cancer Lett. 2005;221:201–211. doi: 10.1016/j.canlet.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 13.Laird PW, Zijderveld A, Linders K, Rudnicki MA, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Divi RL, Beland FA, Fu PP, Von Tungeln LS, Schoket B, Camara JE, Ghei M, Rothman N, Sinha R, Poirier MC. Highly sensitive chemiluminescence immunoassay for benzo[a]pyrene–DNA adducts: validation by comparison with other methods, and use in human biomonitoring. Carcinogenesis. 2002;23:2043–2049. doi: 10.1093/carcin/23.12.2043. [DOI] [PubMed] [Google Scholar]
- 15.Saunders DB, Trapp RG. Association and prediction. In: Dolan J, Lagan C, editors. Basic and Clinical Biostatistics. 2. Appleton and Lange; Norwalk, CT: 1994. pp. 162–187. [Google Scholar]
- 16.Keshava C, Divi RL, Einem TL, Richardson DL, Leonard SL, Keshava N, Poirier MC, Weston A. Chlorophyllin significantly reduces benzo[a]pyrene–DNA adduct formation and alters cytochrome P450 1A1 and 1B1 expression and EROD activity in normal human mammary epithelial cells. Environ Mol Mutagen. 2009;50:134–144. doi: 10.1002/em.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goth-Goldstein R, Stampfer MR, Erdmann CA, Russell M. Interindividual variation in CYP1A1 expression in breast tissue and the role of genetic polymorphism. Carcinogenesis. 2000;21:2119–2122. doi: 10.1093/carcin/21.11.2119. [DOI] [PubMed] [Google Scholar]
- 18.Ma Q, Lu AY. Origins of individual variability in P4501A induction. Chem Res Toxicol. 2003;16:249–260. doi: 10.1021/tx0200919. [DOI] [PubMed] [Google Scholar]
- 19.Dashwood RH. The importance of using pure chemicals in (anti) mutagenicity studies: chlorophyllin as a case in point. Mutat Res. 1997;381:283–286. doi: 10.1016/s0027-5107(97)00221-2. [DOI] [PubMed] [Google Scholar]
- 20.Dashwood R. Chlorophylls as anticarcinogens. Int J Oncol. 1997;10:721–727. doi: 10.3892/ijo.10.4.721. [DOI] [PubMed] [Google Scholar]
- 21.Tachino N, Guo D, Dashwood WM, Yamane S, Larsen R, Dashwood R. Mechanisms of the in vitro antimutagenic action of chlorophyllin against benzo[a]pyrene: studies of enzyme inhibition, molecular complex formation and degradation of the ultimate carcinogen. Mutat Res. 1994;308:191–203. doi: 10.1016/0027-5107(94)90154-6. [DOI] [PubMed] [Google Scholar]