Abstract
Herpesviruses, a family of animal viruses with large (125 – 250 kbp) linear DNA genomes, are highly diversified in terms of host range; nevertheless, their virions conform to a common architecture. The genome is confined at high density within a thick-walled icosahedral capsid with the uncommon (among viruses, generally) but unvarying triangulation number T=16. The envelope is a membrane in which some 11 different viral glycoproteins are implanted. Between the capsid and the envelope is a capacious compartment called the tegument that accommodates ~ 20 – 40 different viral proteins (depending on which virus) destined for delivery into a host cell. A strong body of evidence supports the hypothesis that herpesvirus capsids and those of tailed bacteriophages stem from a distant common ancestor, whereas their radically different infection apparatuses - envelope on one hand and tail on the other - reflect subsequent co-evolution with divergent hosts. Here we review the molecular components of herpesvirus capsids and the mechanisms that regulate their assembly, with particular reference to the archetypal alphaherpesvirus, herpes simplex virus type 1; assess their duality with the capsids of tailed bacteriophages; and discuss the mechanism whereby, once DNA packaging has been completed, herpesvirus nucleocapsids exit from the nucleus to embark on later stages of the replication cycle.
Keywords: herpesvirus, herpes simplex virus type 1, tailed bacteriophage, tegument, nuclear exit, cryo-electron microscopy
1. INTRODUCTION
Herpesviruses infect hosts throughout the animal kingdom, from molluscs to man. Eight family members cause diseases in humans, including two - Epstein-Barr virus and Kaposi’s Sarcoma-associated virus - that are tumorogenic. Here we review current understanding of herpesvirus capsid assembly, a largely conserved process. On the macromolecular scale, these are huge particles - 125 nm in diameter and ~ 200 MDa in mass - and composed, transiently or for the long-term, of ~ 5000 protein molecules of at least seven different kinds. We also discuss the hypothesis that the capsids of herpesviruses and tailed bacteriophages descend from a common ancestor. At first sight, this proposition may appear counter-intuitive, in view of widely differing virion morphologies, disparate hosts, and the implication that elaborate assembly pathways evolved very early, prior to the point at which eukaryotic cells and bacteria diverged. In addition, we summarize some interactions between herpesvirus capsids and teguments, which are attracting increasing research interest. Compared with capsids, however, teguments are less conserved, less ordered, and less understood.
2. HERPESVIRUS CAPSID ASSEMBLY
2.1 Assembly of the procapsid
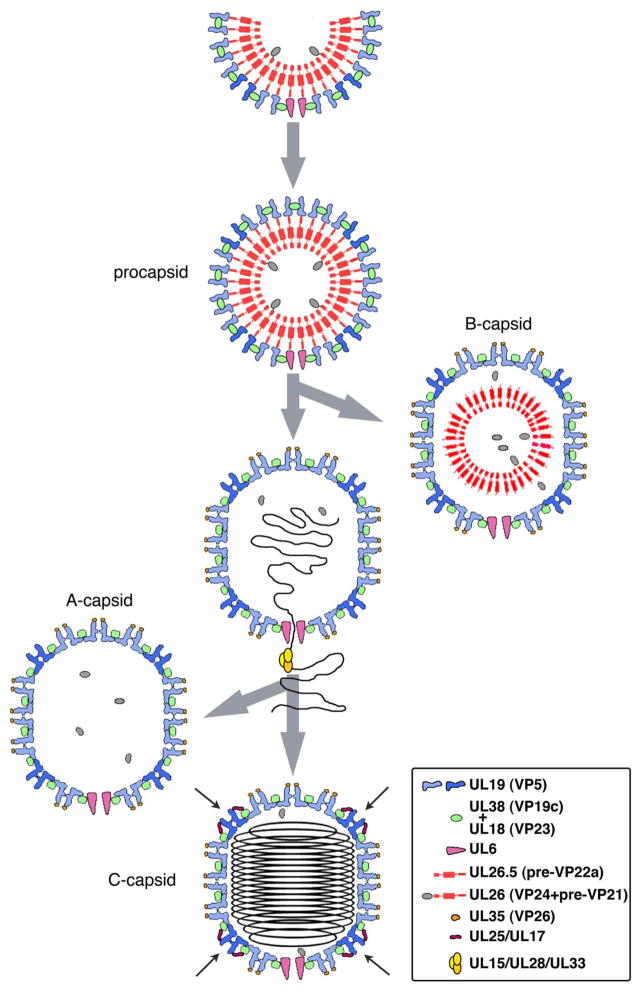
The sequence of events that take place as the herpesvirus nucleocapsid assembles is outlined in Figure 1. First, a procapsid is produced which conforms to T=16 icosahedral symmetry, albeit in a particle that is almost perfectly spherical. One of its 5-fold vertices is occupied by a dodecameric ring of portal protein subunits, giving a distinctive 5:12 symmetry mismatch. At the other 11 vertices are pentamers of UL19, the major capsid protein, with UL19 hexamers elsewhere in the surface lattice. It is plausible but not fully proven that the portal serves as the nucleation complex from which there is outgrowth of co-assembling scaffolding protein and UL19 (Huffman et al. 2008; Yang and Baines 2008). In fact, HSV-1 employs two scaffolding systems - internal and external. The internal scaffold is a 25nm-thick shell of laterally packed dimers of an elongated protein, UL26.5. ~ 10% of these subunits (UL26) have an additional protein, the maturational protease VP24, fused to the N-terminus and positioned on the inside of the scaffold shell (Newcomb et al. 2000). The other end of each dimer, with the C-termini, contacts the inside of the surface shell. The external scaffold is contributed by “triplexes” which are α2β heterotrimers of VP23 and VP19c. Remarkably, adjacent UL19 capsomers do not directly contact each other, but instead are coordinated around the sites of local 3-fold symmetry by their contacts with triplexes (Trus et al. 1995); Fig. 2. Thus the procapsid surface shell consists of one dodecamer of UL6; 150 hexamers and 11 pentamers of UL19; and 320 triplexes.
Figure 1. Assembly and maturation of the HSV-1 nucleocapsid.
Successive states of the capsid are shown schematically in cross-sections. This representation that does not strictly comply with T=16 geometry (see Fig. 6) but is intended to convey key features, such as capsomer protrusions, the floor, triplexes, etc.
Figure 2. Cryo-EM reconstructions of HSV-1 capsids.
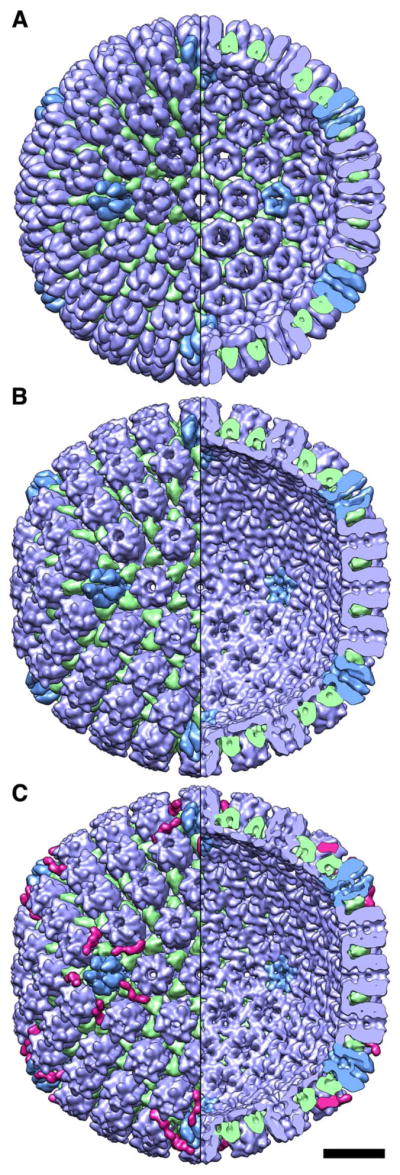
(A) the HSV-1 procapsid; (B) A-capsid; and (C) C-capsid, with DNA computationally removed. Left half of each reconstructon shows the outer surface and right half, the inner surface. Bar, 20 nm.
Biochemical quantitations estimate the scaffolding shell’s complement of dimers of UL26.5 and/or UL26 as ~ 1000 (Newcomb et al. 2000). It has yet to be established whether this shell conforms to a global symmetry - e.g. icosahedral symmetry complementary to that of the outer shell - or whether it is only locally ordered and “micellar” in character. Of note, a similar dilemma persists concerning the packing of the retroviral polyprotein Gag in immature virions (Briggs et al. 2009). However, Gag shells, although also spherical, vary significantly in size whereas the diameter of the herpesvirus scaffold shell is fixed, as it tracks the inner surface of the precisely defined surface shell.
2.2 Procapsid maturation
After the HSV-1 procapsid is complete, the protease is activated and performs two actions; detaching itself from the scaffolding protein and removing a 25-residue peptide from the C-termini of the scaffolding proteins (UL26.5 -> VP22a; UL26 -> VP24 + VP21). (The cytomegalovirus protease also cleaves at a third site (Welch et al. 1993)). It is not clear how the protease is inhibited until the procapsid is complete. One possibility is kinetics, if assembly, once initiated, is fast and protease activation is relatively slow (herpesvirus proteases are known to be slow enzymes - (Tong 2002)). Subsequently, the surface shell undergoes major alteration as it switches into its mature conformation which is more angular (polyhedral) and markedly more robust (Trus et al. 1995; Heymann et al. 2003). Contacts are established between neighboring capsomers via their innermost domains, forming the “floor” of the mature capsid (Fig. 2B, C). This transition is promoted by the packaging of DNA, which applies pressure on the inner surface of the capsid, and by detachment of the C-terminal peptides of the internal scaffolding proteins, which disrupts an interaction that restrains the surface shell from maturing. Concomitant with DNA packaging, the processed scaffolding proteins (VP22a, VP21) are expelled from the capsid but the protease (VP24) is retained. DNA packaging is effected by the viral terminase, a 3-component ATPase complex that translocates DNA from the replicating concatemer into the maturing particle. DNA replication by phages and herpesviruses and, in particular, the roles played by helicases have been reviewed (Marintcheva and Weller 2001).
Two abortive capsids accumulate in the nuclei of infected cells as a result of miscarriage in DNA packaging. B-capsids have a mature surface shell and their internal scaffolding protein is processed but not expelled, shrinking into a smaller and less regular structure. They contain no DNA and it appears that packaging has not been initiated. A-capsids, also with mature surface shells but retaining no scaffold, are viewed as particles on which DNA packaging was initiated but subsequently disrupted with loss of the packaged DNA. A third intranuclear capsid, the C-capsid, contains DNA and is an on-pathway intermediate. In a wild-type infection, the majority of intranuclear particles seen by thin section EM are B-capsids. A-capsids are less abundant, presumably because the event that produces them has lower incidence. Procapsids are few as, once formed, they rapidly convert to A-, B-, or C-capsids. C-capsids are also less abundant than B-capsids, because they exit from the nucleus after packaging is complete. Below, in §3.1, we consider the events that regulate the exit of capsids from the nucleus.
2.3 Parallels in bacteriophage procapsid assembly
Overall, the pathway in Figure 1 is strikingly similar to those followed by tailed bacteriophages (Steven et al. 2005; other chapters in this volume). This resemblance attracted comment as early as 1975 (Friedmann et al. 1975); since then, the number of properties recognized as shared has grown steadily (Table 1). A few aspects appear to be sticking points or at least bear further discussion.
Table 1.
Points of resemblance between phage capsids and herpesvirus capsids
|
Bioinformatics suggest that herpesvirus proteases (4 known structures) and some phage proteases have similar folds. Some phages (e.g. T7, P22) have no protease.
Term refers to non-spherical capsids, such as that of T4 which represents an icosahedron extended along a symmetry axis by insertion of additional rings of hexamers.
All known herpesvirus capsids observe T=16 icosahedral symmetry whereas tailed phage capsids exhibit a variety of T-numbers. This range does include T=16 (Parker et al. 1983; Duda et al. 2006; Weigele et al. 2007; Kuznetsov et al. 2010) and these are the only two families in which this capsid geometry has been documented. In view of the immense number of tailed phages in the biosphere (Hendrix 2003), it is hard to assess the extent to which the T-numbers reported to date afford a representative sampling. Given a common lattice constant of 14 nm arising from a shared capsid protein fold and packing arrangement (see below), it would be a straightforward undertaking to determine - or at least closely approximate - the T-numbers of many more tailed phage viruses by simply measuring capsid diameters by cryo-EM and hence to assess the overall incidence of T=16 geometry.
To date, no phages have been found to have a triplex-like external scaffold. Considering that scaffolding mechanisms are arguably the most diverse aspect of phage capsid assembly (Dokland 1999), it may be only a matter of time before one is discovered. As the HSV procapsid matures, the role of its triplexes switches from morphogenesis to a clamp-like function at the trigonal lattice sites. In this respect, they resemble the trimers of clamping proteins centered on local 3-fold axes of phage capsids, such as those of T4 and lambda. The latter proteins bind to their capsids only after maturation.
A strikingly distinctive behavior of phage procapsids is their maturation transformation, in which capsid protein subunits swivel through large angles and undergo some remodeling (Conway et al. 2001; Gertsman et al. 2009; Ionel et al. 2011; Chen et al. 2011). The main consequence of maturation is a stabilization that enables the capsid to resist the outwards pressure of densely packed DNA; in some systems, further reinforcement follows from the binding of clamping proteins (see above) or the formation of covalent crosslinks (Duda 1998). Maturation is generally accompanied by an increase in diameter by 15–20%, almost doubling the interior volume; hence this transformation is often referred to as expansion. The HSV-1 procapsid also undergoes maturation (see above). Like phage capsids, it changes shape from round to polyhedral but, unlike them, it remains essentially the same size. However this does not represent a fundamental discrepancy: the parts of UL19 subunits that form disconnected rings in the innermost part of the procapsid and which appear to be homologous to phage capsid proteins (Fig. 3A), reorganize so as to establish contacts between adjacent capsomers (Fig. 2). In this transition, the size of the per-subunit footprint in the floor increases by a similar margin as in maturing phage capsids. Thus, much the same allosteric mechanism is employed in both systems.
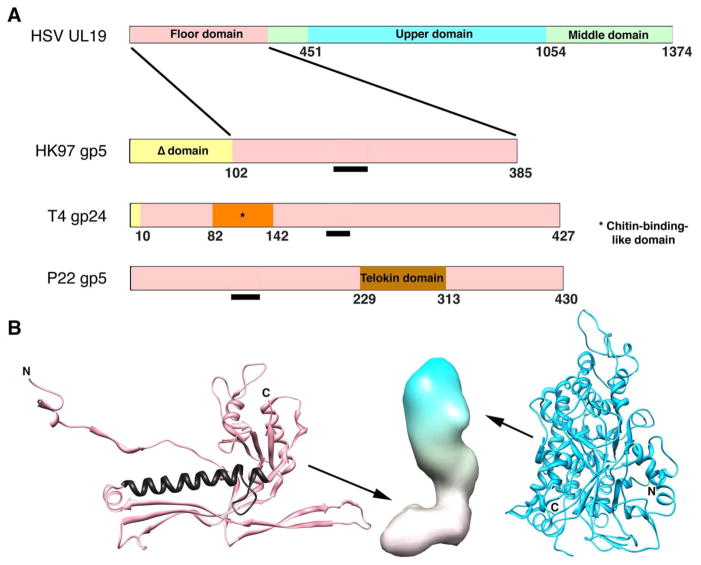
Figure 3. Domain maps of capsid proteins of HSV-1 and three bacteriophages.
(A) The phage proteins are ones for which high resolution structures have been determined. The common core domain is colored pink. The Δ-domain of HK97 is equivalent to an internal scaffolding protein fused to the core domain and is proteolyzed during maturation (gp5 -> gp5*). The T4 capsid protein gp24 forms pentamers at all non-portal vertices of this elongated T=13/Q=20 capsid, whose other capsomers are hexamers of the homolog, gp23. Its short propeptide is removed during maturation (gp24 -> gp24*). P22 does not have a protease. Gp10A of T7 (not shown) appears to have a similar core domain plus an N-terminal domain of ~ 90 residues, which is not removed by proteolysis (Agirrezabala et al. 2007). The black bars mark the location of the long α-helix, a signature feature of this fold. (B) Left: fold of HK gp5* in its mature conformation (Wikoff et al. 2000); PDB: 1OHG. The long helix is colored in black. Middle: low resolution surface rendering of a HSV-1 UL19 subunit in its B-capsid conformation. The floor domain (at bottom) is thought to have a HK97 gp5*-like fold. Right: upper domain of UL19 (Bowman et al. 2003); PDB: 1NO7.
2.4 The building-blocks and their roles in assembly
2.4.1 Major capsid protein
UL19 (150 kDa). UL19 orthologs vary somewhat in size (~ 120 – 150 kDa) among different herpesviruses but in general are much larger than phage capsid proteins (~ 30 – 60 kDa). This discrepancy is attributable to their large C-terminal domains forming the external protrusions that account for the much greater thickness of herpesvirus capsids (~ 15 nm vs. ~ 4 nm). The HSV-1 protrusion consists of two domains, middle and upper (Fig. 3); a crystal structure has been determined for the upper domain (Bowman et al. 2003). The hexagonal lattice spacing in the floor (~ 14 nm) matches that of phage capsids, and cryo-EM data at relatively high resolution support the idea that the floor domain has a phage capsid protein-like fold (Zhou et al. 2000; Baker et al. 2005). The latter proteins consist of a basic core on to which additional domains may be grafted (Fig. 3A). It will be instructive to learn whether herpesvirus floor domains also have such domains. However, it is already clear that floor structures are markedly more conservative than protrusions (Booy et al. 1996; Trus et al. 2001; Davison et al. 2005).
2.4.2 Portal protein
UL6 (74 kDa). When portal proteins of phages or herpesviruses are expressed at high levels, polymorphisms are commonly encountered in the number of subunits in the ring. However, it appears that, in both cases, only 12-mers are assembled into capsids (Lurz et al. 2001; Rochat et al. 2011). Crystal structures of several phage portals, of varying size, have been determined (Simpson et al. 2001; Lebedev et al. 2007; Olia et al. 2011). As yet, the most detailed information on a herpesvirus portal is a cryo-EM reconstruction of the HSV-1 dodecamer at 16Å resolution (Trus et al. 2004) - Fig. 4. The diameter of its wider end is compatible with fitting into the vertex space enclosed by five surrounding UL19 hexamers. There have been differing proposals, based on cryo-ET, as to whether the bulk of the portal complex faces outwards (Cardone et al. 2007) or inwards, as in phages (Deng et al. 2007; Chang et al. 2007; Rochat et al. 2011).
Figure 4. The HSV1 portal protein, UL6.
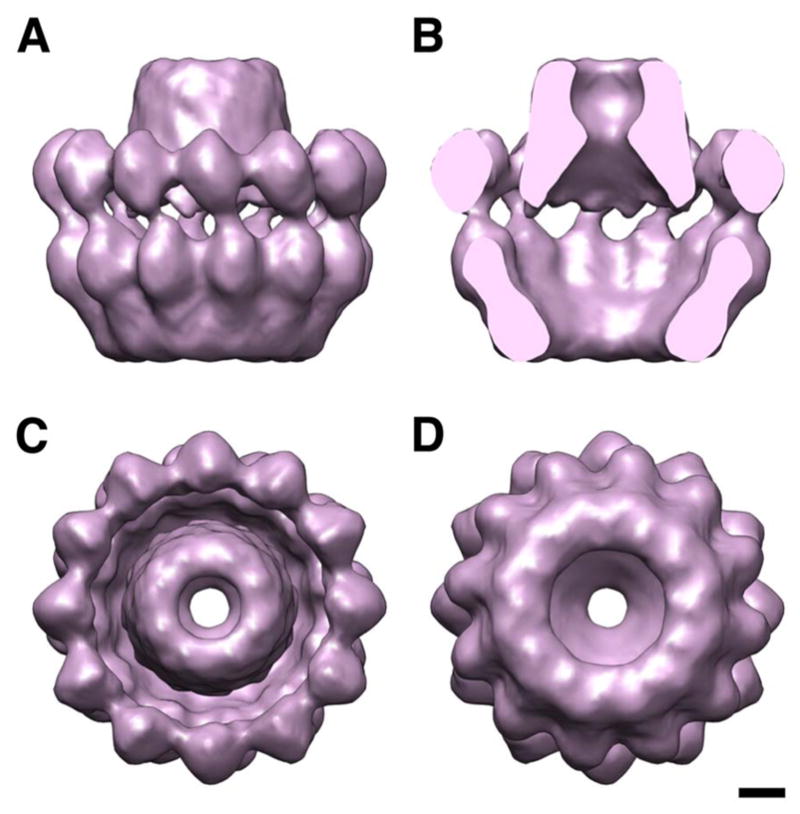
(A) side view; (B) side view, cut; (C) top view; (D) bottom view. Bar, 20 Å.
2.4.3 Maturational protease
VP24 (24 kDa). The structures of four herpesvirus proteases have been determined, exhibiting a common fold (Qiu et al. 1996; Qiu et al. 1997; Hoog et al. 1997). In the absence of crystal structures for phage proteases, bioinformatic analysis has identified a family that is predicted to share the canonical herpesvirus protease fold, with both sets of enzymes having conserved Ser and His residues in their active sites (Cheng et al. 2004). Of note, some phages (e.g. T7, P22) do not require a protease to mature. Thus it would appear that the putatively shared structure represents the ancestral fold, with loss of the protease or its replacement representing more recent evolutionary developments.
2.4.4 Internal scaffolding proteins
UL26.5 (40 kDa) and UL26 (75 kDa). Two forms of this protein are expressed. The minor (less abundant) form, UL26, is initially extended at its N-terminus by the protease and a linker but its C-terminal part is identical to the major form, UL26.5. This fusion provides a convenient mechanism for incorporating that the protease into the assembling procapsid (Sheaffer et al. 2000). Both forms are processed at their C-termini by the protease, during maturation (see §2.2). The shared polypeptide appears to be a flexible α-helix-rich dimer that forms a segmented coiled-coil (Pelletier et al. 1997). The crystal structure of the relatively short scaffolding protein of phage φ29 shows a dimeric coiled coil (Morais et al. 2003).
2.4.5 External scaffolding proteins: triplex proteins
VP23 (33 kDa) and VP19c (50 kDa). VP23 has been expressed in E. coli (Kirkitadze et al. 1998). Assessment of its conformation by biophysical techniques led to the conclusion that the protein was not fully folded but in a “molten globule” state. It is plausible that VP19c is needed for VP23 to fold and enter functional trimers. VP19c has resisted expression until recently (Henson et al. 2011). As yet, there is no high resolution information on any triplex protein but cryo-EM at moderate resolution shows well defined densities, indicative of stably folded subunits. They show a triangular outer surface, underpinned by three equal and symmetrically distributed legs; Fig. 5. It may be, therefore, that the two proteins have similar folds, despite low sequence similarity.
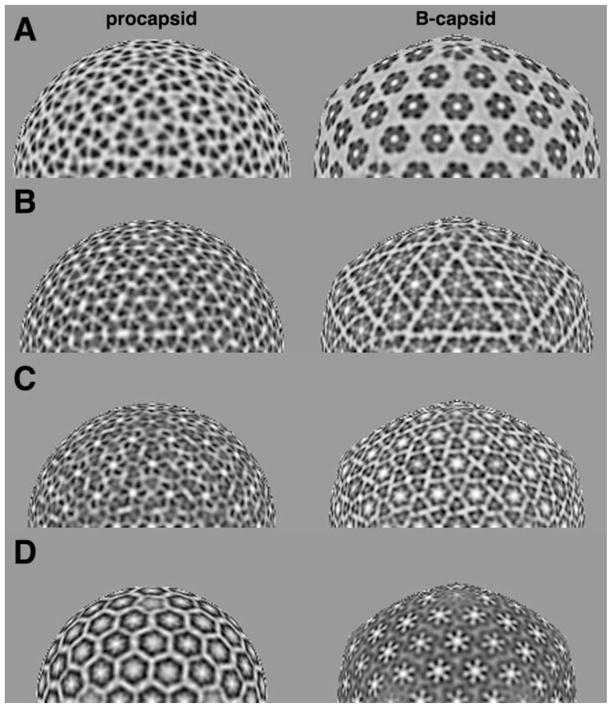
Figure 5. Stratified representations of the HSV-1 procapsid and mature capsid.
The procapsid is sampled on spherical sections one pixel thick, and the B-capsid on similarly sampled icosahedral sections. Protein density is dark. Half-sections are shown. (A) At a radius of 564 Å, the sections pass above the triplexes. The asymmetry of the procapsid hexons contrasts with the 6-fold symmetry of the mature hexons. (B) At 536 Å radius, the triplexes present as triangular densities. (C) At 503 Å radius, the triplexes present as regular triplets of density. (D) At 470Å radius, the floor domains form regular, non-contacting, rings in the procapsid, and the contiguous floor in the mature capsid.
2.4.6 Accessory protein
VP26 (12 kDa). This small protein binds to the mature HSV-1 capsid at sites around the outer tips of hexons but not to pentons (Trus et al. 1992; Zhou et al. 1995) nor to the procapsid (Newcomb et al. 2000). As VP26 is a monomer in solution (Wingfield et al. 1997), its specificity appears to reflect a bipartite binding site requiring appropriate juxtaposition of two adjacent UL19 protrusions. This is achieved in mature hexons but not in pentons nor in immature capsomers, where the protrusions are splayed apart (Figs. 2 and 6). A model has been proposed for part of VP26, based on de novo prediction and cryo-EM data (Baker et al. 2006). VP26 is dispensable for HSV-1 but proteins corresponding to it are needed in some other herpesvirus systems.
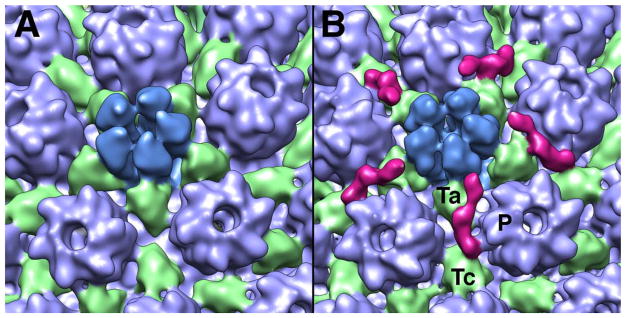
Figure 6. Comparison of the penton-proximal regions of the A-capsid (A) and the C-capsid (B).
The CCSC, a heterodimer of UL15 and UL17, is in magenta. It has multiple points of attachment to the capsid, on the a-triplex Ta, the c-triplex Tc, and the side of the P-hexon.
Survival of protein folds after sequences have diverged to unrecognizably low levels of similarity can provide telling clues as to shared antecedents. Evidence of this kind has provided a basis for the proposed common ancestry of other kinds of bacteriophages and animal viruses (Benson et al. 1999; Bamford et al. 2005; Krupovic and Bamford 2008). However, there is little scope for fold-based comparisons in the present context; of the seven herpesvirus capsid proteins, crystal structures have been determined only for the protease and major fragments of two other proteins, UL25 (Bowman et al. 2006) and the upper domain of UL19 (Bowman et al. 2003) - Fig. 3. As yet, there are no phage protease structures. Herpesvirus protrusions appear to be quite diverse (see above) and are not expected to have counterparts on phage capsid proteins. The same exclusion applies to UL25 which functions downstream from capsid assembly per se in phases of the replication cycle that are not shared by phages. Recognizing the substantial diversity that exists in the assembly strategies of phage capsids, we nevertheless anticipate structural similarity with herpesviruses in the following components: some proteases; portals; (possibly) internal scaffolding proteins; and terminases, for which a tract of significant sequence similarity has been detected between T4 and HSV-1. Further clarification should also come from more detailed information on floor domains.
Nevertheless, other less direct lines of evidence point to common evolutionary origins. There are so many highly distinctive molecular behaviors listed in Table 1, involving quaternary structure and other properties, which are common to phage capsids and herpesvirus capsids but in most cases shared by no other kind of virus, that the a priori probability of their having arisen by chance appears very small.
3. LATER CAPSID-RELATED STEPS IN THE REPLICATION CYCLE
After completion of DNA packaging, the C-capsid has to exit the nucleus by primary envelopment at the inner nuclear membrane, followed by fusion of the membrane thus acquired with the outer nuclear membrane, delivering the nucleocapsid into the cytoplasm. It also has to acquire a tegument, traverse the cytoplasm, and bud into a trans-Golgi-derived compartment (secondary envelopment). Progeny virions are released by fusion of this compartment’s membrane with the host cell plasma membrane. Virions may remain attached to the host cell for a while (“cell-associated virions”) before being released as “extracellular virions”. After entry into a host, the nucleocapsid is transported to the nucleus where it docks on to a nuclear pore complex to discharge its DNA, a mode of genome delivery reminiscent of tailed phages. These processes have been reviewed recently (Baines 2007; Mettenleiter et al. 2009; Guo et al. 2010) and the coverage given here focuses on some structural observations directly related to the nucleocapsid.
3.1 Nuclear exit and the roles of UL25 and UL17
As noted in §2.2, C-capsids migrate out of the nucleus more readily than B-capsids or A-capsids. This barrier is not absolute, as enveloped B-capsids have been observed, albeit rarely (unpublished results). There appears to be a pathway to tegumentation and envelopment on which C-capsids embark with high efficiency, but which can also be entered by other particles, e.g. small membrane vesicles nucleating the formation of L-particles, which resemble capsid-less virions (Szilagyi and Berriman 1994). The efficiency with which C-capsids exit the nucleus correlates with their contents of two viral proteins, UL25 and UL17, which form an elongated heterodimer called the CCSC (C-capsid-specific component) that binds to vertex-proximal sites on C-capsids (Figs. 2 and 6).
The penton-distal portion is contributed by UL25, as initially suggested by docking the crystal structure (Trus et al. 2007), leaving the penton-proximal portion to represent (part of) UL17. The UL25 assignment has since been proven by imaging C-capsids bearing UL25 with an N-terminal GFP fusion, and Fab-labelling with an antibody whose epitope involves residues 99–111 (Conway et al. 2010). The CCSC density furthest from the penton was shown to be the N-terminal region of UL25 which has been shown to be important for capsid binding (Cockrell et al. 2009) but is not represented in the crystal structure, which starts at residue 134. More detailed mapping has assigned the P-hexon interaction to residues 1–26 and residues 28–39 as likely to make the Tc-triplex contact (Cockrell et al. 2011).
Cryo-EM of purified nuclear C-capsids found the CCSC sites to have an average occupancy of ~ 60% (Trus et al. 2007), whereas occupancy is undetectably low (i.e. < 15% or so) on B-capsids and A-capsids. Biochemical studies have reported the presence of these proteins on all three capsids (Ogasawara et al. 2001; Thurlow et al. 2006; Newcomb et al. 2006; Cockrell et al. 2011) but their complements on A- and B-capsids are several-fold lower, and it is possible that these few molecules are scattered over other quasi-equivalent (not vertex-proximal) sites.
UL17 and UL25 are multifunctional proteins. The phenotype of a UL17-null mutant is the accumulation of B-capsids (Salmon and Baines 1998). In view of UL17’s proximity to vertices, this phenotype may reflect the absence of its ability to engage terminase and hence DNA for packaging at the portal vertex. UL17 is also required for UL25 to associate with capsids (Thurlow et al. 2006). UL25 is needed for packaged genomes to remain stably encapsidated (McNab et al. 1998; Stow 2001; Cockrell et al. 2009) and is also involved in the interaction of incoming nucleocapsids with nuclear pore complexes (Preston et al. 2008). Here we focus on its role in nuclear exit (Klupp et al. 2006). Primary envelopment of the C-capsid is a complex process involving remodelling of the karyoskeleton and the inner nuclear membrane (Baines 2007). With that qualification, we envisage the following scenario.
CCSC binding is enhanced by a relatively subtle conformational change in the capsid, affecting its sites on the outer surface (Trus et al. 2007). Aspects of this change appear in a cryo-EM difference map between C-capsids and A-capsids (Fig. 7). Although similar, these structures are not identical as the difference map does not cleanly separate the CCSC; rather, small local changes affect various parts of the shell. Differences affecting the triplexes to which the CCSC binds are arrowed in Figure 7B. Of note, small but significant structural differences have also been detected between DNA-filled capsids of phage HK97 and empty but nominally mature capsids (Duda et al. 2009).
Figure 7. Final switch in HSV-1 capsid maturation.
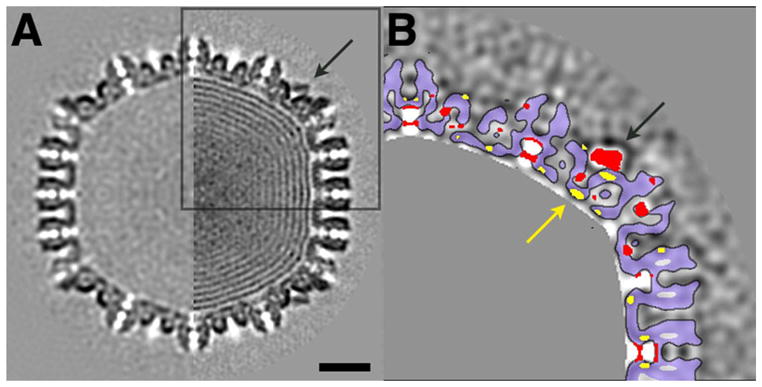
This transition is shown in a comparison of A-capsid and C-capsid. (N.B. The A-capsid is not a direct precursor of the C-capsid; rather it is an abortive capsid resulting from interrupted DNA packaging. We take it to represent the mature capsid prior to the switch induced by late-stage DNA packaging that creates high-affinity sites for the CCSC). A: Half-plane central sections of capsids viewed along a 2-fold axis: left: A-capsid; right: C-capsid. Scale bar, 20 nm. B: Difference map (C-capsid minus A-capsid) of the boxed area in A. Positive (negative) difference densities are dark (light). Positive densities above 1 standard deviation are colored red. Negative densities below 1 standard deviation are yellow. The strongest positive density are the CSSC (arrows in A and B) and density in the mouth of the axial channel through the floor. A single rigid-body shift of a capsid component would register as complementary red and yellow patches of the same size and shape, suitably offset; however, this difference map - and hence this transition - is more complex. The difference map is from density maps filtered at 30 Å. Outlines of capsomers and triplexes are contoured with light blue fill. DNA-related density inside the C-capsid is masked out in gray.
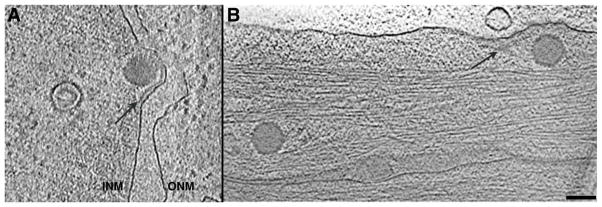
This switch in the C-capsid is presumably elicited by mounting outward pressure as DNA packaging approaches completion. We posit that high occupancy of its CCSC sites enables the C-capsid to bind, via multiple low-affinity interactions, other protein(s) involved in primary envelopment. In this context, cryo-ET observations of vitrified sections of infected cells have detected an additional density layer, between the INM and budding C-capsids (Fig. 8A). If this layer represents viral proteins, UL34 and UL31 are plausible candidates (Baines 2007). If the envisaged interactions between the CCSC and its binding partner(s) are, as we suggest, of such low affinity as to need multivalency for their stable formation, the sparsity of CCSC complexes on the surfaces of A- and B-capsids would disallow embarkation on the nuclear exit pathway.
Figure 8. Snapshots of the herpesvirus life cycle.
(A) Nuclear egress. Tomographic slice from a vitrified section of a NIH 3T3 cell infected with murine cytomegalovirus (for preparation details, see Scrivano et al. 2010). Inside the nucleus are seen a B-capsid and a C-capsid budding into the inner nuclear membrane. A layer of density between them (arrow) is likely to represent a layer of protein. Courtesy of Drs C. Hagen and K. Grünewald. (B) Cell entry. Slice from a tomogram of a PtK2 cell, where two capsids of recently entered HSV-1 virions are visible. The strip of density along the cell membrane (arrow) appears to represent divested tegument. From (Maurer et al. 2008), courtesy of Dr. K. Grünewald. Bar, 100 nm.
3.2 Tegumentation
In addition to delivering their genomes into host cells, tailed phages and herpesviruses both deliver proteins which perform functions needed early in the infection process. However, these proteins and their manner of delivery differ. In phage, they are incorporated into the procapsid and injected from the mature capsid into the host cell via the portal and tail-tube (Casjens and Molineux, this volume; Thomas and Black, this volume). Those of herpesviruses are incorporated into the tegument and released into the host cell cytoplasm when the viral envelope fuses with the host cell membrane. Whereas herpesvirus capsids are conservative in terms of their architecture and molecular components, teguments are more diverse - (reviews: Britt 2007; Guo et al. 2010). Herpesviruses also vary in their teguments’ point of attachment on the capsid; on simian cytomegalovirus, it is to triplexes as well as to capsomer protrusions (simian CMV - Trus et al. 1999; human CMV - Yu et al. 2011) and on HSV-1, to pentons (Chen et al. 2001; Grünewald et al. 2003).
Moreover, teguments are markedly less regular in structure than capsids. Tegument proteins have been operationally distinguished into two classes in terms of their extractability from virions with the detergent Triton X-100 (Wolfstein et al. 2006). Those that are relatively easily extracted are called the outer tegument and those that are more resistant, the inner tegument. The diversity of tegument components raises the challenging question of how they are selected for incorporation into nascent virions. A current working hypothesis is that a subset of inner tegment proteins, including the very large protein UL36 (~ 340 kDa), is anchored on the capsid and serves as a scaffold to which other tegument proteins attach (Coller et al. 2007; Newcomb and Brown 2010).
Cryo-ET of extracellular HSV-1 virions has demonstrated that their teguments are asymmetric, varying in thickness from ~ 5 nm where the capsid is closely apposed to the envelope to ~ 35 nm at the thickest part (Grünewald et al. 2003). Thus the capsid is eccentrically positioned within an approximately spherical virion. Evidence from thin sectioning EM is consistent with this observation while also showing that in cell-associated virions, the tegument is approximately uniform in thickness and the nucleocapsid is centered (Newcomb and Brown 2009). It follows that a substantial reorganization of the tegument takes place, following exocytosis of progeny virions. It is not clear whether the inferred reorganization of tegument is accompanied by a redistribution of glycoproteins in the viral envelope or whether it may correlate with infectivity.
3.3 Capsid-tegument interactions in host cell entry
Snapshots of HSV-1 nucleocapsids immediately after cell entry have been recorded by cryo-ET of PtK2 cells which are thin enough specimens to allow this technique (Maurer et al. 2008). These data suggest that the nucleocapsid is released into the cytoplasm, while the tegument remains largely connected and in contact with the viral envelope (now part of the host cell membrane) - Fig. 8B. This observation suggests that although tegument assembly is templated on the capsid, latterly it is more tightly bound to the envelope. The freed nucleocapsid, eventually retaining a few tegument proteins, is then transported along microtubules towards the nucleus by dynein/dynactin motors.
UL36 has been shown to play a role in the anterograde transport process but appears not to be the only player (Shanda and Wilson 2008): other inner tegument proteins are also involved (Radtke et al. 2010). Once the capsid reaches the nuclear envelope, interactions of UL25 (Pasdeloup et al. 2009) and UL36 (Radtke et al. 2010) with the nucleoporin Nup214 are important for its engagement with a nuclear pore complex.
Acknowledgments
We thank Dr Robert Duda for helpful discussion and Drs Christoph Hagen and Kay Grünewald for the images in Figure 8. This research was supported by the Intramural Research Programs of NIAMS and CIT.
Contributor Information
Giovanni Cardone, Email: cardoneg@mail.nih.gov.
J. Bernard Heymann, Email: Bernard_Heymann@nih.gov.
Naiqian Cheng, Email: chengn@mail.nih.gov.
Benes L. Trus, Email: trusb@mail.nih.gov.
References
- Agirrezabala X, Martin-Benito J, Valle M, Gonzalez JM, Valencia A, Valpuesta JM, Carrascosa JL. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J Mol Biol. 2005;347 (5):895–902. doi: 10.1016/j.jmb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Velazquez-Muriel JA, Gomez-Puertas P, Scheres SH, Carazo JM, Carrascosa JL. Quasi-atomic model of bacteriophage T7 procapsid shell: insights into the structure and evolution of a basic fold. Structure. 2007;15 (4):461–472. doi: 10.1016/j.str.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Baines JD. Envelopment of herpes simplex virus nucleocapsids at the inner membrane. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Chapter 11. Cambridge University Press; Cambridge: 2007. [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79 (23):14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Wedemeyer WJ, Rixon FJ, Baker D, Chiu W. Ab initio modeling of the herpesvirus VP26 core domain assessed by CryoEM density. PLoS Comput Biol. 2006;2 (10):e146. doi: 10.1371/journal.pcbi.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI. What does structure tell us about virus evolution? Curr Opin Struct Biol. 2005;15 (6):655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Benson SD, Bamford JKH, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98 (6):825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like, packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy FP, Trus BL, Davison AJ, Steven AC. The capsid architecture of channel catfish virus, an evolutionarily distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology. 1996;215:134–141. doi: 10.1006/viro.1996.0016. [DOI] [PubMed] [Google Scholar]
- Bowman BR, Baker ML, Rixon FJ, Chiu W, Quiocho FA. Structure of the herpesvirus major capsid protein. EMBO J. 2003;22 (4):757–765. doi: 10.1093/emboj/cdg086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BR, Welschhans RL, Jayaram H, Stow ND, Preston VG, Quiocho FA. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J Virol. 2006;80 (5):2309–2317. doi: 10.1128/JVI.80.5.2309-2317.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. Structure and assembly of immature HIV. Proc Natl Acad Sci USA. 2009;106 (27):11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt B. Maturation and egress. In: Arvin A, Campadelli-Fiume G, Mocarski E, et al., editors. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Chapter 20. Cambridge University Press; Cambridge: 2007. [PubMed] [Google Scholar]
- Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361 (2):426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–280. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81 (4):2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, Baker D, King JA, Chiu W. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci USA. 2011;108 (4):1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DH, Jakana J, McNab D, Mitchell J, Zhou ZH, Dougherty M, Chiu W, Rixon FJ. The pattern of tegument-capsid interaction in the herpes simplex virus type 1 virion is not influenced by the small hexon-associated protein VP26. J Virol. 2001;75 (23):11863–11867. doi: 10.1128/JVI.75.23.11863-11867.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13 (8):2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. Residues of the UL25 Protein of Herpes Simplex Virus That Are Required for Its Stable Interaction with Capsids. J Virol. 2011;85 (10):4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell SK, Sanchez ME, Erazo A, Homa FL. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J Virol. 2009;83 (1):47–57. doi: 10.1128/JVI.01889-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller KE, Lee JI, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81 (21):11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cockrell SK, Copeland AM, Newcomb WW, Brown JC, Homa FL. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol. 2010;397 (2):575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Wikoff WR, Cheng N, Duda RL, Hendrix RW, Johnson JE, Steven AC. Virus maturation involving large subunit rotations and local refolding. Science. 2001;292 (5517):744–748. doi: 10.1126/science.1058069. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Trus BL, Cheng N, Steven AC, Watson MS, Cunningham C, Le Deuff RM, Renault T. A novel class of herpesvirus with bivalve hosts. J Gen Virol. 2005;86 (Pt 1):41–53. doi: 10.1099/vir.0.80382-0. [DOI] [PubMed] [Google Scholar]
- Deng B, O’Connor CM, Kedes DH, Zhou ZH. Direct visualization of the putative portal in the Kaposi’s sarcoma-associated herpesvirus capsid by cryoelectron tomography. J Virol. 2007;81 (7):3640–3644. doi: 10.1128/JVI.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T. Scaffolding proteins and their role in viral assembly. Cell Mol Life Sci. 1999;56 (7–8):580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RL. Protein chainmail: catenated protein in viral capsids. Cell. 1998;94 (1):55–60. doi: 10.1016/s0092-8674(00)81221-0. [DOI] [PubMed] [Google Scholar]
- Duda RL, Hendrix RW, Huang WM, Conway JF. Shared architecture of bacteriophage SPO1 and herpesvirus capsids. Curr Biol. 2006;16 (1):R11–13. doi: 10.1016/j.cub.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Duda RL, Ross PD, Cheng N, Firek BA, Hendrix RW, Conway JF, Steven AC. Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J Mol Biol. 2009;391 (2):471–483. doi: 10.1016/j.jmb.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann A, Coward JE, Rosenkranz HS, Morgan C. Electron microscopic studies on assembly of herpes simplex virus upon removal of hydroxyurea block. JGenVirol. 1975;26:171–181. doi: 10.1099/0022-1317-26-2-171. [DOI] [PubMed] [Google Scholar]
- Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458 (7238):646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald K, Desai P, Winkler DC, Heymann JB, Belnap DM, Baumeister W, Steven AC. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302 (5649):1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- Guo H, Shen S, Wang L, Deng H. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell. 2010;1 (11):987–998. doi: 10.1007/s13238-010-0120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6 (5):506–511. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Henson BW, Johnson N, Bera A, Okoye ME, Desai KV, Desai PJ. Expression of the HSV-1 capsid protein VP19C in Escherichia coli: a single amino acid change overcomes an expression block of the full-length polypeptide. Protein Expr Purif. 2011;77 (1):80–85. doi: 10.1016/j.pep.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat Struct Biol. 2003;10 (5):334–341. doi: 10.1038/nsb922. [DOI] [PubMed] [Google Scholar]
- Hoog SS, Smith WW, Qiu X, Janson CA, Hellmig B, McQueney MS, O’Donnell K, O’Shannessy D, DiLella AG, Debouck C, Abdel-Meguid SS. Active site cavity of herpesvirus proteases revealed by the crystal structure of herpes simplex virus protease/inhibitor complex. Biochemistry. 1997;36 (46):14023–14029. doi: 10.1021/bi9712697. [DOI] [PubMed] [Google Scholar]
- Huffman JB, Newcomb WW, Brown JC, Homa FL. Amino acids 143 to 150 of the herpes simplex virus type 1 scaffold protein are required for the formation of portal-containing capsids. J Virol. 2008;82 (13):6778–6781. doi: 10.1128/JVI.00473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionel A, Velazquez-Muriel JA, Luque D, Cuervo A, Caston JR, Valpuesta JM, Martin-Benito J, Carrascosa JL. Molecular rearrangements involved in the capsid shell maturation of bacteriophage T7. J Biol Chem. 2011;286 (1):234–242. doi: 10.1074/jbc.M110.187211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze MD, Barlow PN, Price NC, Kelly SM, Boutell CJ, Rixon FJ, McClelland DA. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J Virol. 1998;72 (12):10066–10072. doi: 10.1128/jvi.72.12.10066-10072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Granzow H, Keil GM, Mettenleiter TC. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J Virol. 2006;80 (13):6235–6246. doi: 10.1128/JVI.02662-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Bamford DH. Virus evolution: how far does the double beta-barrel viral lineage extend? Nat Rev Microbiol. 2008;6 (12):941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- Kuznetsov YG, Martiny JB, McPherson A. Structural analysis of a Synechococcus myovirus S-CAM4 and infected cells by atomic force microscopy. J Gen Virol. 2010;91 (Pt 12):3095–3104. doi: 10.1099/vir.0.025254-0. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26 (7):1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurz R, Orlova EV, Gunther D, Dube P, Droge A, Weise F, van Heel M, Tavares P. Structural organisation of the head-to-tail interface of a bacterial virus. J Mol Biol. 2001;310 (5):1027–1037. doi: 10.1006/jmbi.2001.4800. [DOI] [PubMed] [Google Scholar]
- Marintcheva B, Weller SK. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog Nucleic Acid Res Mol Biol. 2001;70:77–118. doi: 10.1016/s0079-6603(01)70014-1. [DOI] [PubMed] [Google Scholar]
- Maurer UE, Sodeik B, Grünewald K. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci USA. 2008;105 (30):10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab AR, Desai P, Person S, Roof LL, Thomsen DR, Newcomb WW, Brown JC, Homa FL. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72 (2):1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143 (2):222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol. 2003;10 (7):572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Time-dependent transformation of the herpesvirus tegument. J Virol. 2009;83 (16):8082–8089. doi: 10.1128/JVI.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Brown JC. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol. 2010;84 (18):9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Homa FL, Brown JC. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J Virol. 2006;80 (13):6286–6294. doi: 10.1128/JVI.02648-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Trus BL, Cheng N, Steven AC, Sheaffer AK, Tenney DJ, Weller SK, Brown JC. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74 (4):1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara M, Suzutani T, Yoshida I, Azuma M. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J Virol. 2001;75 (3):1427–1436. doi: 10.1128/JVI.75.3.1427-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18 (5):597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker ML, Ralston EJ, Eiserling FA. Bacteriophage SPO1 structure and morphogenesis. II. Head structure and DNA size. J Virol. 1983;46 (1):250–259. doi: 10.1128/jvi.46.1.250-259.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J Virol. 2009;83 (13):6610–6623. doi: 10.1128/JVI.02655-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier A, Do F, Brisebois JJ, Lagace L, Cordingley MG. Self-association of herpes simplex virus type 1 ICP35 is via coiled-coil interactions and promotes stable interaction with the major capsid protein. JVirol. 1997;71:5197–5208. doi: 10.1128/jvi.71.7.5197-5208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston VG, Murray J, Preston CM, McDougall IM, Stow ND. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J Virol. 2008;82 (13):6654–6666. doi: 10.1128/JVI.00257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Culp JS, DiLella AG, Hellmig B, Hoog SS, Janson CA, Smith WW, Abdel-Meguid SS. Unique fold and active site in cytomegalovirus protease. Nature. 1996;383:275–279. doi: 10.1038/383275a0. [DOI] [PubMed] [Google Scholar]
- Qiu X, Janson CA, Culp JS, Richardson SB, Debouck C, Smith WW, Abdel-Meguid SS. Crystal structure of varicella-zoster virus protease. Proc Natl Acad Sci USA. 1997;94 (7):2874–2879. doi: 10.1073/pnas.94.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010;6 (7):e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat RH, Liu X, Murata K, Nagayama K, Rixon FJ, Chiu W. Seeing the portal in herpes simplex virus type 1 B capsids. J Virol. 2011;85 (4):1871–1874. doi: 10.1128/JVI.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon B, Baines JD. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U(L)15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol. 1998;72 (4):3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrivano L, Esterlechner J, Muhlbach H, Ettischer N, Hagen C, Grunewald K, Mohr CA, Ruzsics Z, Koszinowski U, Adler B. The m74 gene product of murine cytomegalovirus (MCMV) is a functional homolog of human CMV gO and determines the entry pathway of MCMV. J Virol. 2010;84 (9):4469–4480. doi: 10.1128/JVI.02441-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanda SK, Wilson DW. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol. 2008;82 (15):7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer AK, Newcomb WW, Brown JC, Gao M, Weller SK, Tenney DJ. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J Virol. 2000;74 (15):6838–6848. doi: 10.1128/jvi.74.15.6838-6848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Leiman PG, Tao Y, He Y, Badasso MO, Jardine PJ, Anderson DL, Rossmann MG. Structure determination of the head-tail connector of bacteriophage phi29. Acta Crystallogr D Biol Crystallogr. 2001;57 (Pt 9):1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol. 2005;15 (2):227–236. doi: 10.1016/j.sbi.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow ND. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J Virol. 2001;75 (22):10755–10765. doi: 10.1128/JVI.75.22.10755-10765.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilagyi JF, Berriman J. Herpes simplex virus L particles contain spherical membrane-enclosed inclusion vesicles. J Gen Virol. 1994;75 (Pt 7):1749–1753. doi: 10.1099/0022-1317-75-7-1749. [DOI] [PubMed] [Google Scholar]
- Thurlow JK, Murphy M, Stow ND, Preston VG. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J Virol. 2006;80 (5):2118–2126. doi: 10.1128/JVI.80.5.2118-2126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L. Viral proteases. Chem Rev. 2002;102 (12):4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J Virol. 2004;78 (22):12668–12671. doi: 10.1128/JVI.78.22.12668-12671.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Gibson W, Cheng N, Steven AC. Capsid structure of simian cytomegalovirus from cryoelectron microscopy: evidence for tegument attachment sites [published erratum appears in J Virol 1999 May;73(5):4530] J Virol. 1999;73 (3):2181–2192. doi: 10.1128/jvi.73.3.2181-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Heymann JB, Nealon K, Cheng N, Newcomb WW, Brown JC, Kedes DH, Steven AC. Capsid structure of Kaposi’s sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J Virol. 2001;75 (6):2879–2890. doi: 10.1128/JVI.75.6.2879-2890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Homa FL, Booy FP, Newcomb WW, Thomsen DR, Cheng N, Brown JC, Steven AC. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Newcomb WW, Booy FP, Brown JC, Steven AC. Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc Natl Acad Sci USA. 1992;89:11508–11512. doi: 10.1073/pnas.89.23.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26 (4):479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, King J, Hatfull GF, Lawrence JG, Hendrix RW. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. 2007;9 (7):1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- Welch AR, McNally LM, Hall MRT, Gibson W. Herpesvirus proteinase: Site-directed mutagenesis used to study maturational, release, and inactivation cleavage sites of precursor and to identify a possible catalytic site serine and histidine. J Virol. 1993;67:7360–7372. doi: 10.1128/jvi.67.12.7360-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289 (5487):2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Thomsen DR, Homa FL, Booy FP, Trus BL, Steven AC. Hexon-only binding of VP26 reflects differences between the hexon and penton conformations of VP5, the major capsid protein of herpes simplex virus. J Virol. 1997;71:8955–8961. doi: 10.1128/jvi.71.12.8955-8961.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7 (2):227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- Yang K, Baines JD. Domain within herpes simplex virus 1 scaffold proteins required for interaction with portal protein in infected cells and incorporation of the portal vertex into capsids. J Virol. 2008;82 (10):5021–5030. doi: 10.1128/JVI.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shah S, Lee M, Dai W, Lo P, Britt W, Zhu H, Liu F, Zhou ZH. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J Struct Biol. 2011;174 (3):451–460. doi: 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288 (5467):877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, He J, Jakana J, Tatman JD, Rixon FJ, Chiu W. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nature Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]