Abstract
In Drosophila the fat body (FB), a functional analog of the vertebrate adipose tissue, is the 'nutrient sensor' that conveys the nutrient status to the insulin producing cells (IPCs) in the fly brain to release insulin-like peptides (Dilps). Dilp secretion in turn regulates energy balance and promotes systemic growth. We identify Unpaired2 (Upd2), a protein with similarities to type I cytokines, as a secreted factor produced by the FB in the ‘fed’ state. When upd2 function is perturbed specifically in the FB, it results in a systemic reduction in growth and alters energy metabolism. Upd2 activates JAK/STAT signaling in a population of GABAergic neurons that project onto the IPCs. This activation relieves the inhibitory tone of the GABAergic neurons on the IPCs, resulting in the secretion of Dilps. Strikingly, we find that human Leptin, can rescue the upd2 mutant phenotypes, suggesting that Upd2 is the functional homolog of Leptin.
Introduction
Integration of information regarding nutrient status with other physiological processes such as systemic growth, energy expenditure, feeding and reproduction is a complex function performed by multicellular organisms. When this fundamental homeostatic process is disrupted, it can lead to a number of disorders, in particular, obesity, anorexia and diabetes (Morton et al., 2006). In addition, it can have profound effects on cancer and aging (Hursting et al., 2003).
Insulin peptides are key hormones involved in the regulation of carbohydrate and lipid metabolism, tissue growth and longevity (Taguchi and White, 2008). Circulating insulin absorbs nutrients, such as glucose and lipids, and stores them for later use in the form of glycogen and triacylglycerol (TAG). When insulin production from pancreatic beta cells is disrupted in mammals, as in the case of Type I diabetes, the body is unable to utilize the nutrients consumed and instead mounts a starvation response wherein stored lipids and glycogen are broken down to generate energy (Kahn et al., 2005). The production of insulin by pancreatic beta cells is tightly regulated to ensure that appropriate amounts are released into the blood depending on nutrient status and food intake.
The insulin pathway is highly conserved from mammals to Drosophila and plays fundamentally the same physiological functions (Taguchi and White, 2008; Wu and Brown, 2006). A main difference however is that the fly insulin producing cells (IPCs), which are homologous to pancreatic beta cells, are found in the brain (Rulifson et al., 2002). These IPCs lie in the brain median neurosecretory cluster (mNSC) and produce at least three of the eight insulin-like peptides (Dilps 2, 3 and 5) (Brogiolo et al., 2001; Ikeya et al., 2002). A deficiency uncovering multiple Dilps (Dilps1-5) results in flies that, in addition to being smaller, have decreased TAG and increased circulating sugars (Kulkarni et al., 1997). Dilps secreted from the IPCs bind to the insulin receptor in peripheral tissues to promote growth and nutrient utilization.
The FB functions as a key sensor of the nutritional status of the fly and couples systemic growth, metabolism, and stem cell maintenance with nutritional availability. Studies using ex-vivo organ co-cultures of fat bodies and larval brains proposed that the FB secretes growth promoting factors (Britton and Edgar, 1998; Davis and Shearn, 1977). Suppression of amino-acid (AA) transport in the FB, by knocking down the AA transporter slimfast (slif), resulted in flies with systemic growth defects, suggesting that the FB acts as a nutrient sensor that non-autonomously modulates insulin signaling based on nutrient status (Colombani et al., 2003; Geminard et al., 2009). When flies are cultured on rich food, the amount of Dilp accumulation was considerably lower in the IPCs compared to the level of Dilps in IPCs of nutrient-deprived flies. Consistent with this, the level of Dilps in the hemolymph of fed flies was higher than in starved flies (Geminard et al., 2009). Altogether, these results suggest that the regulation of organismal growth in response to nutrient availability involves the control of insulin secretion from brain IPCs by factors originating from the FB.
Here, we identify Upd2, a Drosophila cytokine, as a secreted factor produced by the FB in response to dietary fat and sugars. Upd2 activates JAK/STAT signaling in GABAergic neurons relieving their inhibitory effect on the IPCs in turn resulting in the secretion of Dilps into the hemolymph to promote systemic growth and fat storage. The similarities between Upd2 and vertebrate Leptin are striking as it is secreted from the adipose tissue in mammals under conditions of nutritional surplus in particular fat levels (Zhang et al., 1994). We show that human Leptin can functionally substitute for Drosophila Upd2 and can function as a ligand to the JAK/STAT receptor Dome Drosophila. Altogether, our studies illustrate how a cytokine-mediated pathway regulates the secretion of Insulin to modulate systemic growth according to nutrient availability, in particular dietary fats, and provides evidence for the evolutionary conservation of this signaling module.
RESULTS
Upd2 plays a fat body-specific role to control systemic growth
Cytokines, secreted molecules which communicate intercellular signals, are ideal candidates for remotely signaling the nutritional status (Dinarello and Mier, 1986) and signal through the JAK/STAT pathway. In the Drosophila genome, the three related Upd ligands, Upd1, Upd2 and Upd3, have predicted secondary structures similar to that of type I cytokines (Boulay et al., 2003) and regulate the activity of the JAK/STAT pathway (Wright et al., 2011; Zeidler et al., 2000). They bind to the transmembrane receptor Domeless (Dome)(Brown et al., 2001) which in turn activates the Janus Kinase (JAK) Hopscotch (Hop)( Binari and Perrimon, 1994). Activated Hop regulates the signal transducer and activator of transcription STAT92E/Marelle (Hou et al., 1996). Since cytokines play central roles in mammalian nutrition sensing and metabolic homeostasis, we reasoned that their fly counterparts could be involved in similar processes.
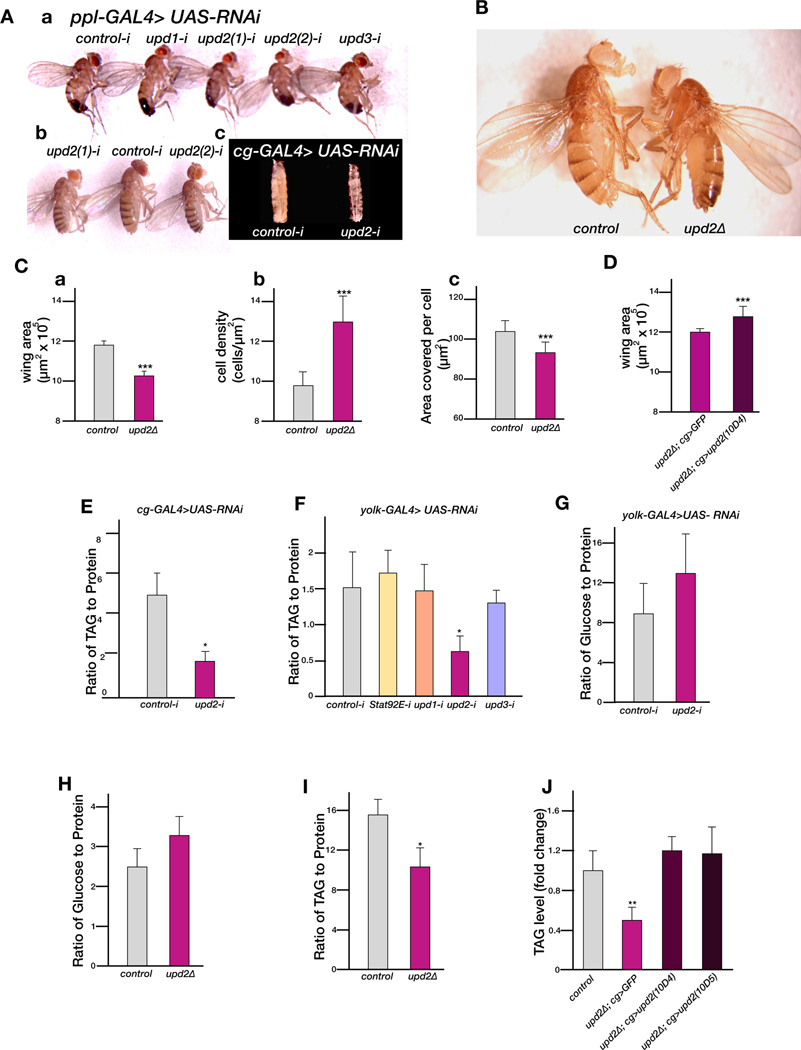
To investigate the role of the Upd cytokines in the FB, and their effects on overall body size, we used RNA interference (RNAi) to knockdown their expression. At least two independent RNAi lines per gene and two different FB GAL4 drivers (ppl-GAL4; and cg-GAL4) were used to ensure that the knockdown was both gene and tissue specific. The efficiency of knockdown of all three genes in the larval FB was comparable (upd1 (80%), upd2 (60%), and upd3 (70%)) as analyzed by qPCR. Interestingly, only FB-specific knockdown of upd2, and not upd1 or upd3, resulted in smaller flies (Figure 1Aa, 1Ab) and larvae (Figure 1Ac), suggesting that this ligand alone plays a FB-specific role in regulating systemic growth. In further support of the model that the effect of Upd2 on systemic growth is specific to the FB, knockdown of upd2 specifically in larval muscles, did not affect body size (Figure S1 A, B).
Figure 1. Upd2 controls systemic growth and metabolism.
(A) Adult flies with fat body (FB)-specific knockdown of upd1, upd2 and upd3 using the ppl-GAL4 driver (ppl-GAL4> upd-RNAi, subpanel a). RNAi of upd2 (upd2-RNAi, indicated as upd2-i in the Figures) using two independent lines (1 and 2) results in size reduction of both adult males (subpanel a) and females (subpanel b). Note that in the subsequent Figures, because upd2(1)-i and upd2(2)-i gave the same results, we used the upd2(1)-i line and simply refer to it as upd2-i. cg-GAL4>upd2-RNAi third instar larvae are smaller and slimmer than control (subpanel c). Control-i is Luciferase-RNAi. (B) Hemizygous adult males for the upd2 deletion allele (upd2Δ) appear smaller and slimmer than y w controls. (C) Quantification of the wing area (subpanel a), cell size (subpanel b), and cell density (subpanel c) of upd2Δ males compared to y w control. (D) Rescue of the wing area phenotype of upd2Δ males by FB-specific expression of upd2 cDNA using cg-GAL4. (E) Quantification of TAG to protein in male larvae with FB-specific knockdown of upd2 using cg-GAL4. (F) The ratio of TAG to protein in adult females with FB-specific knockdown of JAK/STAT pathway components using yolk-GAL4. (G) Circulating sugars in the hemolymph of female adults with FB-specific knockdown of upd2. In (E), (F) and (G) control is white-RNAi. (H) The ratio of glucose to protein in the hemolymph of upd2Δ adult males, compared to y w controls. (I) Ratio of TAG to protein in adult upd2Δ males compared to y w controls (J) Rescue of the TAG phenotype in upd2Δ adult males with FB-specific expression of either 10D4 or 10D5 upd2 cDNA using cg-GAL4. Error bars in the figures represent the standard deviation. p-values were calculated using the Welch’s t-test (*p<0.05, **p<<0.01, ***p<<0.00001). See also Figure S1
Previously, an upd2 homozygous deletion mutant (upd2Δ), that removes the 5’UTR and the first 89 amino acids, was identified and reported to be viable and fertile (Hombria et al., 2005). Examination of the growth phenotypes of upd2Δ flies, compared to an age and population-density matched control, showed that they were significantly slimmer and smaller (Figure 1B), similar to the phenotype generated with FB-specific knockdown of upd2 (Figure 1Ab). Examination of the wings, a tissue that can be easily quantified for growth phenotypes, revealed that upd2 mutant wings have a 10% reduction in overall size (Figure 1Ca), and a significant reduction in both cell number (Figure 1Cb) and cell size (Figure 1Cc). Expression of an upd2 cDNA in the FB of upd2Δ flies was able to rescue the body and the wing size phenotype (Figure 1D), providing further evidence that upd2 plays a FB-specific role in the regulation of systemic growth.
Upd2 plays a fat body-specific role to control metabolism
To test whether Upd2 plays a role in regulating energy metabolism, we measured the levels of triglycerides (TAG) in larvae with FB-specific knockdown of upd2. TAG levels were significantly reduced (Figure 1E). Similarly, upd2Δ larvae also showed a significant reduction in TAG levels (data not shown). Knockdown of upd2 specifically in other tissues such as larval muscles did not affect stored fat levels or body size (Figure S1), consistent with the model that the function of Upd2 is specific to the FB.
In Drosophila organismal growth is restricted to larval stages and genetic manipulations that affect nutrient sensing during adulthood lead to metabolic phenotypes. To assay whether Upd2 plays a specific role in nutrient sensing in adults, we used a GAL4 driver expressed only in the adult FB (yolk-GAL4) (see Experimental Procedures) and measured TAG levels in flies 15 days after eclosion. FB-specific knockdown of upd2 (Figure 1G) was associated with an increase in the amount of circulating sugar in the hemolymph suggesting that Upd2 plays a role in the FB to regulate overall energy metabolism. Consistently, upd2Δ adults displayed an increase in the levels of circulating sugars (Figure 1H), and a significant reduction in TAG levels (Figure 1I), that could be rescued by expressing an upd2-cDNA in the FB (Figure 1J). Interestingly, knockdown of stat92E the transcription factor mediating the JAK/STAT pathway activity, in the FB (efficiency of knockdown in the FB as assayed by qPCR >90%), did not affect TAG storage (Figure 1F), indicating that Upd2 plays a non-autonomous role in regulating fat storage.
Altogether, these results indicate that Upd2 in the FB regulates systemic growth in larvae and energy metabolism in both larvae and adults. Further, the effect of Upd2 is likely FB-non-autonomous as removal of stat92E from the FB is not associated with an effect on systemic growth.
Upd2 signals the fed state and senses fat and sugars
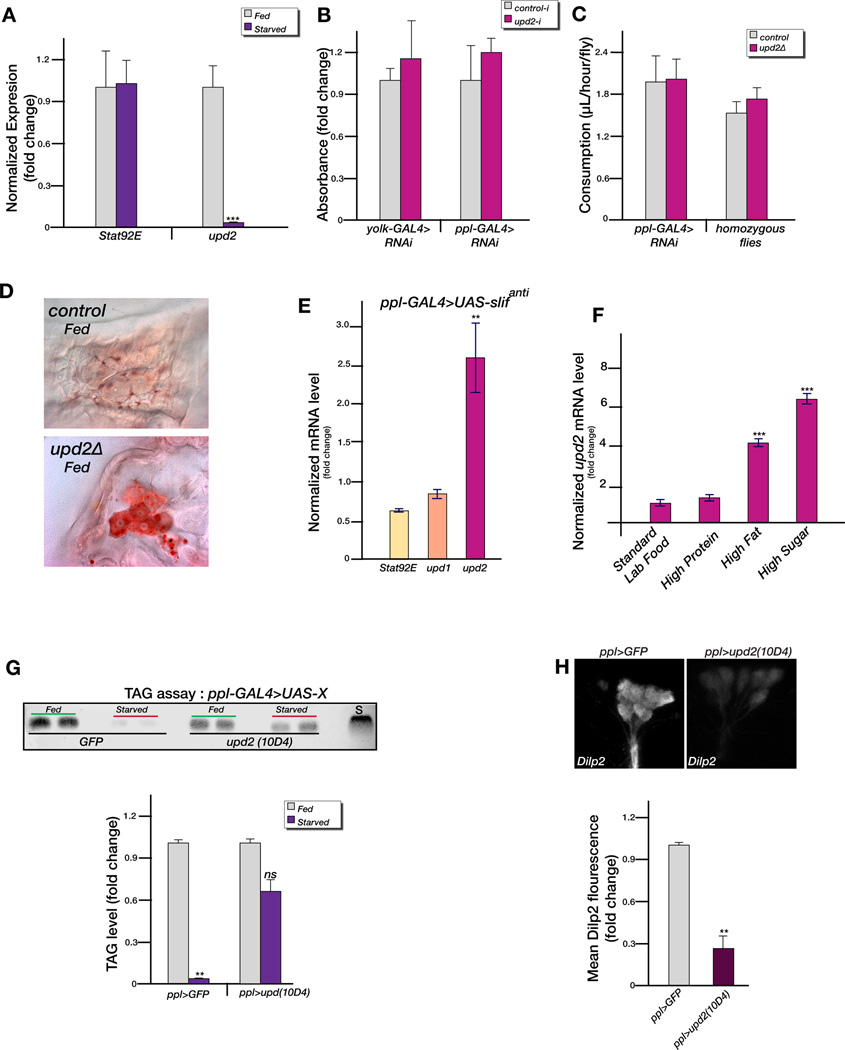
To explore the relationship between Upd2 expression in the FB and the nutritional state, we examined upd2 transcript levels under starvation. Strikingly, we found that upd2 transcripts showed a 98% reduction (Figure 2A) in adult wild-type males starved on 1% sucrose agar for >72 hours. Because a reduction in size and energy metabolism could result from reduced nutritional intake, we measured fly feeding using both the “blue dye” (Xu et al., 2008) and the “capillary feeder (CAFÉ)” assays (Ja et al., 2007). Both tests, performed in three to five replicates on age and population density matched adult male flies, showed that neither FB-specific knockdown of upd2 nor upd2Δ flies had feeding behavioral defects (Figure 2B, C).
Figure 2. Upd2 expression in the fat body signals the fed state.
(A) The steady-state mRNA expression of STAT92E and upd2 was analyzed by qPCR. (B) Blue dye feeding assay, in adult males with FB-specific knockdown of upd2. (C) CAFÉ assay in adult males with FB-specific knockdown of upd2 or upd2Δ. (D) Oil red O staining of oenocytes in third instar male larvae shows that upd2Δ animals accumulate oil red O droplets under fed conditions compared to y w controls. (E) Steady-state mRNA levels in adult males with FB-specific knockdown of slif compared to control (UAS-slifanti). (F) qPCR performed on RNA extracted 5 days after adult males were exposed to different diets. Normalized expression level of upd2 is shown. (G) TLC was used to assay the amount of stored TAG in adult male flies under fed and starved conditions. TAG was extracted from ppl-GAL4>10D4 upd2-cDNA or GFP (control) adult males. S indicates the mobility of coconut oil used as marker in the TLC plate. (H) Dilp2 immunostaining in the IPCs of starved adult male flies, expressing GFP (control) or 10D4 upd2-cDNA using ppl-GAL4. Black error bars represent the standard deviation. Blue error bars represent standard error of mean. p-values were calculated using the Welch’s t-test (*p<0.05, **p<<0.01, ***p<<0.00001).
To assess whether upd2 senses the nutritional state of the organism we analyzed lipid storage in hepatocyte-like cells/oenocytes which accumulate lipid droplets only under conditions of nutritional deprivation and not under a ‘fed’ state (Gutierrez et al., 2007). While control larvae did not accumulate lipid droplets under fed conditions (Figure 2D), upd2Δ larvae showed a striking accumulation of lipid droplets in oenocytes, as assayed by Oil Red O staining, even under conditions of nutritional surplus (Figure 2D). Since upd2Δ flies feed normally this lipid accumulation suggests that Upd2 is required to sense the appropriate nutritional state.
Given that knocking down the amino acid (AA) transporter slimfast (slif) mimics the starvation state (Colombani et al., 2003; Gutierrez et al., 2007), we examined the upd2 steady-state mRNA levels in FB-specific slif knockdown (ppl-GAL4>UAS-slifanti) by qPCR. Strikingly, upd2 level increased by more than two-fold while another JAK/STAT ligand upd1 and the downstream component STAT92E were not significantly altered (Figure 2E). Slif plays a role in transporting AAs, its absence results in a protein-deprived state. Hence the upregulation of upd2 under this state (Figure 2E) suggests that the upstream signal for upd2 expression is not nutrition-derived proteins.
Hence, we assayed upd2 expression in adult males flies subjected to diets rich in protein, fat or sugar and compared upd2 mRNA levels under these conditions with respect to standard lab food. In many independent experiments, we consistently found that 5 days after exposure to high fat and high sugar diets the normalized steady-state level of upd2 mRNAs went up by 4–6 fold, a statistically significant difference compared to levels on standard lab food (Figure 2F). Taken together these results strongly argue that upd2 senses the fed state downstream of fat and sugars.
To address whether Upd2 is sufficient to signal the fed state, we tested whether over-expression of upd2 in wild-type flies can suppress the starvation response associated with nutrient deprivation. We measured the breakdown of stored fat in starved flies over-expressing upd2 in the FB using thin layer chromatography (TLC). TLC was performed on adult flies to measure the fat stores after a period of 24 hours on 1% agar. While in control flies the stored fat levels were nearly depleted after 24 hours (Figure 2G; 98% reduction), the reduction was not significantly affected in flies over-expressing upd2 in their FB (Figure 2G; 35% reduction). Thus, Upd2 can suppress stored fat breakdown under conditions of starvation, indicating that it signals a fed condition even when flies are deprived of nutrients.
Brain IPCs secrete Dilps in response to nutrition and accumulate Dilps under conditions of nutrient deprivation (Geminard et al., 2009). To test if Upd2 over-expression in the FB can suppress accumulation of Dilps in the IPCs under conditions of starvation, age and population density matched adult flies over-expressing Upd2 in the FB were starved for 24 hours on 1% agar. The brains were then dissected and stained for Dilp2 and Dilp5, analyzed by confocal microscopy, and the mean Dilp fluorescence was calculated. Strikingly, Dilp2 and Dilp5 accumulation was significantly lower in brains of starved flies over-expressing Upd2 in their FB (Figure 2H; 78% lesser Dilp2; data not shown for Dilp5), revealing that Upd2 expression in the FB alters Dilp2 and Dilp5 accumulation in the brain IPCs in response to the nutritional state. Note that in all the following experiments the same results were observed for both Dilp2 and Dilp5. For simplicity only the Dilp2 data is shown, and in the text Dilp2 and Dilp5 are referred to as ‘Dilp(s)’.
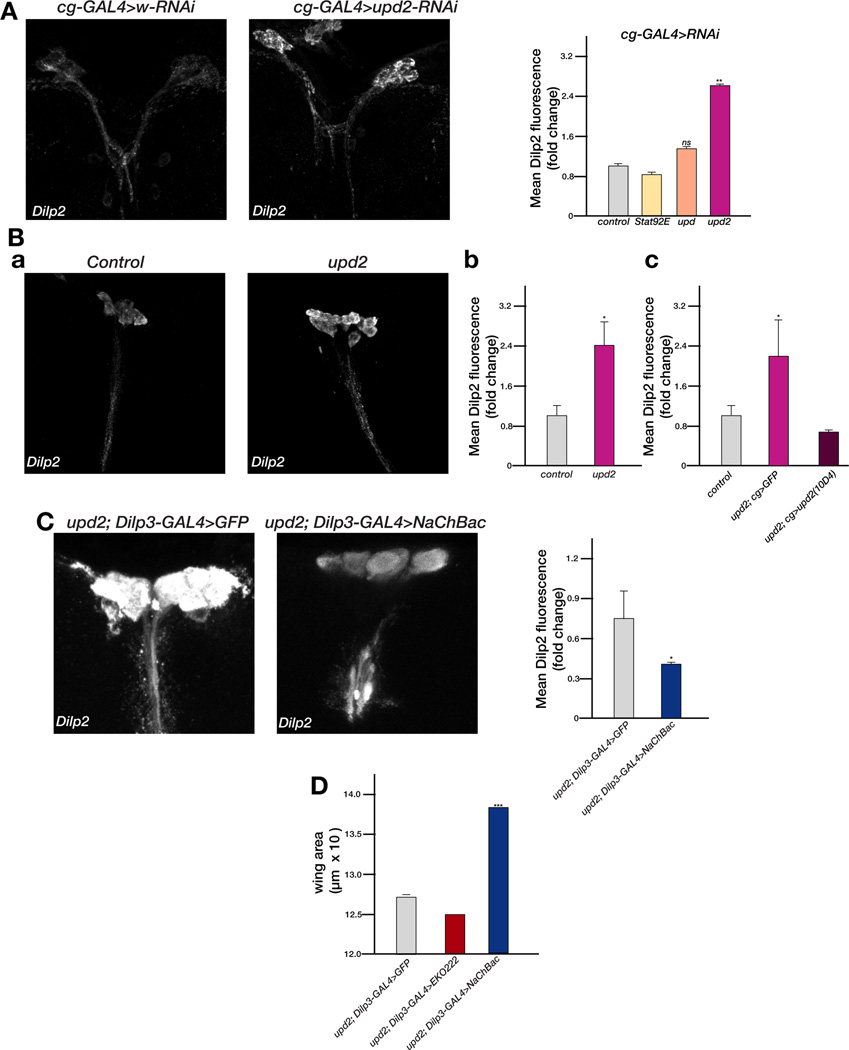
The upd2Δ mutants and FB knockdown of upd2, resemble the dilp1-5 deletion flies with respect to their size and metabolic phenotypes. Hence, we tested whether the primary role of Upd2 is to signal the fed state the IPCs. Two different FB-specific drivers, cg-GAL4 (Figure 3A) and ppl-GAL4 (data not shown), were used for these experiments. RNAi against upd2 in the FB resulted in a statistically significant increase in mean Dilp fluorescence in larval brains (Figure 3A). This increase in Dilp accumulation is most apparent when larvae are actively feeding. We performed qPCR analysis of Dilp in the brains of larvae with FB-specific knockdown of upd2 RNAi to ensure that the increase in mean Dilp fluorescence is not a result of increased transcription. No significant change in Dilp transcription was observed (Figure S2). In addition, FB-specific knockdown of either stat92E or upd1 did not result in increased accumulation of Dilp (Figure 3A), which is consistent with our observations on body size and metabolism (Figure 1). Further, the increase in Dilp accumulation in the IPCs was also observed in both larval (data not shown) and adult upd2Δ homozygous mutant brains (Figure 3B). Finally, the accumulation of Dilp in the IPCs of upd2Δ homozygous mutants could be rescued by expressing upd2 cDNA in the FB (Figure 3Bc), indicating that the inability to release Dilps underlies both the systemic growth and energy metabolism phenotypes of upd2Δ mutants.
Figure 3. Upd2 remotely controls Dilp accumulation in the IPCs.
(A) Maximum intensity projection of cg-GAL4 male larval brains stained with Dilp2 driving expression of either white-RNAi (control) or upd2-RNAi and quantification of the mean Dilp2 immunofluorescence in larval brains of cg-GAL4 animals driving RNAi of JAK/STAT pathway components. (B) Maximum intensity projection of adult male brains stained with Dilp2. The brains were dissected from age-matched upd2Δ and y w control (subpanel a). A significant increase in Dilp2 fluorescence is observed in upd2Δ male brains compared to y w (subpanel b). In subpanel c, rescue of Dilp2 accumulation in upd2Δ flies by FB-specific expression of the 10D4 upd2 cDNA using cg-GAL4. Brains from cg-Gal4 flies were used as positive controls (grey bar). (C) Expression of NaChBac in the IPCs using Dilp3-GAL4 of upd2Δ adult male brains. significantly reduces Dilp2 accumulation in IPCs. (D) Expression of NaChBac in the IPCs of upd2Δ adult male brains using Dilp3-GAL4 significantly increases the wing area of upd2Δ. Note that further inhibition of neuronal activity using the inward rectifying potassium channel (EKO222) does not reduce wing area any further than removing upd2 alone. ns= not significant; error bars represent the standard deviation. p-values were performed using the Welch’s t-test (*p<0.05, **p<<0.01, ***p<<0.00001). See also Figure S2.
IPCs in the Drosophila brain use membrane voltage dependent neurosecretory mechanisms to facilitate Dilp release in response to nutrition (Geminard et al., 2009). Thus, if Upd2 promotes Dilp secretion under fed conditions, then, activating Dilp secretion in upd2Δ homozygous mutants should rescue the upd2Δ growth phenotype. To achieve this, we depolarized the IPCs by expressing the bacterial sodium channel (NaChBac) (Luan et al., 2006; Ren et al., 2001) under the control of the Dilp3-GAL4 driver, which drives expression specifically in the brain IPCs during both larval and adult stages. Under these conditions the level of Dilp accumulation in the IPCs of upd2Δ was significantly reduced (Figure 3C). In addition, this artificial depolarization of the IPCs in an upd2Δ homozygous mutant background rescued the small size of upd2Δ mutants (Figure 3D). Interestingly, hyperpolarizing the neurons by expressing the potassium channel rectifier EKO (White et al., 2001) did not exacerbate the upd2Δ size phenotype (Figure 3D), indicating that in the absence of upd2 the IPCs neurons are already depolarized.
In summary, the reduction in TAG levels, increase in circulating sugars, and the systemic reduction in body size in the absence of upd2, resembles a reduction in insulin signaling, suggesting that the primary role of Upd2 is to remotely control the secretion of Dilps from the IPCs in response to nutrient intake.
A STAT reporter is expressed near the IPCs and is altered in response to starvation
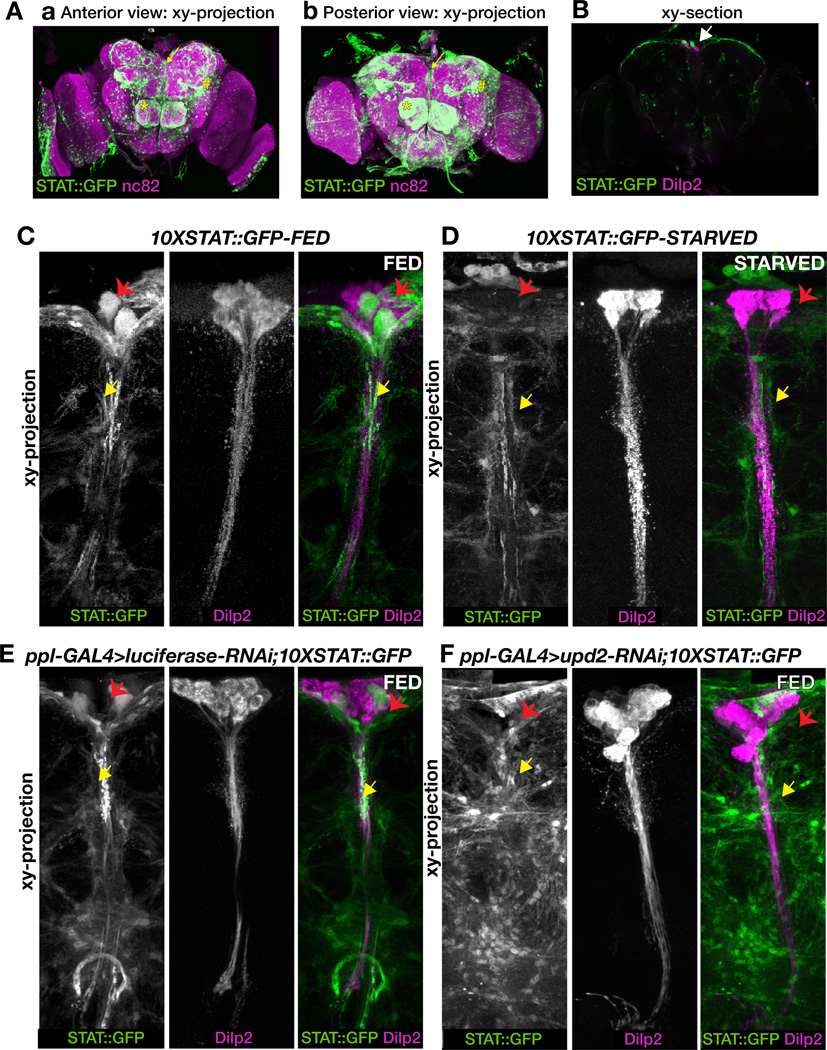
To elucidate how Upd2 regulates Dilp secretion in response to nutrition, we examined the activity of a STAT reporter (10XSTAT::GFP, see Experimental Procedures), previously shown to recapitulate JAK/STAT pathway activation (Bach et al., 2007), in the adult brain. We found that STAT::GFP is widely-expressed throughout the adult brain in both neurons and glia (Figure S3), and more specifically, it is expressed in the olfactory lobe (asterisk, Figure 4A), the optic lobe (hash, Figure 4A), and in the mNSCs (a region which includes the IPCs plus other neurons, see arrow, Figure 4A) of the adult brain. Importantly, analysis of single optical sections revealed that the STAT reporter is expressed in neurons juxtaposed to the IPCs (arrow, Figure 4B) in the mNSC region of the adult brain and in the larval brain (Figure S4).
Figure 4. STAT reporter expression in the adult brain and its response to starvation.
(A) Anterior (subpanel a) and posterior (subpanel b), maximum intensity projections of adult male brains expressing the 10XSTAT::GFP reporter. nc82 (magenta), a presynaptic active zone protein marker, identifies the neuropils. STAT::GFP (green) is widely expressed including in the olfactory lobe (asterisk), the optic lobe (hash), and the median neurosecretory cluster (mNSCs) (arrow). (B) Single optical section along the XY-axis of an adult male brain immunostained for Dilp2 (magenta) and STAT::GFP (green). Note that the cell bodies of the STAT reporter-expressing neurons are located right next to the Dilp2-expressing IPCs (arrow points to the juxtaposition of these neurons). (C–F) 3D projections along XY axis of adult male brains stained for Dilp2 (magenta) and STAT::GFP (green). (C) Under fed conditions the STAT::GFP reporter is expressed in neurons immediately adjacent to the Dilp2-expressing IPCs (red arrow). The STAT reporter is expressed along the neurites that run parallel to the median arborizations of the IPCs (yellow arrow). (D) Under starved conditions, the GFP reporter expression is reduced in the cell body (red arrow) and along the neurites (yellow arrow). Dilp2 fluorescence in the IPCs is increased under starved (D) compared to fed (C) conditions. upd2-RNAi in the FB alters STAT::GFP reporter expression in the brain (F), compared to Luciferase-RNAi control (E). Note that the bouton-like enrichment of the STAT::GFP reporter along the neurites, juxtaposed to the IPCs, is lost in flies expressing FB-specific upd2-RNAi (compare yellow arrow in E with F). STAT::GFP expression in the cell body is reduced in flies expressing FB-specific upd2-RNAi (compare red arrow in E with F). Dilp2 fluorescence is increased in the IPCs of adult males expressing FB-specific upd2-RNAi compared to control (compare E with F). See related Figures S3, S4 and S5.
Next, we addressed whether the expression of the STAT reporter is altered in response to nutritional deprivation. Under fed conditions, the STAT reporter is expressed in the cell body of neurons immediately adjacent to the Dilp-expressing IPCs (red arrow, Figure 4C). In addition, the STAT reporter is expressed along the neurites that run parallel to the median arborizations of the IPCs (yellow arrow, Figure 4C). Along the neurites the reporter is enriched in vesicular structures that co-localize with the pre-synaptic marker Synapsin (Klagges et al., 1996) (Figure S5), suggesting that they may correspond to the sites of synaptic contact with the IPCs. Following nutrient deprivation (adult males kept on 1% sucrose agar at 25°C for more than 72 hours), STAT reporter expression in the neurons juxtaposing the IPCs is altered, and both the expression in the cell body (red arrow, Figure 4D) and the vesicular enrichment along the median branches of the IPCs is reduced or lost (yellow arrow, Figure 4D). Altogether, the alterations in reporter activity in these neurons in response to the nutrient status suggest that they influence Dilp secretion in the fed condition.
If Upd2 controls Dilp levels in the IPCs by activating STAT signaling in the mNSC neurons, then compromising upd2 function should phenocopy the effect of starvation on STAT reporter expression. We analyzed STAT reporter expression in the adult brains of flies that express upd2-RNAi in the FB and compared it to a luciferase-RNAi control. Indeed, while the expression of the STAT reporter in the controls was very similar to that observed in the fed condition (Figure 4E), both the expression of the reporter in the cell body (red arrow, Figure 4F) and the vesicular enrichment along the IPCs (yellow arrow, Figure 4F) was much reduced or lost when upd2 was knocked down in the FB, as observed under starvation.
STAT signaling in mNSC neurons regulates systemic growth and metabolism
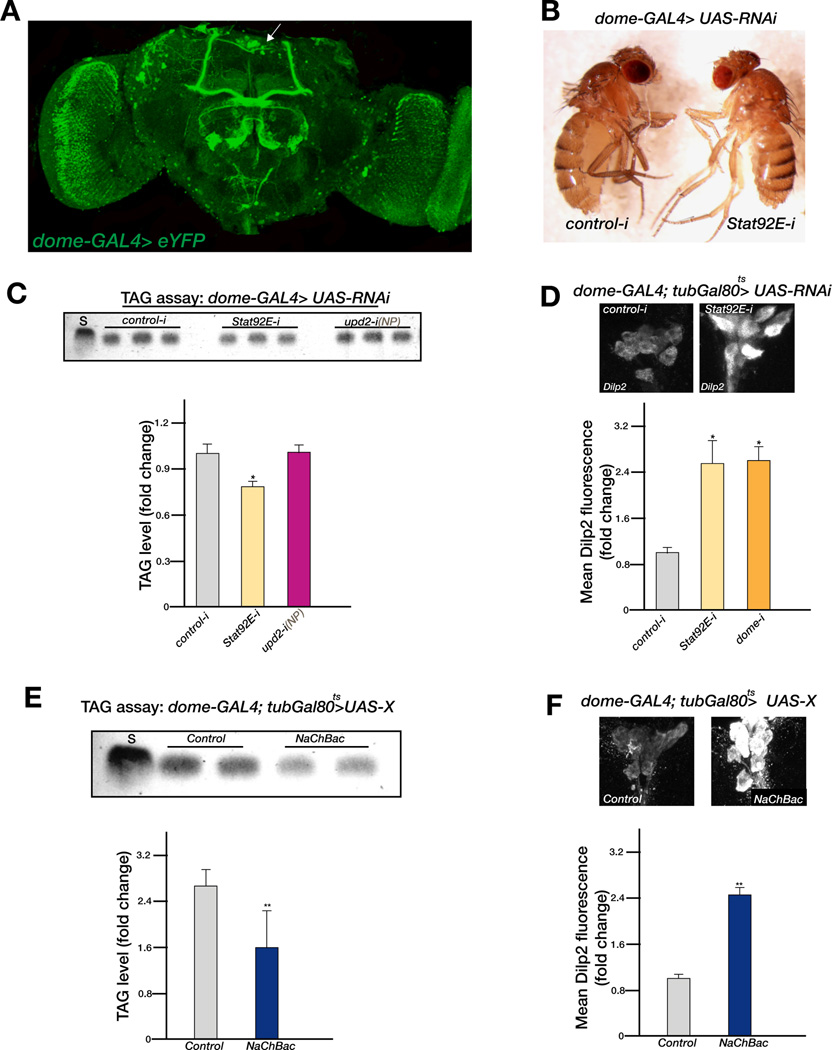
To further test the model that JAK/STAT signaling plays a role in the mNSCs to regulate systemic growth and metabolism, we tested whether knocking down STAT92E in mNSC neurons affected systemic growth and fat storage. To knockdown STAT92E, we used dome-GAL4 (Mandal et al., 2007), which we found drives expression in a number of neurons in the adult brain including the STAT-reporter positive mNSCs (white arrow, Figure 5A). At 25°C, dome-GAL4>stat92E-RNAi results in lethality, presumably because of the role of STAT92E in development. However, when cultured at 18°C, the flies emerged but were significantly smaller than the controls (Figure 5B). We shifted dome-GAL4>stat92E-RNAi adults to 29°C for 7 days to allow for a more significant knockdown of STAT92E expression and assayed for stored fat by performing a TLC assay. Strikingly, the level of stored fat was significantly lower in flies with compromised STAT92E function (Figure 5C). A similar experiment performed with upd2-RNAi flies failed to reveal a role for Upd2 in the dome-GAL4 neurons, as there was no effect on stored fat (Figure 5C) or body size (data not shown).
Figure 5. STAT signaling in mNSC neurons regulates systemic growth and metabolism.
(A) Maximum intensity projection of adult female brain expressing enhanced YFP (eYFP) under the control of dome-GAL4. dome-GAL4 is widely expressed and its expression domain includes a cluster of neurons in the mNSC region (white arrow). (B) Expression of STAT92E-RNAi in the dome-GAL4 expression domain results in small and slim flies. (C) The amount of TAG measured by TLC is significantly reduced in adult females expressing STAT92E-RNAi with dome-GAL4 but not with upd2-RNAi. (D) Dilp2 fluorescence is significantly increased in both STAT92E-RNAi and dome-RNAi in the IPCs of adult females using dome-Gal4. Control in (B), (C) and (D) is GFP-RNAi. (E) The amount of TAG is significantly reduced in adult females expressing NaChBac with dome-Gal4. (F) The expression of NaChBac in dome-Gal4 neurons significantly increases Dilp2 accumulation in the IPCs of adult females. Control in (E) and (F) is GFP overexpression. Note that in (D), (E) and (F), Gal80ts was used to allow transgene expression only in adults. Error bars represent the standard deviation. p-values were calculated using the Welch’s t-test (*p<0.05, **p<<0.01, ***p<<0.00001).
To examine if compromising the function of the receiving end of the JAK/STAT pathway results in increased Dilp accumulation in the IPCs, we used dome-GAL4; tub-GAL80ts to express stat92E-RNAi or dome-RNAi only during adulthood (see Experimental Procedures). We assayed the expression of Dilp2 in the IPCs of these flies 7 days post transfer at the restrictive temperature and compared it to a GFP-RNAi control. Dilp2 accumulation significantly increased when stat92E or dome function was compromised in adults (Figure 5D), suggesting that the JAK/STAT pathway plays to promote Dilp2 secretion from the IPCs.
The dome-GAL4 driver is expressed not only in the brain but in other tissues of the fly as well (eg., gut, muscle). Hence, to address whether JAK/STAT signaling in the mNSC neurons activates/depolarizes or inhibits/hyperpolarizes neuronal activity, we expressed the bacterial sodium channel (NaChBac) using dome-GAL4 at the restrictive temperature (dome-GAL4; tub-GAL80ts>NaChBac). This forced depolarization resulted in reduced fat storage (Figure 5E) and increased Dilp2 accumulation (Figure 5F) suggesting that activation of the JAK/STAT pathway in mNSC neurons prevents their neuronal firing.
JAK/STAT plays a role in neurons expressing vesicular GABA transporter to modulate systemic growth and metabolism
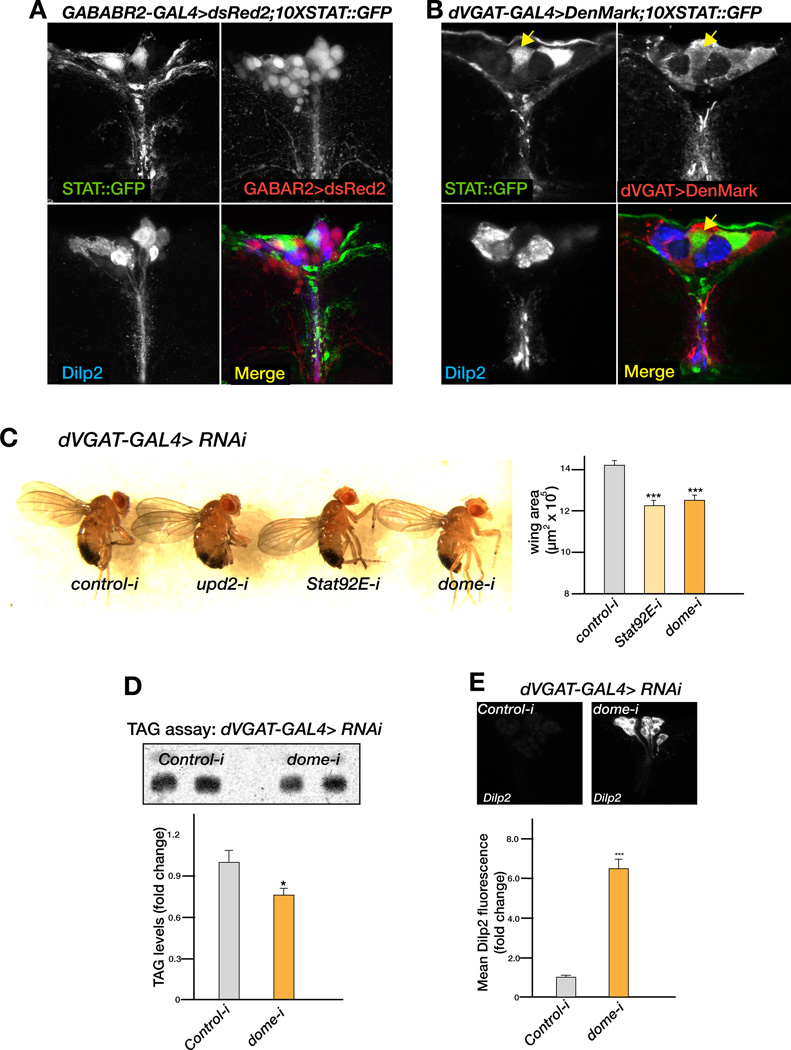
IPCs express receptors for serotonin and GABA, and both these neurotransmitters exert an inhibitory effect to prevent Dilp release (Enell et al., 2010; Luo et al., 2012). Thus, we examined whether the cells in the mNSCs that express the STAT reporter could exert their effects through those neurotransmitter systems. However, while neurons that express the STAT reporter do not express the GABA receptor (Figure 6A), a subset of them expressed the Drosophila vesicular GABA transporter (dVGAT) (yellow arrows, Figure 6B) that is required for GABA release (Fei et al., 2010). We found that reducing the activity of dome and STAT in dVGAT neurons affects systemic growth at 25°C (Figure 6C). Next, we assayed the level of stored fat in dVGAT-GAL4>dome-RNAi adult flies at 29°C (high levels of lethality were observed for dVGAT-GAL4>STAT-RNAi at this temperature so we analyzed dome flies in the TAG and Dilp assays). The knock down of dome in dVGAT neurons results in a significant decrease in stored fat (Figure 6D) and an increase in Dilp2 accumulation in the IPCs (Figure 6E), indicating that JAK/STAT signaling in GABAergic neurons relieves their inhibitory effect on IPCs. Consistent with our relay model, loss of JAK/STAT components in the Dilp neurons themselves did not affect systemic growth (data not shown) or metabolism (as assayed by stored fat, Figure S6).
Figure 6. Role of the JAK/STAT pathway in GABAergic neurons affects systemic growth and metabolism by influencing Dilp2 accumulation in the IPCs.
(A) XY-slice of the mNSC region of 10XSTAT::GFP male adult brains expressing dsRed2 under the control of GABA-B-R2-GAL4. Immunostaining for Dilp2 (blue), GFP (green, STAT::GFP expression) and dsred2 (red, GABA-B-R2 expression domain) shows that Dilp2 IPCs are positive for the GABA receptor, and that STAT reporter neurons do not overlap with the GABA-B-R2-GAL4 expression domain. (B) XY-slice of the mNSC region of 10XSTAT::GFP male adult brains expressing the membrane associated dendritic marker DenMark under the control of the dVGAT-GAL4 driver. Immunostaining for Dilp2 (blue), GFP (green, STAT::GFP expression) and DenMark (red, dVGAT expression domain). STAT reporter neurons are positive for the vesicular GABA transporter dVGAT (yellow arrow).(C) Knockdown of dome and STAT in dVGAT neurons results in small and slim flies, as quantified by the reduction in wing area. (D) The amount of TAG measured by TLC in adult males is significantly reduced when dome-RNAi is expressed with dVGAT-GAL4. (E) Dilp2 fluorescence in IPCs is significantly increased when dome expression is compromised in the GABAergic neurons of adult males. In (C), (D) and (E) control is GFP-RNAi. Error bars represent the standard deviation. p-values were calculated using the Welch’s t-test (*p<0.05, ***p<<0.00001). See related Figure S6.
Human Leptin can functionally substitute for Upd2 by signaling via the JAK/STAT receptor Dome
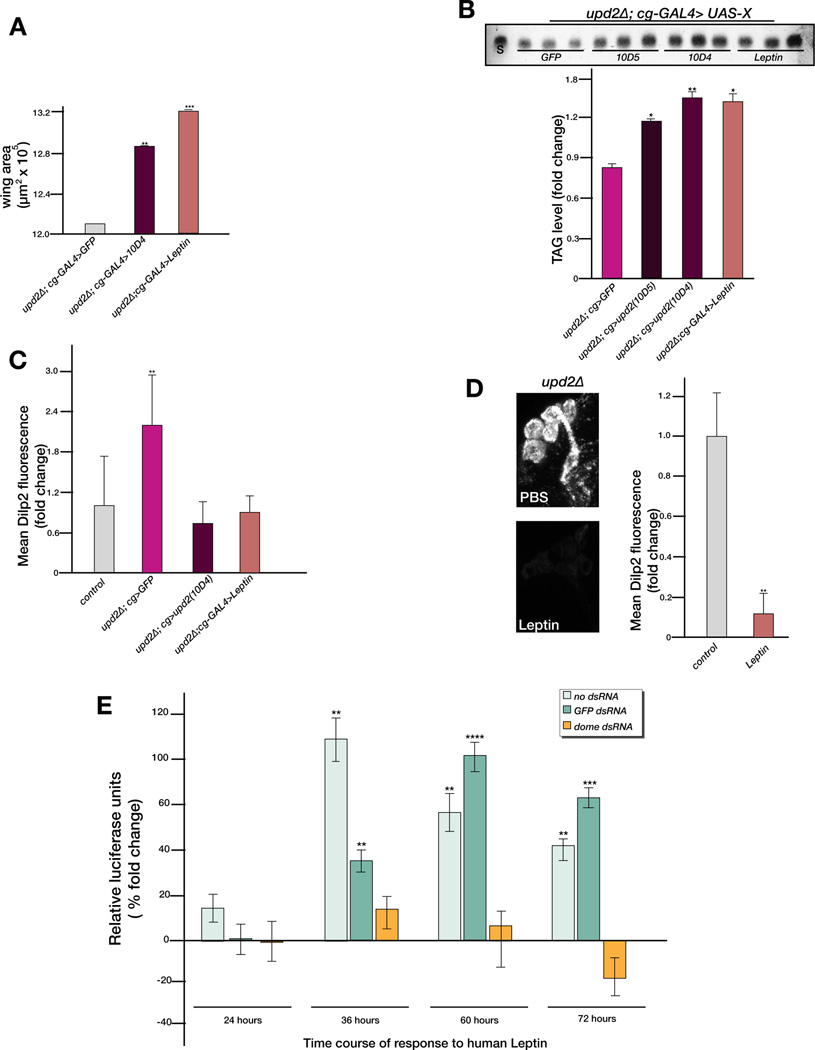
Upd2 encodes a type I cytokine that presents structural features to Leptin (Boulay et al., 2003). Because our finding that Upd2 influences insulin secretion by impinging on STAT signaling in GABAergic neurons in order to ‘dis-inhibit’ Dilp neurons is reminiscent to a recent finding that Leptin activates STAT signaling in GABAergic neurons and in turn results in the de-repression of a neuronal group called POMC (propiomelanocortin) neurons allowing them to fire (Vong et al., 2011), we tested the possibility that Leptin can substitute for Upd2. We generated transgenic flies that expressed human Leptin under the control of the UAS promoter (see Experimental Procedures) and expressed this transgene in the FB of upd2Δ flies. Strikingly, similar to flies expressing upd2 cDNA in the FB, human Leptin cDNA could rescue the growth (Figure 7A), fat storage (Figure 7B) and the Dilp2 accumulation phenotypes (Figure 7C) of upd2Δ flies. In addition, as documented in previous studies in mice (Caton et al., 2011; Ni et al., 2008; Rodgers and Shearn, 1977), injecting physiologically relevant dose of recombinant human Leptin (see Experimental Procedures) into upd2Δ flies significantly reduced the accumulation of Dilp2 in upd2Δ brains (Figure 7D) and solved the primary defect underlying a loss of Upd2 function. These results suggest that exogenous Leptin signals through the same pathway as Upd2.
Figure 7. Human Leptin can rescue the Upd2Δ and signals through Domeless.
(A) Quantification of wing area in upd2Δ males with FB-specific expression of upd2 cDNA (10D4) and human Leptin cDNA (B) The amount of TAG measured by TLC is significantly reduced in adult males expressing human Leptin cDNA in the FB. (C) Quantification Dilp2 accumulation in FB-specific expression of the 10D4 upd2 cDNA and human Leptin cDNA in upd2Δ background. Brains from cg-Gal4 flies were used as positive controls (grey bar). (D) Maximum intensity projection of upd2Δ adult male brains stained with Dilp2 after injection of human Leptin or control (PBS). The difference in Dilp2 fluorescence is quantified. (E) Response of the 10XSTATLuc reporter in Kc cells on incubation with 4nM human Leptin. The ratio of Renilla to firefly luciferase activity is measured at regular intervals from 24–72 hours and quantified as a percent fold change in relative light units with respect to cells incubated with media alone in the presence of no dsRNA, control or dome dsRNA. Error bars represent the standard deviation. p-values were calculated using the t-test (*p<0.05, ** p<<0.01, ***p<0.001, ****p<0.00001). See related Figure S7.
In order to test whether human Leptin is a bona fide ligand of the JAK/STAT pathway receptor Dome, we used a well-established JAK/STAT reporter assay (10XSTATLuc) in Drosophila Kc cells (Baeg et al., 2005; Hombria et al., 2005; Wright et al., 2011). Drosophila Kc cells were transfected with 10XSTATLuc and Actin-promoter driving Renilla luciferase (Act-Renilla) together with dsRNAs against dome or control (GFP) or without dsRNA treatment. The cells were then incubated with 4nM recombinant human Leptin, a concentration at which Upd ligands effectively activate the 10XSTATLuc reporter in cell-culture (Wright et al., 2011). We measured the ratio of firefly to Renilla luciferase activities (relative luciferase units (RLU)) at different time points after incubation with human Leptin and compared it to the RLU of cells stimulated with media lacking Leptin. A significant increase in RLU in cells stimulated with Leptin over time was observed with control dsRNA or in cells not treated with dsRNAs (Figure 7E). However, cells treated with dsRNA against dome (two independent dome dsRNAs were used) were unable to respond to Leptin (Figure 7E). By plotting the luciferase signal as a function of the amount of human Leptin added, we find that the binding affinity (Km-Michaelis constant) of human Leptin for Dome is 2.37nM (Figure S7A), which is comparable to the Km of 1nM reported for the human Leptin receptor (DasGupta et al., 2005). These results provide strong evidence that human Leptin is able to signal through the JAK/STAT pathway in Drosophila cells by engaging the JAK/STAT receptor Dome.
Discussion
Previous studies have postulated the existence of secreted factors, produced by the FB, that stimulate systemic growth by stimulating cell proliferation and that the FB-the fly nutrient sensor- couples Dilp secretion from the brain IPCs depending on the nutritional status (Britton and Edgar, 1998; Colombani et al., 2003; Davis and Shearn, 1977; Geminard et al., 2009). Here, we show that the JAK/STAT ligand Upd2, a type 1 cytokine signal, is involved in the inter-organ communication between the FB and the brain IPCs. We demonstrate that human Leptin can rescue the upd2 mutant phenotypes, implying that an invertebrate model system is suited to address questions pertaining to Leptin biology.
Upd2 plays a fat body-specific role in communicating the fed state to the brain IPCs
Upd proteins have secondary structures predicted to have α-helices similar to that of type I cytokines belonging to the IL-6 family, and sequence alignments suggest that they show some similarity to vertebrate Leptins (Boulay et al., 2003; Harrison et al., 1998). Among the three Upd ligands that activate the Dome receptor, only Upd2 plays a significant role in communicating the nutritional status from the FB. This is somewhat surprising as all three Upd proteins are secreted JAK/STAT pathway agonists and are able to activate the JAK/STAT pathway non-autonomously in-vivo. However, the signal sequences of the different Upds appear to confer them with different biophysical properties, as illustrated by tissue culture assays showing that, while Upd1 and Upd3 associate primarily with the extracellular matrix, Upd2 is easily detectable in the media (Wright et al., 2011). In addition, secretion assays showed that Upd2 is able to condition tissue culture media more potently than either Upd1 or Upd3. Altogether, these results suggest that Upd2 activates JAK/STAT signaling at greater distances than Upd1 or Upd3.
As evidenced by the growth and metabolic phenotypes of FB-specific knockdown, Upd2 seems to be required only in the FB but the reason for this tissue specificity is unclear. A previous study, which analyzed the Upd2 protein using a hidden Markov model, suggested that Upd2 is probably not secreted via the ‘classical’ Golgi-ER machinery (Hombria et al., 2005) because it lacks a signal peptide. In fact, other type I cytokines involved in inter-organ cross-talk also lack the signal peptide and are secreted by unconventional secretory pathways (Haas et al., 2011). Thus, one possible reason for the tissue specificity of Upd2 could be that the FB is the only tissue that can secrete this protein in an active form. Future work, contingent on the development of techniques and reagents to detect Upd2 in the fly hemolymph, will clarify this issue.
Existence of nutrient specific FB-derived signals
The identification of Upd2 as a nutrient regulated signal from the FB that does not depend on AAs but is produced in response to dietary fats and sugars reveals that different nutrient-specific secreted factors exist in the fly. Interestingly, the upregulation of upd2 levels in FB knockdown of slif suggests the existence of a homeostatic feedback loop whereby Upd2, in the context of low protein, promotes utilization of fat and carbohydrate energy sources. High sugar diets in flies have been shown to trigger a lipogenesis program akin to high fat diets in mammals (Musselman et al., 2011; Zhou et al., 2005), suggesting that Upd2 is most likely downstream of signals that are produced by increased fat stores. This is a highly significant finding given that it questions a broadly prevailing view that one dominant secreted factor downstream of AA’s governs nearly all aspects of systemic growth and metabolism in flies. Our findings support the model that the fly FB secretes numerous factors that regulate systemic growth and metabolism downstream of various components of the fly diet.
JAK/STAT activation in GABAergic neurons and parallels with the adipose-hypothalamic circuit in mammals
Our results indicate that STAT activation in GABAergic neurons inhibits their firing. Previous work has implied that the GABA-B-receptors in Dilp neurons inhibit Dilp release (Enell et al., 2010). Given that these GABAergic neurons are pre-synaptic to the IPCs, we propose that activation of STAT in GABAergic neurons relieves the IPCs from repression, thus resulting in Dilp release. This is reminiscent to the observation that first order-neurons responding to adipose-derived Leptin are the inhibitory GABAergic neurons expressing LepRs (Vong et al., 2011). When LepRs are activated by Leptin they regulate Stat3 phosphorylation which, by an unknown mechanism, inhibits the firing of the GABAergic neurons. This in turn relieves the repression on a neuronal group called POMC (propiomelanocortin) neurons allowing them to fire (Vong et al., 2011). Thus, this circuit module is strikingly reminiscent to what we observe in the fly.
There are many outstanding questions yet to be resolved regarding the signaling mechanisms by which the JAK/STAT pathway regulates GABAergic neurons. The target(s) of the JAK/STAT pathway in regulating neuronal firing in mammalian GABAergic neurons remains to be identified (Vong et al., 2011). It has been suggested that Leptin activation of STAT signaling may be required for the long-term effects of Leptin’s action on energy homeostasis rather than for acute effects of Leptin (Karsten et al., 2006), and that the acute effects of Leptin on the membrane potential of certain neuronal groups require activation of PI3-K signaling rather than STAT (Schober et al., 2005). However, the role of JAK/STAT versus PI3-K signaling in modulating the electrophysiology of the presynaptic GABAergic neurons is yet to be clarified (Vong et al., 2011), especially as previous studies were done on non-GABAergic neuronal groups. Altogether, further investigations into the role of JAK/STAT signaling in modulating neurotransmission in GABAergic neurons will be necessary to clarify how JAK/STAT signaling regulates their activities. Importantly, based on the similarity of the circuits and the conservation of the signaling pathways, studies in the fly are likely to provide insights relevant to mammalian neurophysiology.
Parallels between Upd2 and Leptin
The physiology of Leptin signaling in vertebrates is undoubtedly more complex and different from the physiology of flies. upd2Δ mutant flies are smaller and leaner whereas mutations in Leptin in mammals are associated with obesity. There is however some striking parallels. We find that under starvation upd2 mRNA steady-state levels drop significantly (Figure 2A), and there is a significant increase of upd2 mRNA expression under high fat diets (Figure 2F). This is similar to the alteration in Leptin levels during starvation and high fat diets (Ahima et al., 1996). Examination of the role of Leptin in the physiology of starvation, by providing mice with exogenous Leptin during periods of nutrient restriction, revealed that the primary physiological role of Leptin is to regulate the neuroendocrine system during starvation. Leptin reduced the reproductive capacity and increased stress hormone levels, which in turn increases the survival capacity of the organism under adverse nutrient conditions (Ahima et al., 1996). Consistent with this, flies with ablated IPCs, which are unable to produce insulin, perform much better under starvation conditions and increased stress conditions (Broughton et al., 2005). Given that the role of Upd2 is to promote insulin secretion, the reduction of Upd2 levels during starvation decreases Dilp secretion and increases the chances of survival under starvation (upd2Δ mutants are more starvation resistant than the wild-type controls; Figure S7B). Hence, in this context, the primary physiological role of Upd2 and Leptin converge.
EXPERIMENTAL PROCEDURES
Drosophila strains and Diets
Details of fly strains, transgenic fly construction, standard lab food composition and temperatures for specific crosses can be found in Extended Experimental procedures
Triglycerides measurements
Triglyceride assays were performed as previously described (Al-Anzi et al., 2009; Armknecht et al., 2005). Further details can be found in Extended Experimental procedures.
Hemolymph glucose measurements
Quantification of glucose concentration in the hemolymph was done as described in (Geminard et al., 2009).
qPCR
Total RNA was prepared from triplicates of 15, fed (standard lab food) or starved (>72 hours on 1% sucrose agar), age-matched adult males at 25C. cDNA was prepared using 1µg RNA and qPCR was performed using iQ SYBR Green Supermix. alphatubulin and rp49 were used to normalize the RNA levels. Relative quantification of mRNA levels was calculated using the comparative CT method. See Extended Experimental Procedures for information on oligos.
Feeding assays
Blue dye assay
Feeding assay was adapted from (Xu et al., 2008).
CAFÉ assay
The CAFÉ assay was performed as described in (Ja et al., 2007).
Oil red O staining
The oil red O staining was performed as previously described (Gutierrez et al., 2007; Palanker et al., 2009).
Immunostaining, confocal imaging and analysis
Immunostaining of larval and adult brains were performed based on protocols from (Pfeiffer et al., 2010). Images were captured using Leica SP2 and Zeiss LSM 780 confocal systems and analyzed using Zeiss ZenLite 2009, Leica LAS AF lite and Image J. To calculate the intensity of Dilp staining, mean gray value was calculated from maximum intensity projections of a similar number of confocal stacks using Image J. For details on antibodies see Extended Experimental Procedures.
Leptin Injections
Human recombinant Leptin (Sigma Aldrich, # L4146-1MG) was dissolved as per package instructions in a solution of Hcl and NaOH to a final concentration of 1µg/µL to prepare the human Leptin stock solution which was then frozen in aliquots at −20C. Fresh aliquots were thawed on ice before each experiment. The working solution was made by diluting the human Leptin stock solution in PBS to a final concentration of 0.001µg/µL and injected into 10 days old adult males of y w and upd2Δ. PBS was injected in mock controls. The injections were done by inserting the needle at the junction of the thorax and abdomen close to the haltere, using a microinjector ( Eppendorf, Femtoget) at a pressure of 305kPa using needles pulled in a pipette puller ( Kopf vertical pipette puller, model 720) at heater setting 14.1. At least 25-20 flies were injected per condition. Flies were allowed to recover for 24 hours, and then the brains were dissected and stained, analyzed and Dilp2 levels were quantified as outlined in the previous section. A minimum of 3–5 brains were analyzed per experiment. The experiments were performed three independent times.
Tissue-culture and luciferase assays
Drosophila Kc cells were maintained in Schneider’s medium (GIBCO), 10% heat-inactivated FBS (SIGMA) and 5% Pen-Strep (GIBCO) at 25°C. Experiments were run in 96 well plates (in six replicates per condition), 150ng/uL of the appropriate dsRNA were seeded in the wells prior to the start of experiment. Each well was transfected with 0.05ng 10XSTATLuc, 14ng Act-Renilla, 106 ng pAC-PL (used as carrier DNA). 96 hours post transfection, media supplemented with 4nM human recombinant Leptin or control media supplemented with no Leptin was added to the wells. Luciferase activity was measured using DualGlo reagents (Promega) as per kit instructions and measured using the Analyst GT plate reader. For details of amplicons used for dsRNA production refer to Extended Experimental Procedures.
Statistical analysis
Statistical analysis was performed using Welch’s T-test using Excel.
Supplementary Material
Research Highlights.
Upd2 is produced by the Drosophila fat body in response to dietary fats and sugars.
Dome activation by Upd2 in GABAergic neurons disinhibits insulin producing cells.
This results in Insulin secretion, promoting systemic growth and fat storage.
Human Leptin can signal via Drosophila JAK/STAT receptor and substitute for Upd2.
Acknowledgments
We thank M. Zeidler, E. Bach, B. Hassan, B. White, B. Al-Anzi, M. Pankratz, P. Shen, J. Wang, D. Krantz, J. Simpson, A. Gould, E. Hafen, H. Stocker, P. Leopold, G. Technau and E. Olson for reagents. We are thankful to the Transgenic RNAi facility (TRiP), NIG-fly stock center at Japan and the Bloomington Stock Center for flies, the Developmental studies hybridoma bank for monoclonal antibodies and the Drosophila RNAi screening center (DRSC) at the Harvard Medical school for dsRNA reagents and plate reader equipment. C. Villalta performed embryonic microinjections for creating the human Leptin transgenic flies. We thank F. Demontis and C. Pitsouli for critical comments on the manuscript. We are grateful to Y. Kwon for advice on the STAT luciferase assay and I. Droujinine for discussions and help with the diet experiments. We appreciate P. Raghavan and F .Wirtz-Peitz for insightful comments during the course of the work. This work was supported by 5P01CA120964 and 5R01DK088718 from the NIH. N.P is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahima RS, Prabakaran D, Mantzoros C, Qu D, Lowell B, Maratos-Flier E, Flier JS. Role of leptin in the neuroendocrine response to fasting. Nature. 1996;382:250–252. doi: 10.1038/382250a0. [DOI] [PubMed] [Google Scholar]
- Al-Anzi B, Sapin V, Waters C, Zinn K, Wyman RJ, Benzer S. Obesityblocking neurons in Drosophila. Neuron. 2009;63:329–341. doi: 10.1016/j.neuron.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armknecht S, Boutros M, Kiger A, Nybakken K, Mathey-Prevot B, Perrimon N. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods in enzymology. 2005;392:55–73. doi: 10.1016/S0076-6879(04)92004-6. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes & development. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binari R, Perrimon N. Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes & development. 1994;8:300–312. doi: 10.1101/gad.8.3.300. [DOI] [PubMed] [Google Scholar]
- Boulay JL, O'Shea JJ, Paul WE. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity. 2003;19:159–163. doi: 10.1016/s1074-7613(03)00211-5. [DOI] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MD, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Caton PW, Kieswich J, Yaqoob MM, Holness MJ, Sugden MC. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia. 2011;54:3083–3092. doi: 10.1007/s00125-011-2288-0. [DOI] [PubMed] [Google Scholar]
- Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Boutros M, Perrimon N. Drosophila Wnt/Fz pathways. Science's STKE : signal transduction knowledge environment. 2005;2005:cm5. doi: 10.1126/stke.2832005cm5. [DOI] [PubMed] [Google Scholar]
- Davis KT, Shearn A. In vitro growth of imaginal disks from Drosophila melanogaster. Science. 1977;196:438–440. doi: 10.1126/science.403606. [DOI] [PubMed] [Google Scholar]
- Dinarello CA, Mier JW. Interleukins. Annual review of medicine. 1986;37:173–178. doi: 10.1146/annurev.me.37.020186.001133. [DOI] [PubMed] [Google Scholar]
- Enell LE, Kapan N, Soderberg JA, Kahsai L, Nassel DR. Insulin signaling, lifespan and stress resistance are modulated by metabotropic GABA receptors on insulin producing cells in the brain of Drosophila. PLoS One. 2010;5:e15780. doi: 10.1371/journal.pone.0015780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei H, Chow DM, Chen A, Romero-Calderon R, Ong WS, Ackerson LC, Maidment NT, Simpson JH, Frye MA, Krantz DE. Mutation of the Drosophila vesicular GABA transporter disrupts visual figure detection. J Exp Biol. 2010;213:1717–1730. doi: 10.1242/jeb.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geminard C, Rulifson EJ, Leopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 2009;10:199–207. doi: 10.1016/j.cmet.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Haas Y, Windig JJ, Calus MP, Dijkstra J, Haan M, Bannink A, Veerkamp RF. Genetic parameters for predicted methane production and potential for reducing enteric emissions through genomic selection. Journal of dairy science. 2011;94:6122–6134. doi: 10.3168/jds.2011-4439. [DOI] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombria JC, Brown S, Hader S, Zeidler MP. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288:420–433. doi: 10.1016/j.ydbio.2005.09.040. [DOI] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annual review of medicine. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn CR, King GL, Moses AC, Weir GC, Jacobson AM, Smith RJ. Joslin's Diabetes Mellitus. 14 edn. Boston, MA: 2005. [Google Scholar]
- Karsten P, Plischke I, Perrimon N, Zeidler MP. Mutational analysis reveals separable DNA binding and trans-activation of Drosophila STAT92E. Cell Signal. 2006;18:819–829. doi: 10.1016/j.cellsig.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO, Reifegerste R, Reisch D, Schaupp M, Buchner S, Buchner E. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci. 1996;16:3154–3165. doi: 10.1523/JNEUROSCI.16-10-03154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RN, Wang ZL, Wang RM, Hurley JD, Smith DM, Ghatei MA, Withers DJ, Gardiner JV, Bailey CJ, Bloom SR. Leptin rapidly suppresses insulin release from insulinoma cells, rat and human islets and, in vivo, in mice. J Clin Invest. 1997;100:2729–2736. doi: 10.1172/JCI119818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Becnel J, Nichols CD, Nassel DR. Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5-HT(1A) receptor. Cell Mol Life Sci. 2012;69:471–484. doi: 10.1007/s00018-011-0789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal L, Martinez-Agosto JA, Evans CJ, Hartenstein V, Banerjee U. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446:320–324. doi: 10.1038/nature05585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton GJ, Cummings DE, Baskin DG, Barsh GS, Schwartz MW. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Disease models & mechanisms. 2011;4:842–849. doi: 10.1242/dmm.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. Drosophila HNF4 regulates lipid mobilization and beta-oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltagegated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Rodgers ME, Shearn A. Patterns of protein synthesis in imaginal discs of Drosophila melanogaster. Cell. 1977;12:915–921. doi: 10.1016/0092-8674(77)90155-6. [DOI] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Schober M, Rebay I, Perrimon N. Function of the ETS transcription factor Yan in border cell migration. Development. 2005;132:3493–3504. doi: 10.1242/dev.01911. [DOI] [PubMed] [Google Scholar]
- Taguchi A, White MF. Insulin-like signaling, nutrient homeostasis, and life span. Annu Rev Physiol. 2008;70:191–212. doi: 10.1146/annurev.physiol.70.113006.100533. [DOI] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S, Jr, Lowell BB. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BH, Osterwalder TP, Yoon KS, Joiner WJ, Whim MD, Kaczmarek LK, Keshishian H. Targeted attenuation of electrical activity in Drosophila using a genetically modified K(+) channel. Neuron. 2001;31:699–711. doi: 10.1016/s0896-6273(01)00415-9. [DOI] [PubMed] [Google Scholar]
- Wright VM, Vogt KL, Smythe E, Zeidler MP. Differential activities of the Drosophila JAK/STAT pathway ligands Upd, Upd2 and Upd3. Cell Signal. 2011;23:920–927. doi: 10.1016/j.cellsig.2011.01.020. [DOI] [PubMed] [Google Scholar]
- Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler MP, Bach EA, Perrimon N. The roles of the Drosophila JAK/STAT pathway. Oncogene. 2000;19:2598–2606. doi: 10.1038/sj.onc.1203482. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liu KY, Bradley P, Perrimon N, Wong ST. Towards automated cellular image segmentation for RNAi genome-wide screening. Medical image computing and computer-assisted intervention : MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. 2005;8:885–892. doi: 10.1007/11566465_109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.